Abstract
Ubiquitination is essential for ubiquitin/proteasome-mediated protein degradation in plant development and defense. Here, we identified a novel E3 ubiquitin ligase RING1 gene, CaRING1, from pepper (Capsicum annuum). In pepper, CaRING1 expression is induced by avirulent Xanthomonas campestris pv vesicatoria infection. CaRING1 contains an amino-terminal transmembrane domain and a carboxyl-terminal RING domain. In addition, it displays in vitro E3 ubiquitin ligase activity, and the RING domain is essential for E3 ubiquitin ligase activity in CaRING1. CaRING1 also localizes to the plasma membrane. In pepper plants, virus-induced gene silencing of CaRING1 confers enhanced susceptibility to avirulent X. campestris pv vesicatoria infection, which is accompanied by compromised hypersensitive cell death, reduced expression of PATHOGENESIS-RELATED1, and lowered salicylic acid levels in leaves. Transient expression of CaRING1 in pepper leaves induces cell death and the defense response that requires the E3 ubiquitin ligase activity of CaRING1. By contrast, overexpression of CaRING1 in Arabidopsis (Arabidopsis thaliana) confers enhanced resistance to hemibiotrophic Pseudomonas syringae pv tomato and biotrophic Hyaloperonospora arabidopsidis infections. Taken together, these results suggest that CaRING1 is involved in the induction of cell death and the regulation of ubiquitination during the defense response to microbial pathogens.
Plants continuously confront pathogen attacks using a variety of defense responses, including callose deposition (Gómez-Gómez et al., 1999), oxidative burst (Apel and Hirt, 2004), synthesis of pathogenesis-related (PR) proteins (van Loon et al., 2006), and localized cell death via the hypersensitive response (HR; Hwang and Hwang, 2011; Kim and Hwang, 2011). Hypersensitive cell death is a genetically controlled process that eliminates infected and neighboring cells using the product of a resistance (R) gene that can recognize a pathogen carrying the corresponding avirulence (Avr) effector (Dangl and Jones, 2001). During pathogen infection, plants mount a defense response by altering physiological and biological systems via proteomic plasticity through protein synthesis (transcription and translation) as well as remodeling and degradation processes (posttranslational modification; Stone and Callis, 2007). There are many mechanisms for protein modification, including phosphorylation, methylation, acetylation, myristoylation, glycosylation, and ubiquitination (Kwon et al., 2006). Among these, ubiquitination is essential for ubiquitin/proteasome-mediated protein degradation. In some cases, ubiquitin-mediated protein degradation can be as influential in proteome structure as protein production and modification (Callis and Vierstra, 2000; Dreher and Callis, 2007). The ubiquitination pathway is involved in plant development (e.g. embryogenesis, photomorphogenesis, hormone regulation, and senescence; Moon et al., 2004; Dreher and Callis, 2007) as well as in defense (Zeng et al., 2006). Ubiquitin contains 76 amino acids and it is covalently conjugated to Lys residues in substrate proteins (Smalle and Vierstra, 2004; Dreher and Callis, 2007). There are several types of ubiquitination with different outcomes. Polyubiquitination is required for degradation of the protein via the proteasome. Ubiquitin is attached to target proteins in a stepwise conjugation cascade. First, a ubiquitin-activating enzyme (E1) activates ubiquitin in an ATP-dependent manner. Second, the activated ubiquitin is then transferred to a ubiquitin-conjugating enzyme (E2) by E1 enzyme. Finally, E2 enzyme interacts with a specific E3 ubiquitin ligase (E3), leading to ubiquitination of the substrate protein or autoubiquitination of E3. In this pathway, E3 ubiquitin ligase plays a central role in selecting candidate proteins for ubiquitination (Ciechanover, 1998).
E3 ubiquitin ligases are classified into two groups based on the presence of a HECT (for Homology to E6-AP C Terminus) or RING (for Really Interesting New Gene)/U box domain, which have different subunit compositions and mechanisms of action (Pickart, 2001). The RING domain has a consensus sequence containing Cys and His residues (Cys-X2-Cys-X9-39-Cys-X1-3-His-X2-3-Cys/His-X2-Cys-X4-48-Cys-X2-Cys), which functions as a binding site for the ubiquitin-E2 intermediate that has a zinc-binding domain formed by conserved Cys and His residues. The RING domains of RING finger proteins can be divided into two types, C3-H-C4 and C3-H2-C3, according to the presence of Cys or His in the fifth position (Borden and Freemont, 1996). The E3 ubiquitin ligase activity of RING finger proteins can be detected through in vitro autoubiquitination. Among the 1,300 E3 ubiquitin ligase genes in the Arabidopsis (Arabidopsis thaliana) genome, more than 400 are predicted to belong to the RING finger type (Stone et al., 2005). However, the functions of only a few RING-type proteins have been characterized in vivo.
In plants, ubiquitination regulates endogenous signals in response to pathogen attack. Although plants contain many RING finger proteins, only some of them have been shown to be induced by biotic stresses (Dreher and Callis, 2007; Hong et al., 2007). These RING finger proteins are specifically induced in plants by pathogen attack and play an essential role in plant defense (Zeng et al., 2006). In Arabidopsis, the ARABIDOPSIS TOXICOS EN LEVADURA (ATL) gene family members ATL2 and ATL6 are rapidly up-regulated in response to chitin, a basal defense elicitor associated with fungal cell walls and insect exoskeletons (Salinas-Mondragón et al., 1999). Defense-related genes such as NON-EXPRESSER OF PATHOGENESIS-RELATED GENES1, PHENYLALANINE AMMONIA-LYASE, and CHALCONE SYNTHASE are induced constitutively under normal growth conditions in Arabidopsis mutants that constitutively express ATL2 (Serrano and Guzmán, 2004). The fungal elicitor N-acetylchitooligosaccharide induces the expression of rice (Oryza sativa) EL5 RING E3 ligase, which possesses in vitro ubiquitination activity (Takai et al., 2002). In Arabidopsis cell cultures and seedlings, the general bacterial flagellin peptide elicitor (flg22), which also stimulates basal defense responses, can induce a 2.5-fold change in the expression levels of over 250 genes (Felix et al., 1999). Among these up-regulated genes were genes encoding 10 putative RING finger E3 ligases, including RING-H2 FINGER A3b (RHA3b), RHA1b, RING MEMBRANE-ANCHOR1, and ATL6 (Navarro et al., 2004). RING finger proteins, such as the F-box proteins AVR9/CF-9 RAPIDLY ELICITED GENES189 (ACRE189; van den Burg et al., 2008) and ACRE132 (Durrant et al., 2000), are involved in gene-for-gene resistance-initiated responses. U-box E3 ligases are variants of RING finger proteins, and some have been implicated in plant disease resistance (Zeng et al., 2004; González-Lamothe et al., 2006; Yang et al., 2006), including ACRE74 and ACRE276 from tobacco (Nicotiana tabacum), which are essential for positive regulation of plant defense responses. By contrast, the pathogen-induced C3-H-C4-type RING finger protein CaRFP1 is associated with disease susceptibility and osmotic stress tolerance in pepper (Capsicum annuum; Hong et al., 2007). The Arabidopsis E3 ubiquitin ligases PLANT U-BOX PROTEIN22 (PUB22), PUB23, and PUB24 also play roles in the negative regulation of pathogen-associated molecular patterns-triggered immunity (Trujillo et al., 2008).
In this study, we identified CaRING1, which encodes a novel E3 ubiquitin ligase RING1 protein in pepper. CaRING1 expression is induced by avirulent Xanthomonas campestris pv vesicatoria (Xcv) infection. We show that a RING-type protein is essential for HR production and resistance to infection with virulent (Ds1) and avirulent (Bv5-4a) strains of Xcv. CaRING1 encodes a low-Mr protein that contains an N-terminal transmembrane (TM) domain and a C-terminal RING-H2 domain. CaRING1 displays in vitro E3 ubiquitin ligase activity and localizes to the plasma membrane. Virus-induced gene silencing (VIGS) and Agrobacterium tumefaciens-mediated transient expression in pepper were used to characterize the activities of CaRING1 in plant defense. CaRING1-silenced pepper plants are highly susceptible to infection by avirulent Xcv and show compromised or delayed reactive oxygen species (ROS) induction, HR, PR gene expression, and accumulation of free salicylic acid (SA) during infection. By contrast, transient CaRING1 expression in pepper leaves triggers the hypersensitive cell death response, which requires the E3 ubiquitin ligase activity of CaRING1. Transgenic Arabidopsis plants overexpressing CaRING1 also exhibit enhanced bacterial and fungal disease resistance, accompanied by the induction of SA-responsive AtPR1 and ethylene/jasmonic acid (JA)-responsive AtPDF1.2 (for PLANT DEFENSIN1.2) during Pseudomonas syringae pv tomato (Pst) infection. These findings suggest that during the defense response to microbial pathogens, CaRING1 is involved in the activation of cell death via autoubiquitination and the regulation of posttranslational modification.
RESULTS
CaRING1 Encodes a Protein Containing a Conserved TM Domain and a RING Domain
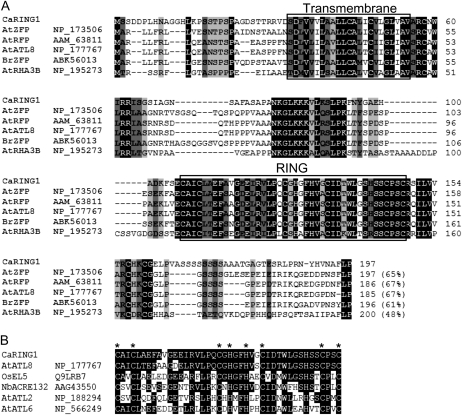
CaRING1 cDNA (GQ359822) was isolated from a cDNA library constructed from pepper leaves infected with the avirulent Xcv strain Bv5-4a, according to a macro-cDNA array method (Jung and Hwang, 2000). The CaRING1 cDNA sequence contains 985 bp, with a predicted open reading frame of 591 bp (Supplemental Fig. S1). The predicted CaRING1 protein comprises 197 amino acids and has a calculated molecular mass of 20.566 kD and a pI of 8.68. The PROSITE and SMART programs (http://www.expasy.ch/prosite/ and http://smart.embl-heidelberg.de/, respectively) revealed a putative TM domain for subcellular localization at the N terminus (residues 33–55) and a RING domain in the C-terminal region (residues 107–148; Fig. 1A). The RING domain belongs to the conserved C3H2C3-type RING-H2 group, which are essential for E3 ubiquitin ligase activity in the ubiquitin/26S proteasome system. CaRING1 shows relatively high amino acid sequence identity (48%–65%) to zinc finger or RING finger proteins in Arabidopsis and field mustard (Brassica rapa; Fig. 1A), which all contain highly conserved TM and RING domains. The RING domain of CaRING1 shares 55% to 90% identity with other plant defense-related RING finger proteins and contains eight conserved zinc-coordinating Cys and/or His residues that are essential for E3 ubiquitin ligase activity (Fig. 1B; Stone et al., 2005). Thus, domain analysis of the predicted CaRING1 sequence indicates that this protein may play a role in plant defense by modifying the proteome via ubiquitination.
Figure 1.
Amino acid sequence analysis of pepper CaRING1 and the RING-H2 finger domain. A, Alignment of deduced amino acid sequences of pepper CaRING1 and other RING-H2 finger proteins from Arabidopsis (accession nos. NP_173506, NP_177767, AAM_63811, and NP_195273) and field mustard (accession no. ABK56013). Identical amino acids are shown in black boxes, and conserved amino acids are shaded in gray. Black boxes indicate the TM and RING domains. B, Alignment of RING domains. Sequences include CaRING1, Arabidopsis ATL8 (accession no. NP_177767), rice EL5 (accession no. Q9LRB7; Takai et al., 2002), Nicotiana benthamiana ACRE132 (accession no. AAG43550; Durrant et al., 2000), Arabidopsis ATL2 (accession no. NP_188294; Serrano and Guzmán, 2004), and Arabidopsis ATL6 (accession no. NP_566249). Identical amino acids are shown in black boxes. Conserved Cys (C) and His (H) residues are indicated by asterisks.
Avirulent Xcv Infection Induces CaRING1 Expression in Pepper Leaves
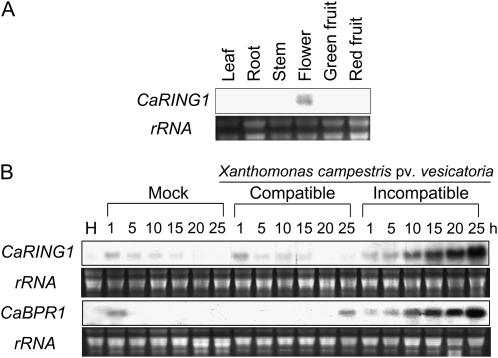
To ascertain whether CaRING1 is expressed in specific plant tissues, RNA gel-blot analysis was used to examine CaRING1 transcript levels in different plant tissues. CaRING1 transcripts were detected in the flowers of healthy pepper plants but not in the leaves, stems, roots, or green/red fruits (Fig. 2A). This pattern indicates that, with the exception of flowers, CaRING1 is not constitutively expressed in healthy plant organs.
Figure 2.
RNA gel-blot analysis of CaRING1 expression in pepper plants. Samples were hybridized with a [32P]dCTP-labeled CaRING1 probe. Equal loading of total RNA (10 μg) was verified by visualizing rRNA on a gel stained with ethidium bromide. A, Organ-specific expression of CaRING1 in pepper plants. B, Expression of CaRING1 and CaBPR1 in pepper leaves at various time points after inoculation at the six-leaf stage with 109 cfu mL−1 of the virulent Xcv strain Ds1 (compatible) or the avirulent Xcv strain Bv5-4a (incompatible) or 10 mm MgCl2. H, Healthy leaves; Mock, Leaves treated with 10 mm MgCl2.
To investigate CaRING1 transcript levels induced during compatible and incompatible interactions with Xcv, pepper leaves were inoculated with virulent (compatible) and virulent (incompatible) strains (Ds-1 and Bv5-4a, respectively). RNA gel-blot analysis revealed rapid and strong CaRING1 induction in leaves inoculated with Xcv Bv5-4a, with maximum levels achieved 25 h after inoculation (Fig. 2B). In contrast, mock-treated and virulent strain Ds1-inoculated leaves showed only negligible up-regulation of CaRING1. CaBPR1, which was used as a defense marker gene of pepper, was distinctly up-regulated during the incompatible interactions with Xcv (Fig. 2B). These findings indicate that CaRING1 is induced specifically during the incompatible interaction between pepper plants and Xcv.
CaRING1 Functions as an E3 Ubiquitin Ligase
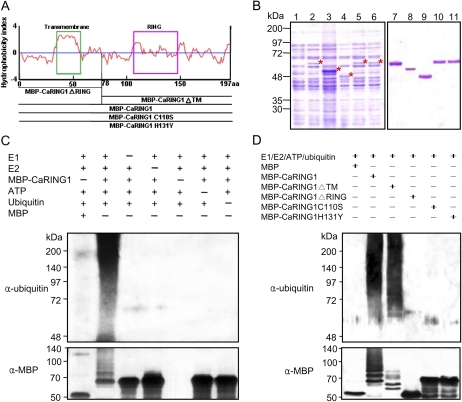
RING domain-containing proteins function as E3 ubiquitin ligases (Zhang et al., 2007; Lin et al., 2008), and the C terminus of CaRING1 contains a conserved C3H2C3-type RING-H2 domain (amino acids 107–148). The putative hydrophobicity of CaRING1 and a scheme for the construction of mutants are shown in Figure 3A. To test whether CaRING1 possesses E3 ubiquitin ligase activity, full-length CaRING1 was expressed in Escherichia coli (BL21) as a fusion protein with maltose-binding protein (MBP; Fig. 3, A and B). Following cell lysis, affinity chromatography was used to purify MBP-CaRING1 from the soluble fraction containing total proteins. Yeast E1 (ubiquitin-activating enzyme 1 [UBA1]) and Arabidopsis E2 (ubiquitin-conjugating enzyme 10 [UBC10]) were used for the in vitro E3 ubiquitin ligase activity assay (Fig. 3C). Ubiquitination activity was detected using anti-ubiquitin antibody and anti-MBP antibody. MBP-CaRING1 displayed E3 ubiquitin ligase activity (polyubiquitinated smear ladders), whereas MBP did not demonstrate in vitro ubiquitination. MBP-CaRING1 did not display E3 ubiquitin ligase activity in the absence of E1, E2, E3, ATP, or ubiquitin. These data indicate that CaRING1 protein functions as an E3 ubiquitin ligase.
Figure 3.
E3 ubiquitin ligase activity of CaRING1. A, Hydrophobicity index and expression constructs used for the in vitro ubiquitination assay. Recombinant fusion proteins are as follows: MBP-CaRING1, containing the full-length CaRING1; MBP-CaRING1ΔRING and MBP-CaRING1ΔTM mutants, with deletion of the C-terminal RING-H2 domain and N-terminal TM domain, respectively; and MBP-CaRING1C110S and MBP-CaRING1H131Y substitution mutants, with replacement of Cys-110 with Ser and His-131 with Tyr, respectively. The green box indicates the TM domain. B, SDS-PAGE of the recombinant MBP-CaRING1 and mutant variant proteins. Fusion constructs were transformed into E. coli BL21 (DE3). Cells were grown in LB medium, and expression of recombinant proteins was induced with 300 μm isopropyl-β-d-thiogalactopyranoside. Lane 1, Soluble fraction of uninduced E. coli MBP-CaRING1 extract. Lanes 2 to 6, Soluble fractions of induced E. coli MBP-CaRING1, CaRING1ΔTM, CaRING1ΔRING, CaRING1C110S, and CaRING1H131Y, respectively. Induced proteins are indicated by asterisks. Lanes 7 to 11, Purified MBP-CaRING1, CaRING1ΔTM, CaRING1ΔRING, CaRING1C110S, and CaRING1H131Y, respectively. C, E3 ubiquitin ligase activity assay of CaRING1. Recombinant MBP-CaRING1 fusion protein was incubated in the presence or absence of E1 (ScUBA1), E2 (AtUBC10), ATP, and/or ubiquitin. The reactions were analyzed with immunoblots using anti-ubiquitin antibodies (top panel) and anti-MBP antibodies (bottom panel). E3 ubiquitin ligase activity of MBP-CaRING1 was only detected in the presence of E1, E2, ATP, and ubiquitin. D, E3 ubiquitin ligase activity assay of CaRING1 mutant proteins. MBP-CaRING1 protein was used as a positive control. Only the mutant MBP-CaRING1ΔTM exhibited E3 ubiquitin ligase activity, indicating that the RING-H2 domain is essential for enzyme activity. [See online article for color version of this figure.]
N- and C-terminal deletion mutants (MBP-CaRING1ΔTM and MBP-CaRING1ΔRING, respectively) as well as substitution mutants with changes to individual conserved amino acids were generated to determine whether the E3 ubiquitin ligase activity of CaRING1 requires a complete RING domain (Fig. 3D). Among all the MBP-CaRING1 mutants generated, only MBP-CaRING1ΔTM exhibited E3 ubiquitin ligase activity. This result indicates that a complete RING domain is essential for E3 ubiquitin ligase activity in CaRING1. Interestingly, however, MBP-CaRING1ΔTM displayed a decreased polyubiquitinated smear pattern on the immunoblot using the anti-MBP antibody (Fig. 3D; Supplemental Fig. S2), indicating that the TM region of CaRING1 also partly supports E3 ubiquitin ligase activity or may be ubiquitinated. The UbPred programs (http://ubpred.org/) predict that the TM region of CaRING1 contains a putative ubiquitination site at residue Lys-15. Thus, we further used the MBP-CaRING1K15R mutant to determine if the TM region of CaRING1 is ubiquitinated (Supplemental Fig. S2). The MBP-CaRING1K15R mutant exhibited ubiquitination patterns similar to MBP-CaRING1 on the immunoblots using both anti-ubiquitin and anti-MBP antibodies, indicating that Lys-15 is not required for E3 ubiquitin ligase activity.
CaRING1 Localizes to the Plasma Membrane
CaRING1 is predicted to contain a TM domain (amino acids 33–55) in the N-terminal region. Fusion proteins between CaRING1 and soluble-modified (sm)GFP were used to determine the subcellular localization of CaRING1, and expression of the fusion proteins was driven by the cauliflower mosaic virus 35S promoter (Fig. 4A). Following particle bombardment into onion (Allium cepa) epidermal cells, transient expression of 35S:CaRING1:smGFP showed that CaRING1 localizes specifically to the plasma membrane (Fig. 4B). Plasmolysis confirmed that CaRING1 localizes to plasma membranes rather than cell walls.
Figure 4.
Subcellular localization of CaRING1 and its mutants using transient expression in onion epidermal cells. A, Schematic structure of CaRING1 and the constructs used for subcellular localization analysis. Lines indicate deleted regions. The smGFP gene was fused to the 3′ region of constructs. TM, TM domain; RING, RING-H2 finger domain. B, Transient expression of smGFP or smGFP-tagged constructs in onion epidermal cells 24 h after biolistic transformation, as detected by confocal laser-scanning microscopy. Constructs are as follows: smGFP (control), CaRING1:smGFP (wild type), CaRINGΔTM:smGFP (TM domain-deleted mutant), and CaRINGΔRING:smGFP (RING domain-deleted mutant). Arrows indicate the plasma membrane in a plasmolyzed cell. Bars = 200 μm.
Expression vectors containing genes encoding smGFP fusions to TM and RING domain deletion mutants (35S:CaRING1ΔTM:smGFP and 35S:CaRING1ΔRING, respectively) were used to test the prediction that the putative TM domain functions in the subcellular localization of CaRING1 (Fig. 3A). Fluorescent signals from CaRING1ΔTM:smGFP were detected in the cytosol and nuclei, whereas signals from CaRING1ΔRING:smGFP were found only at the plasma membrane (Fig. 4B). These results indicate that the TM domain of CaRING1 is essential for subcellular localization to the plasma membrane.
Transient Expression of CaRING1 Induces Cell Death in Pepper Leaves
Since avirulent Xcv infection strongly induces CaRING1 expression in pepper leaves during HR, further transient expression experiments were performed to investigate the potential relationship between CaRING1 expression and induction of cell death. Agrobacterium-mediated transient expression of CaRING1 (35S:CaRING1) induced a much stronger cell death response in pepper leaves compared with transient expression of the control vector (35S:00; Fig. 5A). UV-fluorescing phenolic compounds accumulated in pepper leaves expressing CaRING1 but were barely detected in leaves expressing the empty control vector. In tobacco, such UV-fluorescing compounds indicate an accumulation of the phenolic phytoalexins, such as scopoletin and its aglycone (Costet et al., 2002), which represent an indicator of the HR (Gachon et al., 2004; Fig. 5A). To test whether ubiquitination functions in inducing cell death, site-directed mutants 35S:CaRING1C110S and 35S:CaRING1H131Y, in which a Ser was substituted for a conserved Cys and a His was changed by Tyr in the RING domain of CaRING1, respectively, were transiently expressed in pepper leaves. These RING domain mutants exhibited distinctly decreased cell death phenotypes compared with the wild-type 35S:CaRING1 (Fig. 5B). Compromised cell death phenotypes and accumulation of UV-fluorescing phenolic compounds were associated with the expression of these mutations. In agroinfiltrated pepper leaves, 35S:CaRING1-induced cell death led to a significant increase in electrolyte leakage compared with empty vector controls (Fig. 5C). In contrast, the CaRING1 mutants did not generate high levels of electrolyte leakage and ROS. Reverse transcription (RT)-PCR and immunoblot analyses showed that 35S:CaRING1 and the site-directed mutants 35:CaRING1C110S and 35S:CaRING1H131Y were transiently expressed at both transcript and protein levels in pepper leaves 24 and 48 h after agroinfiltration (Fig. 5, D and E). No significant differences between 35S:CaRING1 and the mutants were found in the transient expression levels of their transcripts and proteins. Taken together, these results indicate that expression and ubiquitination of CaRING1 are required to induce cell death effectively in pepper leaves.
Figure 5.
Induction of cell death by transient expression of CaRING1 in pepper leaves. A, Cell death phenotypes in pepper leaves transiently expressing 35S:CaRING1 at 7 d after Agrobacterium-mediated transformation with the concentrations indicated (OD600 = 1.0, 0.5, and 0.1). UV-fluorescing phenolic compounds associated with cell death are indicated in the same leaves. B, Site-directed CaRING1 mutations cause reduced cell death. Reduced-cell-death phenotypes are shown in pepper leaves infiltrated with Agrobacterium strains (OD600 = 1.0) carrying the 35S:CaRING1C110S or 35S:CaRING1H131Y mutant. The marked regions on each leaf indicate the area infiltrated with Agrobacterium strains (OD600 = 1.0). Photographs were taken 3 d after infiltration. UV-fluorescing phenolic compounds on the same leaf are shown in the right panels. C, Quantification of electrolyte leakage from pepper leaf discs transiently expressing empty vector control (35S:00), 35S:CaRING1, 35S:CaRING1C110S, or 35S:CaRING1H131Y at various time points after infiltration (OD600 = 1.0). The data represent means ± sd from three independent experiments. D, Transient expression of empty vector control (35S:00) and 35S:CaRING1, 35S:CaRING1C110S, or 35S:CaRING1H131Y in pepper leaves 24 and 48 h after infiltration (OD600 = 1.0). Gene expression was analyzed by quantitative RT-PCR. E, Immunoblot analysis of 35S:CaRING1 and 35S:CaRING1C110S expression in pepper leaves 24 and 48 h after infiltration with Agrobacterium carrying 35S:CaRING1:GFP, 35S:CaRING1C110S:GFP, or 35S:CaRING1H131Y:GFP. Total protein was extracted from mature leaves and used to detect GFP-tagged protein. Coomassie blue (CBB) staining confirmed equal protein loading. H, Healthy leaves.
CaRING1 Is Required for Resistance to Xcv Infection in Pepper Plants
The incompatible interaction with Xcv avirulent strain Bv5-4a rapidly and strongly induced CaRING1 expression in pepper leaves. To determine the role played by CaRING1 in the basal defense response of pepper, VIGS was performed in pepper plants using Tobacco rattle virus (TRV; Liu et al., 2002; Choi et al., 2007). A 385-bp fragment from the 3′ end of the CaRING1 open reading frame was cloned into the TRV vector (TRV:CaRING1). Empty TRV vector (TRV:00) was used as a negative control. In comparison with pepper plants expressing the control vector, CaRING1-silenced (TRV:CaRING1) plants were highly susceptible to virulent Xcv infection, which was accompanied by an extensive suite of disease symptoms 6 d after inoculation. Little necrotic disease development was observed in CaRING1-silenced (TRV:CaRING1) pepper plants. However, the HR induced by avirulent Xcv infection was much weaker and delayed in CaRING1-silenced (TRV:CaRING1) pepper plants compared with plants expressing the empty TRV vector (TRV:00). Consistent with observations of transient CaRING1 expression in pepper leaves, UV-fluorescing phenolic compounds were induced as part of the defense response in plants expressing the empty TRV vector (TRV:00; Fig. 6A). This HR marker was substantially reduced in CaRING1-silenced leaves. CaRING1-silenced leaves showed significantly higher levels of bacterial growth compared with leaves expressing the empty vector control (Fig. 6B). Electrolyte leakage was used to quantify differences in cell death between plants expressing the empty TRV vector (TRV:00) and CaRING1-silencing (TRV:CaRING1) vector during the HR. Avirulent Xcv infection induced earlier and more substantial levels of electrolyte leakage in leaves expressing empty TRV vector (TRV:00) than in CaRING1-silenced (TRV:CaRING1) leaves (Fig. 6E). Similarly, leaves expressing empty vector (TRV:00) showed stronger trypan blue staining than leaves in which CaRING1 was silenced (Fig. 6C). In addition, CaRING1-silenced (TRV:CaRING1) plants exhibited significantly reduced accumulation of ROS (hydrogen peroxide [H2O2]) at 12 h after inoculation compared with plants with empty vector (Fig. 6D). Reduced H2O2 production in CaRING1-silenced (TRV:CaRING1) plants was also visualized by 3,3-diaminobenzidine staining (Fig. 6C). These results indicate that CaRING1 expression plays a significant role in the induction and signal transduction of the HR during avirulent Xcv infection. However, the gene knockdown strategy such as VIGS may silence genes with sequence similar to the target gene. To reduce this nonspecific silencing effect, a 299-bp fragment from the 3′ end of the CaRING1 untranslated region (UTR) was cloned into the TRV2 vector (TRV2:CaRING1UTR) to specifically silence the CaRING1 gene. Like CaRING1-silenced pepper plants (Fig. 6), CaRING1 UTR-silenced (TRV:CaRING1UTR) pepper plants exhibited enhanced susceptibility to virulent and avirulent Xcv infection, which was accompanied by increased bacterial growth as well as reduced H2O2 accumulation, cell death, and electrolyte leakage (Supplemental Fig. S3).
Figure 6.
Enhanced susceptibility of CaRING1-silenced pepper plants to infection with the virulent Xcv strain Ds1 and the avirulent Xcv strain Bv5-4a. A, Disease symptoms developed on empty vector control (TRV:00) or CaRING1-silenced (TRV:CaRING1) pepper leaves 6 and 3 d after inoculation with the virulent (Ds1; compatible) and avirulent (Bv5-4a; incompatible) strains, respectively. Highlighted areas of leaves show areas inoculated with the bacterial concentrations indicated. Infected leaves with UV-fluorescing phenolic compounds associated with susceptibility to infection and HR cell death are shown in the right panels. B, Growth of the virulent Xcv strain Ds1 and avirulent Xcv strain Bv5-4a in empty vector control (TRV:00) or CaRING1-silenced (TRV:CaRING1) pepper leaves at 0 or 3 d after inoculation (5 × 104 cfu mL−1). Data represent means ± sd from three independent experiments. Asterisks indicate significant differences between empty vector control and CaRING1 gene-silenced pepper plants as determined by Student’s t test (P < 0.05). C, Reduced H2O2 production and cell death in CaRING1-silenced leaves infected with the avirulent Xcv strain Bv5-4a (107 cfu mL−1). Xcv-infected leaves were harvested and stained with 3,3-diaminobenzidine (DAB) and trypan blue at 12 and 24 h after inoculation, respectively. Bars = 500 μm. D, Production of H2O2 in leaf discs from empty vector control (TRV:00) and CaRING1-silenced (TRV:CaRING1) pepper leaves at different time points after inoculation with 107 cfu mL−1 of the avirulent Xcv strain Bv5-4a. Data represent means ± sd from three independent experiments. Asterisks indicate significant differences between empty vector control and CaRING1-silenced pepper plants as determined by Student’s t test (P < 0.05). E, Cell death was monitored by electrolyte leakage (conductivity) from empty vector control (TRV:00) and CaRING1-silenced (TRV:CaRING1) pepper leaves challenged with the avirulent Xcv strain. Data represent means ± sd from three independent experiments. Asterisks indicate significant differences between empty vector control and CaRING1-silenced pepper plants as determined by Student’s t test (P < 0.05).
Using quantitative RT-PCR, PR marker gene transcript levels were assayed in empty vector control (TRV:00) and CaRING1-silenced pepper leaves before and 12 and 24 h after infection with virulent and avirulent strains of Xcv (Fig. 7A). CaRING1 expression was very low in CaRING1-silenced pepper leaves, suggesting that this gene was silenced effectively. Three pepper PR genes were assayed: CaBPR1 (basic PR1; SA-dependent marker), CaDEF1 (defensin), and CaPO2 (peroxidase; positive regulator of HR). In comparison with plants expressing the empty vector control (TRV:00), significantly lower levels of CaBPR1 induction were detected in CaRING1-silenced (TRV:CaRING1) leaves 24 h after infection with the avirulent Xcv strain. This finding suggests that CaRING1 may play a positive regulatory role in CaBPR1 expression. Similarly, induction of the pepper PR genes CaDEF1 and CaPO2 was also reduced by silencing of CaRING1 (Fig. 7A). Like CaRING1-silenced pepper leaves, CaRING1 UTR-silenced leaves exhibited significantly reduced induction of defense-related genes CaBPR1, CaDEF1, and CaPO2 during Xcv infection compared with empty vector control leaves, especially in the incompatible interactions (Supplemental Fig. S4).
Figure 7.
Expression of pepper defense-related maker genes and SA accumulation in CaRING1-silenced pepper leaves infected with Xcv. A, Expression of CaRING1 and pepper defense-related marker genes in empty vector control (TRV:00) and CaRING1-silenced (TRV:CaRING1) pepper plants 12 and 24 h after inoculation with virulent and avirulent strains of Xcv. Gene expression was analyzed using quantitative RT-PCR and normalized using constitutively expressed 18S rRNA. Data represent means ± sd from three independent experiments. Asterisks indicate significant differences between empty vector control and CaRING1-silenced pepper plants as determined by Student’s t test (P < 0.05). H, Healthy leaves. B, Free SA and total SA levels in empty vector control (TRV:00) or CaRING1-silenced (TRV:CaRING1) pepper leaves 12 and 24 h after inoculation (107 cfu mL−1) with the virulent Xcv strain Ds1 (compatible) and the avirulent Xcv strain Bv5-4a (incompatible). Data represent means ± sd from three independent experiments. Asterisks indicate significant differences between empty vector control and CaRING1-silenced pepper plants as determined by Student’s t test (P < 0.05). FW, Fresh weight.
Since CaBPR1 expression was reduced in CaRING1-silenced pepper plants, it is possible that the synthesis and/or recognition of SA is also affected. Free SA and glucoside-conjugated SA (SAG) levels were determined in CaRING1-silenced (TRV:CaRING1) plants infected with virulent and avirulent strains of Xcv. Significant accumulation of free SA was detected in plants expressing the empty TRV vector (TRV:00), whereas CaRING1-silenced (TRV:CaRING1) plants showed, on average, a 2-fold reduction in free SA accumulation 24 h after infection with the avirulent Xcv strain (Fig. 7B). By contrast, CaRING1-silenced (TRV:CaRING1) plants exhibited reduced free SA but highly elevated SAG accumulation during infection with the avirulent strain of Xcv (Fig. 7B). CaRING1-silenced (TRV:CaRING1) leaves accumulated approximately 2-fold more SAG than wild-type leaves at 24 h after avirulent Xcv infection. These findings indicate that CaRING1 negatively regulates the transformation of SAG to SA. In addition, the reduction in free SA levels in CaRING1-silenced leaves supports the hypothesis that CaRING1 modulates the expression of SA-dependent CaBPR1 as well as resistance to Xcv infection in pepper.
Overexpression of CaRING1 Enhances Disease Resistance in Arabidopsis
Since transformation of pepper plants is difficult, CaRING1 was overexpressed in Arabidopsis to assess the in vivo effects of CaRING1. Among 16 T2 lines of CaRING1-overexpressing (OX) plants, lines 5, 13, and 16 displayed strong CaRING1 expression levels. These lines were selected for further study after confirmation of CaRING1 transcript levels using RT-PCR (Fig. 8A). No apparent phenotypic differences were observed between the wild type and CaRING1-OX lines.
Figure 8.
CaRING1-OX transgenic Arabidopsis plants exhibit enhanced resistance to Pst DC3000 infection. A, RT-PCR analysis of CaRING1 expression in wild-type (WT) and 35S:CaRING1 transgenic lines. Expression of the ubiquitin (UBQ) gene was used as a control. B, Disease symptoms on the leaves of wild-type or transgenic plants 6 d after infection with Pst DC3000 (106 cfu mL−1). C, Bacterial growth in wild-type and transgenic plant leaves at 0 or 3 d after inoculation with Pst DC3000 (105 cfu mL−1). Data represent means ± sd from three independent experiments. Statistically significant differences between means were determined using Fisher’s lsd test (P < 0.05). D, Expression of CaRING1, AtPR1, AtPDF1.2, and AtRD29a in wild-type and transgenic plants (T3) at 24 and 48 h after infiltration with Pst DC3000. Quantitative analysis was performed using RT-PCR, and relative gene expression levels were normalized using the constitutively expressed gene AtACT1. Data represent means ± sd from three independent experiments. Asterisks indicate significant differences between empty vector control and CaRING1-silenced pepper plants as determined by Student’s t test (P < 0.05). E, Free SA and SAG levels in wild-type and transgenic plants 24 and 48 h after infiltration with Pst DC3000. Data represent means ± sd from two independent experiments. Asterisks indicate significant differences between wild-type and transgenic plants as determined by Student’s t test (P < 0.05). FW, Fresh weight. [See online article for color version of this figure.]
To determine the role played by CaRING1 in pathogen resistance, wild-type and CaRING1-OX lines were inoculated with Pst DC3000 and disease development was observed (Fig. 8B). Six days after inoculation of leaves with Pst DC3000, wild-type plants showed severe chlorotic symptoms, whereas CaRING1-OX transgenic lines exhibited only slight chlorotic symptoms. Consistent with a reduction in visible disease symptoms, CaRING1 transgenic lines exhibited significantly lower bacterial growth 3 d after inoculation than wild-type plants (Fig. 8C). In Arabidopsis, enhanced disease resistance is often associated with elevated expression of several marker PR genes. Quantitative RT-PCR was used to examine the expression of AtPR1, which is associated with the SA-dependent pathway, AtPDF1.2, which is related to the ethylene/JA-dependent pathway, and AtRD29a, a known regulator of abscisic acid metabolism (Fig. 8D). Pst DC3000 infection induced stronger expression of AtPR1 and AtPDF1.2 in transgenic plants than in the wild type. In particular, CaRING1-OX transgenic plants exhibited a close relationship between AtPR1 and AtPDF1.2 induction upon Pst DC3000 infection. After inoculation with Pst DC3000, the expression levels of AtPR1 and AtPDF1.2 transcripts in CaRING1-OX transgenic plants peaked at 24 and 48 h, respectively. These results indicate that CaRING1 may effectively regulate cross talk between SA- and ethylene/JA-dependent pathways in the defense response of Arabidopsis. However, levels of the abscisic acid-responsive transcript AtRD29a were reduced in wild-type and CaRING1-OX transgenic plants 24 h after inoculation with Pst DC3000, but the down-regulation was more pronounced in the CaRING1-OX line compared with the wild-type plants. As compared with the wild-type plants inoculated with Pst DC3000, accumulation of both free SA and SAG was significantly enhanced in CaRING1-OX transgenic plants 24 and 48 h after inoculation with Pst DC3000 (Fig. 8E). These findings support the possibility that the increased expression of AtPR1 in transgenic plants is intimately associated with the induced accumulation of SA for resistance to Pst DC3000 infection.
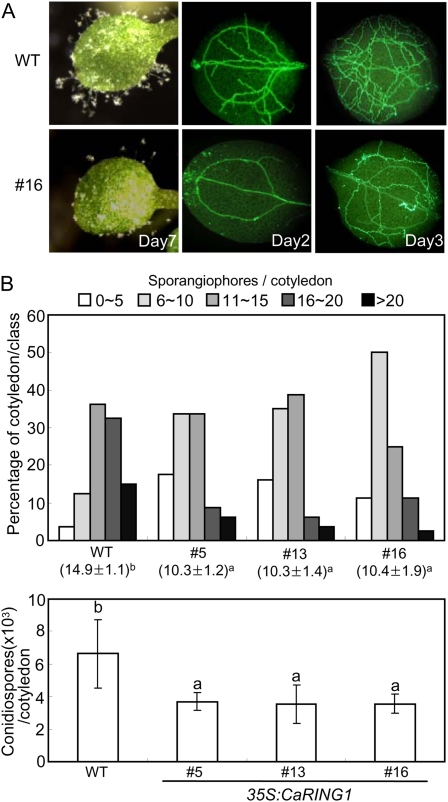
To determine whether CaRING1 overexpression alters resistance to a biotrophic oomycete pathogen, 7-d-old cotyledons of transgenic CaRING1-OX plants were sprayed with a conidiospore suspension (5 × 104 mL−1) of Hyaloperonospora arabidopsidis isolate Noco2 (Fig. 9). Transgenic CaRING1-OX plants not only exhibited reduced formation of sporangiophores compared with wild-type plants but also showed retarded hyphal growth in cotyledons (Fig. 9A). Consistent with these visually observed phenotypes, the number of sporangiophores and spores was reduced significantly in CaRING1 transgenic lines compared with wild-type plants (Fig. 9B). These observations indicate that CaRING1 overexpression suppresses H. arabidopsidis sporulation and induces a defense response against the infection. Taken together, these results suggest that CaRING1 overexpression enhances basal defense to hemibiotrophic bacterial and biotrophic oomycete pathogens in Arabidopsis plants.
Figure 9.
CaRING1-OX transgenic Arabidopsis plants show enhanced resistance to H. arabidopsidis isolate Noco2 infection. A, Disease symptoms and aniline blue-stained mycelia on cotyledons of wild-type (WT) and transgenic Arabidopsis plants at 7 d and 2 to 3 d after spray inoculation with H. arabidopsidis isolate Noco2 (5 × 104 spores mL−1), respectively. B, Production of sporangiophores per cotyledon of wild-type and transgenic plants 6 d after inoculation with H. arabidopsidis. Single cotyledons from over 60 seedlings per genotype were analyzed with the microscope and categorized into one of five categories of sporangiophore production (0–5, 6–10, 11–15, 16–20, or >20, depending on the number of sporangiophores observed). Average numbers of newly formed sporangiophores on cotyledons of wild-type and transgenic lines are indicated below each of the lines analyzed. C, Sporulation of H. arabidopsidis isolate Noco2 per cotyledon at 8 d after inoculation. Each experiment contained average spore counts from 50 inoculated cotyledons of wild-type and transgenic lines. Statistically significant differences between means were determined using Fisher’s lsd test (P < 0.05). [See online article for color version of this figure.]
DISCUSSION
In this study, we have found that the pepper E3 ubiquitin ligase RING1 (CaRING1) gene is required for disease resistance and for the HR cell death against Xcv infection. CaRING1 contains an N-terminal TM domain and C-terminal region RING domain (Fig. 1). The presence of a RING domain in CaRING1 indicates that this protein may possess E3 ubiquitin ligase activity (i.e. the ability to transfer ubiquitin to specific substrates in the ubiquitination machinery; Fig. 3). Ubiquitination is a unique eukaryotic posttranslational modification system that has been shown to play a significant role in the recognition and induction of various signals by modulating the stability of proteins involved in signal perception or responses (Smalle and Vierstra, 2004; Kwon et al., 2006; Dreher and Callis, 2007). In plants, various physiological and biochemical pathways are regulated by ubiquitin-mediated degradation of specific target proteins in response to biotic and abiotic stresses (Kwon et al., 2006; Dreher and Callis, 2007). To date, RING domains have been identified in several defense-related proteins induced by pathogen infection (Hong et al., 2007; Stone and Callis, 2007; Liu et al., 2008). The E3 ubiquitin ligase activity of RING-type proteins can be predicted by in silico analysis of amino acid sequences (Stone et al., 2005). This study has shown that bacterial pathogens can induce CaRING1 expression and that CaRING1 is important for production of the HR and restriction of pathogen growth, both of which are controlled by E3 ubiquitin ligases (Devoto et al., 2003).
MBP-CaRING1 displayed E3 ubiquitin ligase activity, in which E1, E2, and ATP were required for in vitro ubiquitination. Analysis of deletion and site-directed mutants revealed that the CaRING1 RING domain is essential for in vitro ubiquitination (Fig. 3). However, immunoblotting with anti-MBP antibody determined that the E3 ubiquitin ligase activity of MBP-CaRING1ΔTM was significantly lower than that of MBP-CaRING1 (Fig. 3D). Previously, it has been suggested that MBP is ubiquitinated by E3 ubiquitin ligases (Matsuda et al., 2001; Takai et al., 2002). Therefore, the reduced level of ubiquitination may be due to deletion of the TM domain (residues 1–78), and UbPred programs (http://ubpred.org/) reveal that this region of CaRING1 contains a predicted ubiquitination site (residue 15). BCA1, a RING-type E3 ubiquitin ligase in human cells, is known to regulate its own stability by autoubiquitination (Amemiya et al., 2008). Unexpectedly, however, the mutation of CaRING1 at the Lys-15 residue in the TM region did not abrogate the ubiquitination of CaRING1 K15R (Supplemental Fig. S2). However, the MBP-CaRING1ΔTM deletion mutant was impaired, suggesting an involvement of this TM domain at least in in vitro ubiquitination. Transfer of ubiquitins from the E2 to substrates, which is mediated by RING E3s, is known to require the substrate-docking site on RING E3s, which is located many angstroms from the predicted location of the docked E2-bound ubiquitin (Zheng et al., 2000; Deshaies and Joazeiro, 2009). Thus, decreased ubiquitination by MBP-CaRING1ΔTM may be associated with a change in space between the MBP (substrate) and the E2 on CaRING1 caused by the TM deletion. Taken together, these results suggest that the TM region of CaRING1 may be involved in the transfer of ubiquitins from the E2 to substrates on CaRING1, leading to the normal ubiquitination. However, the precise mechanism underlying the ubiquitination of CaRING1 remains to be investigated.
Following transient expression of CaRING1 in onion epidermal cells, CaRING1 localized to the plasma membrane and its TM domain was essential for this subcellular localization (Fig. 4). The E3 ubiquitin ligases Arabidopsis RING1 and rice EL5 are rapidly induced as part of the disease resistance process, and these proteins have recently been detected in the plasma membrane (Koiwai et al., 2007; Lin et al., 2008). In particular, Arabidopsis RING1 is associated with plasma membrane lipid rafts, and since similar assemblages have been implicated in signaling pathways and apoptosis of animal lymphocytes, this protein may be induced for programmed cell death in Arabidopsis (Lin et al., 2008). These data suggest that localization of CaRING1 to the plasma membrane may be important for signal transduction of hypersensitive cell death during infection.
Agrobacterium-mediated CaRING1 expression significantly induced a cell death phenotype in pepper leaves (Fig. 5). Markers for hypersensitive cell death include the accumulation of UV-fluorescing phenolic compounds, a ROS (H2O2) burst, and increased electrolyte leakage. These characteristic markers were detected at areas of localized cell death in leaves transiently expressing CaRING1. The findings described above strongly support the notion that CaRING1 expression is required for HR-like cell death. The E3 ubiquitin ligase activities of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are involved in cell death and defense (Yang et al., 2006). ACRE276 was identified as a potential E3 ubiquitin ligase, since it was rapidly induced in R gene Cf9-carrying tomato plants by infection with the fungal pathogen Cladosporium fulvum expressing the elicitor Avr9 (Avr9/Cf9). Although ACRE276 does not encode a RING-type E3 ubiquitin ligase, such as CaRING1, ACRE276 knockdown compromised HR triggered by various elicitors (Yang et al., 2006). More importantly, Agrobacterium-mediated expression of the site-directed mutants 35S:CaRING1C110S and 35S:CaRING1H131Y led to the distinctly decreased cell death and early defense responses compared with the wild-type 35S:CaRING1 (Fig. 5B). Together with E3 ubiquitin ligase activity assay data of these CaRING1 mutant proteins (Fig. 3D), these findings strongly support the notion that E3 ubiquitin ligase activity of CaRING1 is required for the induction of cell death in pepper plants.
CaRING1-silenced pepper plants exhibited enhanced susceptibility to virulent and avirulent Xcv infection (Figs. 6 and 7). This finding suggests that CaRING1 induction confers enhanced resistance to Xcv infection in pepper plants. CaRING1-silenced pepper plants displayed reduced induction of phenolic compounds, ROS burst, PR gene expression, and the HR, all of which are crucial components for effective resistance to avirulent Xcv infection. Expression of the SA-dependent CaBPR1, which is an essential PR gene during the HR, is severely compromised in the response of CaRING1-silenced pepper plants to avirulent Xcv infection (Fig. 8). Free SA and total SA were quantified to determine whether the reduced CaBPR1 expression correlated with SA accumulation. Consistent with the previous finding, CaRING1-silenced leaves showed an approximately 2-fold reduction in avirulent Xcv-induced free SA. However, SAG levels were significantly higher in CaRING1 gene-silenced leaves. In many plants, free SA represents a key compound for the activation of plant defense, and its accumulation is necessary for the induction of the HR and the expression of marker PR genes (Lee et al., 1995; Alvarez, 2000). In general, enhanced production of free SA induces SAG accumulation in the defense response, whereas reduced expression of free SA is accompanied by decreased accumulation of SAG (Zhou et al., 1998; Lee et al., 2007). Since high concentrations of free SA are phytotoxic, a large proportion of the SA in plants is present in conjugated, inactive forms that are glucosylated for storage. Moreover, SAG is not exuded from infected leaves and does not induce PR gene expression (Lee et al., 1995). These data support the suggestion that the decrease in free SA in CaRING1 gene-silenced leaves enhances plant susceptibility to avirulent Xcv infection. Thus, it is possible that CaRING1 may not modulate the production of SA synthesis but rather the conversion of SA to SAG, since the accumulation of total SA was similar in CaRING1-silenced and empty vector control plants. It is clear that the relationship between CaRING1 expression and the mechanisms underlying the conversion of SA to SAG during infection with avirulent Xcv strains remains to be elucidated.
CaRING1-OX Arabidopsis plants exhibited significantly enhanced resistance to Pst DC3000 infection, which was accompanied by rapid induction of AtPR1, increased accumulation of SA and AtPDF1.2, but not AtRD29a (Fig. 8). These enhanced defense responses may be activated by various signal transduction pathways, which can be modulated by plant hormones such as SA and JA/ethylene (Lee and Hwang, 2005; Pieterse et al., 2009; Choi and Hwang, 2011). In Arabidopsis, AtPR1 induction depends upon the SA-dependent pathway (Glazebrook et al., 1996; Rogers and Ausubel, 1997), whereas AtPDF1.2 is a JA/ethylene-responsive marker (Penninckx et al., 1998). Our findings support the suggestion that CaRING1 is necessary for the coordination of SA- and JA/ethylene-responsive gene expression. However, further study will be required to develop a full understanding of the mechanisms underlying cross talk between SA- and JA/ethylene-dependent signaling pathways in CaRING1-OX Arabidopsis plants during the defense response. In addition, CaRING1 overexpression also enhanced resistance to infection with the biotrophic pathogen H. arabidopsidis (Fig. 9). Similar to our findings, enhanced resistance to TMV and P. syringae pv tabaci infection has recently been demonstrated in tobacco plants overexpressing the E3 ubiquitin ligase OsBIRF1 (Liu et al., 2008).
In conclusion, it is clear that the E3 ubiquitin ligase function of pepper CaRING1 is critical for the plant defense response. Moreover, overexpression of CaRING1 can confer nonhost resistance to infection with bacterial and fungal pathogens. Taken together, our results suggest that CaRING1 is a positive regulator of cell death in pepper and requires E3 ubiquitin ligase activity for its function. It is also likely that CaRING1 functions as a novel positive regulator of cell death by degrading negative regulators of cell death. Thus, CaRING1 may be important for timely activation of the plant defense response, accompanied by HR. Further identification of CaRING1 substrate proteins will be required for complete elucidation of the roles played by this E3 ubiquitin ligase in the plant defense response.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Pepper (Capsicum annuum ‘Nockwang’) seeds were planted in a plastic tray (55 × 35 × 15 cm) containing soil mix (perlite:vermiculite:loam soil, 1:1:3, v/v/v) and grown at 25°C under a 16-h day and light intensity of 65 μmol photons m−2 s−1. At the two-leaf stage, three seedlings were transplanted to a plastic pot (5 × 15 × 10 cm) containing the same soil mix. Pepper plants at the six-leaf stage were used for pathogen inoculation or other treatments.
Seeds of Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) and CaRING1-OX lines were surface sterilized, planted on growth medium (1× Murashige and Skoog [MS] and 1% [w/v] Suc), and kept at 4°C for a minimum of 3 d to overcome dormancy. Seedlings at the two-leaf stage were transplanted to pots containing vermiculite, perlite, and loam soil (1:1:2, v/v/v) and then grown in a climate chamber at 24°C under long-day conditions (16 h of light per day; 130 μmol photons m−2 s−1) and 60% humidity.
Pathogens and Inoculation Procedures
The bacterial strains Xanthomonas campestris pv vesicatoria Ds1 (virulent) and Bv5-4a (avirulent) were grown overnight in yeast-nutrient broth (5 g L−1 yeast extract, 8 g L−1 nutrient broth) at 28°C. A needleless syringe was used to infiltrate bacterial suspensions into the abaxial side of fully expanded leaves at the six-leaf stage. Infected leaves were harvested at various time points to determine bacterial growth and perform RNA gel-blot analysis. Pseudomonas syringae pv tomato DC3000 was grown overnight in King’s B medium containing 50 mg mL−1 rifampicin and 50 mg mL−1 kanamycin. To measure bacterial growth, leaves from 4-week-old wild-type and CaRING1-OX Arabidopsis transgenic T3 plants were infiltrated with 105 colony-forming units (cfu) mL−1 Pst DC3000 using a needleless syringe. The infected plants were incubated at 26°C and harvested at various time points to determine bacterial growth. One-week-old seedlings of wild-type (Col-0) and CaRING1-OX transgenic plants were spray inoculated with conidiospores (5 × 104 spores mL−1) of Hyaloperonospora arabidopsidis isolate Noco2. The infected plants were incubated at 17°C in a controlled-environment chamber. Sporangiophores and conidiospores were counted at 6 and 8 d after inoculation, respectively.
Isolation and Sequence Analysis of CaRING1
Previously, leaves were inoculated with the avirulent Xcv strain Bv5-4a, and following isolation of total mRNA, cDNAs were amplified for use as probes to screen a cDNA library established from elicited pepper cells (Jung and Hwang, 2000). Differential hybridization was then used to isolate the 594-bp full-length CaRING1 cDNA. Among the cDNA clones tested, CaRING1 cDNA hybridized strongly to cDNA probes from leaves infected with the avirulent Xcv strain Bv5-4a.
Sequence analysis was performed using BLAST (Altschul et al., 1997; http://blast.ncbi.nlm.nih.gov/), and homologous proteins were identified by searching with the CaRING1 protein sequence. TMHMM (Krogh et al., 2001) and SMART (http://smart.embl-heidelberg.de/) Web servers were used for the identification of protein domains. A phylogenetic tree was constructed by the neighbor-joining method using ClustalW (http://www.ch.embnet.org/software/ClustalW.html).
Expression and Purification of Recombinant Proteins
The vector pMAL-c4X (New England Biolabs) was used to construct expression vectors containing fusions with CaRING1, RING-H2 domain-deleted CaRING1ΔRING, TM domain-deleted CaRING1ΔTM, and the site-directed mutants CaRING1K15R, CaRING1C110S, and CaRING1H131Y. The expression constructs were transformed into Escherichia coli BL21 (DE2) cells to produce MBP fusion proteins. As a negative control, pMAL-c4x vector was used to express MBP alone. Overnight cultures grown at 37°C were used to inoculate fresh Luria-Bertani (LB) medium (50 μg mL−1 kanamycin), and then cells were incubated at 37°C until the optical density at 600 nm (OD600) was 0.4 to 0.6. Expression was then induced by the addition of 300 μm isopropyl-β-d-thiogalactopyranoside, and cultures were grown for a further 3 h at 37°C. Cells were harvested and resuspended in column buffer (20 mm Tris-HCl [pH 7.4] and 0.2 m NaCl), followed by sonication and centrifugation at 13,000g for 15 min at 4°C. Expressed MBP fusion proteins were purified by amylase affinity chromatography, according to the manufacturer’s instructions (New England Biolabs).
A clone encoding the full-length Arabidopsis UBC10 (At5g53300) was obtained from the Arabidopsis Biological Resource Center (http://www.Arabidopsis.org), and the fragment encoding UBC10 was ligated into pET28a (Invitrogen). The 6xHis-UBC10 fusion protein was transformed into E. coli BL21 (DE2) and expressed as described above. Harvested cells were resuspended in 1× native purification buffer (50 mm NaH2PO4 [pH 8.0] and 0.5 m NaCl), and 6xHis-UBC10 was purified by affinity chromatography using nickel-nitrilotriacetic acid agarose resin, according to the manufacturer’s instructions (Invitrogen).
E3 Ubiquitin Ligase Activity Assay
For the E3 ubiquitin ligase activity assay, each reaction (30 μL final volume) contained 10 μg of bovine ubiquitin (Sigma-Aldrich), 0.1 μg of yeast E1 (Boston Biochem), 0.2 μg of purified Arabidopsis E2 6xHis-UBC10, and 0.2 μg of purified E3 (MBP-CaRING1, CaRING1ΔTM, CaRING1ΔRING, CaRING1K15R, CaRING1C110S, or CaRING1H131Y) in ubiquitination buffer (40 mm Tris-HCl [pH 7.5], 5 mm MgCl2, 2 mm ATP, and 2 mm dl-dithiothreitol). After incubation at 30°C for 3 h, the reaction was stopped with 2× SDS-PAGE loading buffer (20 mm Tris-HCl [pH 7.5], 20% glycerol, 5% SDS, 2 mm EDTA, 200 mm dl-dithiothreitol, and 0.02% bromphenol blue) at 80°C for 7 min. Aliquots (10 μL) for each reaction were separated by electrophoresis using 8% SDS-PAGE gels. Immunoblot analyses were performed using anti-ubiquitin antibody (Sigma-Aldrich) and anti-MBP antibody (New England Biolabs) raised in rabbits.
Subcellular Localization of CaRING1
For transient expression in onion (Allium cepa) epidermal cells, the cauliflower mosaic virus 35 promoter was used to drive the expression of gene fusions (CaRING1-smGFP, the RING domain deletion CaRING1ΔRING-smGFP, or the TM domain deletion CaRING1ΔTM-smGFP) carried in the binary vector p326GFP. Plasmids were purified using Qiagen plasmid maxi kits, and particle bombardment assays were performed using a Bio-Rad He/1000 particle delivery system. Bombarded cells were incubated for 24 h on 1× MS agar, and GFP fluorescence was observed using a LSM 5 Exciter microscope (Carl-Zeiss) with a 488-nm filter. To confirm that smGFP was anchored to the cell membrane, plasmolyzed cells were observed.
RNA Gel-Blot Analysis
TRIzol (Invitrogen) was used to isolate total RNA from healthy pepper tissues (leaves, stems, roots, flowers, and fruits) and leaves infected with the virulent Xcv strain Ds1 and the avirulent Xcv strain Bv5-4a. The RNA was separated by electrophoresis on 1.2% formaldehyde-agarose gels, blotted onto Hybond N+ (Pall), and then incubated at 65°C for 1 h in hybridization buffer (5% [w/v] dextran sulfate, 0.25 m disodium phosphate [pH 7.2], 7% [w/v] SDS, and 1 mm EDTA). EcoRI restriction was used to release the cDNA fragment containing CaRING1 from the TOP blunt vector. The fragment was then randomly labeled using [γ-32P]dCTP, heat denatured, added to hybridization buffer, and incubated with blocked membranes overnight at 65°C. Membranes were washed once with 2× SSC, 0.1% SDS at room temperature, twice with 0.1× SSC, 0.1% SDS at 65°C, and then exposed to x-ray film.
Virus-Induced Gene Silencing
In this study, the TRV vectors pTRV1 and pTRV2 were used for VIGS (Liu et al., 2002). TOP blunt vector (Enzynomix) containing full-length CaRING1 cDNA (385 bp) was digested with EcoRI, and the resulting fragment was inserted into the same site in pTRV2. The 3′ region of the CaRING UTR was PCR amplified using specific primers of CaRING1UTR (forward, 5′-GAATTCGAGTTGCCGGTAGCTAGCTCCTCAT-3′; reverse, 5′-GAATTCACAAATTCACATTAATCCAAAC-3′) and inserted into pTRV2 to generate pTRV2:CaRING1UTR. pTRV1, pTRV2:00, pTRV2:CaRING1, and pTRV2:CaRING1UTR constructs were transformed into Agrobacterium tumefaciens strain GV3101. Cultures of Agrobacterium strain GV3101 containing each construct were grown for 16 h at 28°C. Agrobacterium cultures were pelleted, resuspended in infiltration buffer (10 mm MES, 10 mm MgCl [pH 5.7]), and adjusted to an OD600 of 1.0. Cells were incubated in 200 μm acetosyringone at room temperature for 1 to 2 h before use. An equal volume of pTRV1 Agrobacterium culture was mixed with one of the pTRV2 cultures before infiltration (OD600 = 0.2). Agrobacterium cultures were infiltrated into the cotyledons of pepper seedlings (Hong et al., 2008). After 24 to 48 h at 17°C in a growth chamber, infected plants were grown under a 16/8-h light/dark cycle at 25°C. At 4 and 5 weeks after infiltration, the upper leaves of the infected plants were used for RT-PCR analysis and various disease assays.
Measurement of SA and SAG
SA extraction and quantification were performed as described by Aboul-Soud et al. (2004) and Choi et al. (2011). Pepper and Arabidopsis leaf tissues (0.5 g) were ground to a powder using liquid nitrogen and extracted in 1 mL of 90% methanol. 3-Hydroxy benzoic acid (50 μg) in 100% methanol was added to each sample as an internal standard. Samples were vortexed, sonicated for 15 min, and centrifuged at 15,000g for 10 min at 4°C. The supernatant was transferred to a tube, and the remaining pellet was reextracted in 1 mL of absolute methanol. The supernatant fractions were combined and dried in a speed vacuum under heat. The pellet was resuspended in 1 mL of 5% TCA and sonicated for 10 min. Organic extraction of free SA was performed by adding 1 mL of ethylacetate:cyclopentane:isopropanol (50:50:1). The upper phase containing free SA was transferred to a tube, and the aqueous phase was then reextracted. Supernatants were dried under nitrogen gas and then suspended in 0.5 mL of the HPLC mobile phase (55% acetonitrile:45% deionized water with 4% [v/v] acetic acid). The resulting solution was filtered and separated on a C18 analytical column (J’sphere ODS-H80, 150 × 4.6 mm; YMC) using HPLC (Waters) and detected with a fluorescence detector (excitation at 305 nm, emission at 405 nm; Waters). The HPLC was programmed for isocratic conditions (80% methanol) and a flow rate of 1.0 mL min−1. The aqueous phase containing SAG was acidified to pH 1 with HCl and then boiled for 30 min to release SA from any acid-labile conjugated forms. The released SA was then extracted with the organic mixture and treated as described above. SA and SAG were quantified by area integration of HPLC peaks.
Generation of Transgenic Arabidopsis Plants
Full-length CaRING1 cDNA was PCR amplified in the sense and antisense directions using the following primers: CaRING1 forward (5′-TCTAGAATGAGTAGTGATGATCCTTTACAT-3′) and reverse (5′-GAGCTCTTACGGCAGAAATGC-3′). Fragments were cloned into the TOP blunt vector (Enzynomix). The resulting plasmid was digested with XbaI/SacI, and then the CaRING1 gene fragment was ligated into pBIN35S. To generate transgenic Arabidopsis plants, the construct was transformed into Agrobacterium strain GV3101 using the floral dipping method (Clough and Bent, 1998). Putative transgenic Arabidopsis plants harboring the 35S:CaRING1 construct were selected on MS plates (Duchefa) containing 50 mg L−1 kanamycin.
Protein Gel-Blot Analysis
Pepper leaves were infiltrated with Agrobacterium carrying CaRING1-GFP, CaRING1C110S-GFP, or CaRING1H131Y-GFP. Total proteins were extracted from the pepper leaves transiently expressing these constructs. For immunoblotting, harvested pepper leaves (0.5 g) were ground in liquid nitrogen and homogenized in 500 μL of extraction buffer (50 mm Tris [pH 7.5], 150 mm NaCl, 4 m urea, 0.2% Nonidet P-40, and plant protease inhibitor cocktail [Roche]). The lysates were centrifuged at 15,000g for 30 min at 4°C. Supernatants were collected and quantified by Bradford assay. Proteins were analyzed using immunoblots probed with anti-GFP antibody (Santa-Cruz Biotechnology).
RT-PCR and Real-Time RT-PCR
TRIzol reagent (Invitrogen) was used to extract total RNA from the leaves of pepper and Arabidopsis plants, according to the manufacturer’s recommendations. First-strand cDNA was synthesized from total RNA (2 μg) using Moloney murine leukemia virus reverse transcriptase (Enzynomics). Real-time PCR was performed using the SYBR Green Supermix (Bio-Rad). The following gene-specific primer pairs were used: for CaRING1, forward (5′-ATGAGTAGTGATGATCCTTTACATAACG-3′) and reverse (5′-TTACGGCAGAAATGCATTTACAT-3′); for Arabidopsis UBQ, forward (5′-CAAGACAGGAGAAATATGTCTCG-3′) and reverse (5′-ATCCTTTCTTAGGCATAG-3′); for CaBPR1, forward (5′-CAGGATGCAACACTCTGGTGG-3′) and reverse (5′-ATCAAAGGCCGGTTGGTC-3′); for CaDEF1, forward (5′-CAAGGGAGTATGTGCTAGTGAGAC-3′) and reverse (5′-TGCACAGCACTATCATTGCATAC-3′); for CaPO2, forward (5′-GCAGAAACGGATCTCCCTG-3′) and reverse (5′-CTCCCATTCTAATCATGGCAG-3′); and for pepper 18S rRNA, forward (5′-AAACGGCTACCACATCCAAG-3′) and reverse (5′-ACCCATCCCAAGGTTCAACT-3′).
Agrobacterium-Mediated Transient Expression in Pepper Leaves
For analysis of cell death caused by CaRING1 overexpression, Agrobacterium strain GV3101 containing empty control vector (35S:00) or binary vectors expressing CaRING1 (35S:CaRING1) or the site-directed CaRING1 mutants (35S:CaRING1C110S and 35S:CaRING1H131Y) was infiltrated into pepper leaves at the eight-leaf stage (Choi and Hwang, 2011). The development of cell death was observed at 3 and 7 d after infiltration. Agrobacterium cultures were grown overnight at 28°C in 10 mL of LB medium containing 50 μg mL−1 kanamycin and 50 μg mL−1 rifampicin. Cultures were pelleted and resuspended in induction medium (10 mm MES [pH 5.6], 10 mm MgCl2 and 200 mm acetosyringone) at OD600 = 2 and then left at room temperature for 2 to 3 h prior to infiltration. Agrobacterium cultures (OD600 = 0.1–1.0) were infiltrated into leaves between the lateral veins, and then plants were incubated and observed at room temperature for 2 weeks. Autofluorescence was detected under UV illumination.
Histochemical Staining
For visualization of H2O2, inoculated leaves were detached at various time points and dipped into a solution containing 1 mg mL−1 3,3-diaminobenzidine solution. Samples were stained for 15 h and then cleared in 95% ethanol. For detection of cell death, infected leaves were sampled at the time points indicated and then stained with lactophenol-trypan blue solution (10 mL of lactic acid, 10 mL of glycerol, 10 g of phenol, and 10 mg of trypan blue in 10 mL of distilled water). Leaves were boiled briefly in staining solution and then cleared in chloral hydrate solution (2.5 g mL−1 chloral hydrate). Samples were mounted in 60% glycerol, and representative phenotypes were photographed with a light microscope (Olympus).
Measurement of Electrolyte Leakage
To assay cell death, electrolyte leakage was measured from six leaf discs. Leakage was compared between wild-type plants and lines transiently expressing CaRING1 or empty vector control as well as CaRING1-silenced plants. Leaf discs were immersed in 30 mL of nonionic distilled water and then shaken slowly at room temperature. After incubation, the conductivity of the bathing solution was measured using a Crison conductivity meter (Hach).
Quantification of H2O2 Using the Xylenol Orange Assay
H2O2 production was measured in wild-type and CaRING1-OX transgenic plants using freshly prepared xylenol orange reagents. Two reagents were prepared: reagent A [25 mm FeSO4, 25 mm (NH4)2SO4, and 2.5 m H2SO4] and reagent B (125 μm xylenol orange [Sigma-Aldrich] and 100 mm sorbitol). The xylenol orange reaction mixture comprised 0.1 mL of reagent A and 10 mL of reagent B. Pepper leaves were infiltrated with 107 cfu mL−1 avirulent Xcv strain Bv5-4a. Leaf discs were removed using a cork borer and then floated on 1 mL of distilled water. To quantify the H2O2, 100 μL of supernatant was added immediately to 1 mL of xylenol orange mix and then incubated for 30 min at room temperature. H2O2 production was determined spectrophotometrically by measuring A560. A standard H2O2 solution was used for calibration.
Sequences for genes used in this study can be found in the GenBank/EMBL data libraries under the following accession numbers: GQ359822 (CaRING1), AF053343 (CaBPR1), AF442388 (CaDEF1), DQ489711 (CaPO2), At2G14610 (AtPR1), At5g44420 (AtPDF1.2), D13044 (AtRD29a), and At3g62250 (AtUBQ5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide and predicted amino acid sequences of pepper CaRING1 cDNA, which encodes a RING-H2 finger protein.
Supplemental Figure S2. SDS-PAGE and E3 ubiquitin ligase activity assay of the recombinant MBP-CaRING1 and mutant variant proteins.
Supplemental Figure S3. Enhanced susceptibility of CaRING1UTR-silenced pepper plants to Xcv infection.
Supplemental Figure S4. Expression of CaRING1 and pepper defense-related marker genes in empty vector control (TRV:00) and CaRING1UTR-silenced (TRV:CaRING1UTR) pepper plants 12 and 24 h after inoculation with the virulent (Ds1; compatible) and avirulent (Bv5-4a; incompatible) strains of Xcv.
Acknowledgments
We thank Dr. S.P. Dinesh-Kumar (Yale University) for the pTRV1 and pTRV2 vectors and Dr. U. Bonas (Martin-Luther-Universitaet) for the Agrobacterium strain GV3101.
References
- Aboul-Soud MAM, Cook K, Loake GJ. (2004) Measurement of salicylic acid by a high-performance liquid chromatography procedure based on ion-exchange. Chromatographia 59: 129–133 [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez ME. (2000) Salicylic acid in the machinery of hypersensitive cell death and disease resistance. Plant Mol Biol 44: 429–442 [DOI] [PubMed] [Google Scholar]
- Amemiya Y, Azmi P, Seth A. (2008) Autoubiquitination of BCA2 RING E3 ligase regulates its own stability and affects cell migration. Mol Cancer Res 6: 1385–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Borden KL, Freemont PS. (1996) The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol 6: 395–401 [DOI] [PubMed] [Google Scholar]
- Callis J, Vierstra RD. (2000) Protein degradation in signaling. Curr Opin Plant Biol 3: 381–386 [DOI] [PubMed] [Google Scholar]
- Choi DS, Hwang BK. (2011) Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23: 823–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HW, Kim YJ, Hwang BK. (2011) The hypersensitive induced reaction and leucine-rich repeat proteins regulate plant cell death associated with disease and plant immunity. Mol Plant Microbe Interact 24: 68–78 [DOI] [PubMed] [Google Scholar]
- Choi HW, Kim YJ, Lee SC, Hong JK, Hwang BK. (2007) Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol 145: 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. (1998) The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J 17: 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Costet L, Fritig B, Kauffmann S. (2002) Scopoletin expression in elicitor-treated and tobacco mosaic virus-infected tobacco plants. Physiol Plant 115: 228–235 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Devoto A, Muskett PR, Shirasu K. (2003) Role of ubiquitination in the regulation of plant defence against pathogens. Curr Opin Plant Biol 6: 307–311 [DOI] [PubMed] [Google Scholar]
- Dreher K, Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JDG. (2000) cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12: 963–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Gachon C, Baltz R, Saindrenan P. (2004) Over-expression of a scopoletin glucosyltransferase in Nicotiana tabacum leads to precocious lesion formation during the hypersensitive response to tobacco mosaic virus but does not affect virus resistance. Plant Mol Biol 54: 137–146 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T. (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- González-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JDG. (2006) The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JK, Choi HW, Hwang IS, Hwang BK. (2007) Role of a novel pathogen-induced pepper C3-H-C4 type RING-finger protein gene, CaRFPI, in disease susceptibility and osmotic stress tolerance. Plant Mol Biol 63: 571–588 [DOI] [PubMed] [Google Scholar]
- Hong JK, Choi S, Kim SH, Yi SY, Kim YJ, Hwang BK. (2008) Distinct roles of the pepper pathogen-induced membrane protein gene CaPIMP1 in bacterial disease resistance and oomycete disease susceptibility. Planta 228: 485–497 [DOI] [PubMed] [Google Scholar]
- Hwang IS, Hwang BK. (2011) The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol 155: 447–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HW, Hwang BK. (2000) Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 13: 136–142 [DOI] [PubMed] [Google Scholar]
- Kim DS, Hwang BK. (2011) The pepper receptor-like cytoplasmic protein kinase CaPIK1 is involved in plant signaling of defense and cell death responses. Plant J 66: 642–655 [DOI] [PubMed] [Google Scholar]
- Koiwai H, Tagiri A, Katoh S, Katoh E, Ichikawa H, Minami E, Nishizawa Y. (2007) RING-H2 type ubiquitin ligase EL5 is involved in root development through the maintenance of cell viability in rice. Plant J 51: 92–104 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kwon SJ, Choi EY, Choi YJ, Ahn JH, Park OK. (2006) Proteomics studies of post-translational modifications in plants. J Exp Bot 57: 1547–1551 [DOI] [PubMed] [Google Scholar]
- Lee HI, León J, Raskin I. (1995) Biosynthesis and metabolism of salicylic acid. Proc Natl Acad Sci USA 92: 4076–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- Lee SC, Hwang BK. (2005) Induction of some defense-related genes and oxidative burst is required for the establishment of systemic acquired resistance in Capsicum annuum. Planta 221: 790–800 [DOI] [PubMed] [Google Scholar]
- Lin SS, Martin R, Mongrand S, Vandenabeele S, Chen KC, Jang IC, Chua NH. (2008) RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. Plant J 56: 550–561 [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang H, Yang Y, Li G, Yang Y, Wang X, Basnayake BM, Li D, Song F. (2008) Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses. Plant Mol Biol 68: 17–30 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Matsuda N, Suzuki T, Tanaka K, Nakano A. (2001) Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J Cell Sci 114: 1949–1957 [DOI] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD. (2004) The transcriptional innate immune response to flg22: interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135: 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF. (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM. (1997) Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9: 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Mondragón RE, Garcidueñas-Piña C, Guzmán P. (1999) Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in Arabidopsis thaliana. Plant Mol Biol 40: 579–590 [DOI] [PubMed] [Google Scholar]
- Serrano M, Guzmán P. (2004) Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics 167: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Stone SL, Callis J. (2007) Ubiquitin ligases mediate growth and development by promoting protein death. Curr Opin Plant Biol 10: 624–632 [DOI] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai R, Matsuda N, Nakano A, Hasegawa K, Akimoto C, Shibuya N, Minami E. (2002) EL5, a rice N-acetylchitooligosaccharide elicitor-responsive RING-H2 finger protein, is a ubiquitin ligase which functions in vitro in co-operation with an elicitor-responsive ubiquitin-conjugating enzyme, OsUBC5b. Plant J 30: 447–455 [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. (2008) Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Tsitsigiannis DI, Rowland O, Lo J, Rallapalli G, Maclean D, Takken FL, Jones JD. (2008) The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 20: 697–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LCM, Rep M, Pieterse CM. (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Yang CW, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JDG, Sadanandom A. (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Vega-Sánchez ME, Zhu T, Wang GL. (2006) Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res 16: 413–426 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102: 533–539 [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]