Abstract
Imbibed Arabidopsis (Arabidopsis thaliana) seeds are encapsulated by mucilage that is formed of hydrated polysaccharides released from seed coat epidermal cells. The mucilage is structured with water-soluble and adherent layers, with cellulose present uniquely in an inner domain of the latter. Using a reverse-genetic approach to identify the cellulose synthases (CESAs) that produce mucilage cellulose, cesa5 mutants were shown to be required for the correct formation of these layers. Expression of CESA5 in the seed coat was specific to epidermal cells and coincided with the accumulation of mucilage polysaccharides in their apoplast. Analysis of sugar composition showed that although total sugar composition or amounts were unchanged, their partition between layers was different in the mutant, with redistribution from adherent to water-soluble mucilage. The macromolecular characteristics of the water-soluble mucilage were also modified. In accordance with a role for CESA5 in mucilage cellulose synthesis, crystalline cellulose contents were reduced in mutant seeds and birefringent microfibrils were absent from adherent mucilage. Although the mucilage-modified5 mutant showed similar defects to cesa5 in the distribution of sugar components between water-soluble and adherent mucilage, labeling of residual adherent mucilage indicated that cesa5 contained less cellulose and less pectin methyl esterification. Together, the results demonstrate that CESA5 plays a major and essential role in cellulose production in seed mucilage, which is critical for the establishment of mucilage structured in layers and domains.
Seeds of a number of species, such as Arabidopsis (Arabidopsis thaliana), accumulate polysaccharides in the epidermal cells of their seed coat. On imbibition, the hydrated polysaccharides expand, rupture the outer tangential cell wall, and form a viscous mucilage that encapsulates the seed. The physiological role of seed mucilage remains to be determined, but functions in the control of germination or seed dispersal have been proposed (Witztum et al., 1969; Young and Evans, 1973; Gutterman and Shem-Tov, 1996; Penfield et al., 2001; Arsovski et al., 2009). In Arabidopsis, the major component of seed mucilage is the pectin rhamnogalacturonan I (RG I; Goto, 1985; Western et al., 2000, 2001, 2004; Penfield et al., 2001, Usadel et al., 2004; Macquet et al., 2007a). Detailed characterization of the structure and composition of seed mucilage in the Columbia (Col-0) accession have shown that it is not uniform but is composed of two layers, termed water soluble and adherent. The inner adherent layer is tightly associated with the seed and is composed of two domains (Macquet et al., 2007a). The water-soluble layer of mucilage is almost exclusively unbranched RG I in a slightly expanded random-coil conformation. In contrast, RG I in adherent mucilage has a small number of arabinan and/or galactan ramifications (Dean et al., 2007; Macquet et al., 2007a, 2007b; Arsovski et al., 2009). There are also minor amounts of the pectin homogalacturonan (HG) within the adherent mucilage (Willats et al., 2001), and its degree of methyl esterification differs, being higher in the outer compared with the inner domain (Macquet et al., 2007a). The inner domain of adherent mucilage also contains cellulose (Windsor et al., 2000; Willats et al., 2001; Blake et al., 2006; Macquet et al., 2007a).
The complex structure of Arabidopsis seed mucilage implies a high degree of organization in the temporal and spatial deposition of its components. The accumulation of mucilage polysaccharides in the apoplast of epidermal cells of the seed coat occurs during seed development. The seed coat is a maternal tissue, and the epidermal cell layer is one of two that constitute the outer integument of the ovule. The complex morphological changes that occur during the differentiation of seed coat epidermal cells have been characterized (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). After an initial phase of cell expansion, polysaccharides are synthesized and secreted into the apoplast in a polarized manner between the radial and outer tangential cell walls. This is correlated with the formation of a central column of cytoplasm, an increase in Golgi stacks (Young et al., 2008), and the accumulation of starch granules. This column and the apical surface of the cytoplasm then accumulate secondary cell wall material, leading to programmed cell death. The resulting polygonal cells have a central volcano-shaped structure, called a columella, connected to reinforced radial cell walls, thus forming a moat around the columella that is filled with dehydrated polysaccharides under a primary cell wall.
The identification of maternally inherited mutants affected in seed mucilage production has led to the characterization of a number of genes coding either transcription regulators or polysaccharide metabolism enzymes. The majority of the transcription regulators identified (APETALA2, ENHANCER OF GLABRA3, GLABRA2 [GL2], MYB5, MYB61, TRANSPARENT TESTA8, TRANSPARENT TESTA GLABRA1, TRANSPARENT TESTA GLABRA2) regulate the differentiation of seed coat cells and are required for normal epidermal cell morphology and mucilage production (Jofuku et al., 1994; Rerie et al., 1994; Walker et al., 1999; Nesi et al., 2000; Penfield et al., 2001; Johnson et al., 2002; Zhang et al., 2003; McAbee et al., 2006; Gonzalez et al., 2009; Li et al., 2009). An exception is the recently characterized LEUNIG_HOMOLOG1, where mutants are only affected in mucilage extrusion (Bui et al., 2011; Walker et al., 2011). This transcription regulator appears to modulate the expression of the β-d-galactosidase MUCILAGE-MODIFIED2 (MUM2).
Mutants affected in genes coding enzymes involved in polysaccharide metabolism present more diverse phenotypes, in accord with the complexity of the structure and composition of seed mucilage. RHAMNOSE SYNTHASE2/MUM4 produces UDP-l-Rha, one of the subunits of the RG I backbone (Usadel et al., 2004; Western et al., 2004; Oka et al., 2007), and the putative galacturonsyltransferase GAUT11 could potentially synthesize mucilage RG I or HG pectin domains from UDP-d-GalA (Caffall et al., 2009). The RG I deposited in the apoplast of seed coat epidermal cells appears to be synthesized in a more branched form that is subsequently matured by enzymes in the apoplast; the β-d-galactosidase MUM2 removes Gal residues from RG I galactan branches, and the bifunctional β-d-xylosidase/α-l-arabinofuranosidase BXL1 removes Ara from side chains (Dean et al., 2007; Macquet et al., 2007a; Arsovski et al., 2009). HG is synthesized in a highly methyl esterified form and is then demethyl esterified by pectin methylesterases. Mutants defective for the subtilisin-like Ser protease AtSBT1.7 are affected in both mucilage polysaccharide and outer tangential cell wall properties (Rautengarten et al., 2008). This protease could target a pectin methylesterase or activate a pectin methylesterase inhibitor, as methyl esterification was reduced in the mutant. The genes defective in the mutants mum3 and mum5 remain to be identified, but they affect the composition or structural properties of released mucilage (Western et al., 2001; Macquet et al., 2007a).
Cellulose microfibrils are synthesized from UDP-Glc by a multiprotein complex within the plasma membrane containing cellulose synthase (CESA) catalytic subunits. In Arabidopsis, 10 CESA genes have been identified, but functional analysis of mutants has not yet associated all of them with physiological roles. Mutants in CESA4, CESA7, CESA8, and CESA9 are affected in secondary cell wall cellulose (Taylor et al., 1999, 2000, 2003; Stork et al., 2010), and a role in cellulose synthesis in primary cell walls is favored for the remainder. CESA1 and CESA3 null mutants are gametophytic lethal and have been proposed to be unique components of primary wall CESA complexes (Persson et al., 2007). Mutants with point mutations such as cesa1rsw1 or cesa3je5 have shown that CESA1 and CESA3 function is required in a variety of tissues (Desnos et al., 1996; Fagard et al., 2000a; Williamson et al., 2001; Burn et al., 2002). CESA6 null mutants have relatively mild growth phenotypes that have been related to partial redundancy with CESA2 and CESA5, based on double mutant phenotypes (Desprez et al., 2007; Persson et al., 2007). Although cesa9 mutants exhibit defects in secondary cell wall formation in seed coat epidermal cells, to date no mutant affecting the production of cellulose in seed mucilage has been described.
Here, using a reverse genetic approach, we show that CESA5 is required for the production of seed mucilage cellulose, in agreement with its expression in the epidermal layer of the seed coat. Furthermore, the mum3 mutant, isolated in a forward genetic screen, is shown to be a cesa5 mutant allele. A detailed functional analysis of the contribution of CESA5 to mucilage structure and composition is presented and shows a key role for cellulose in the adhesion of mucilage pectin components to the seed. Comparison of cesa5 and mum5 seed mucilage indicates that cellulose must interact with another component of seed mucilage to create the strong adhesion of the inner layer to the seed.
RESULTS
Seed Mucilage Staining Is Modified in cesa5 Mutants
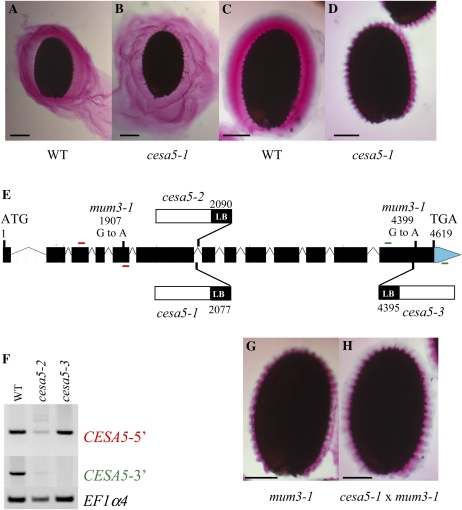
A previous study of Arabidopsis seed coat mucilage had shown, using cytochemical probes and cellulase digestion, that the inner domain of adherent mucilage contains cellulose (Macquet et al., 2007a). In order to determine which CESAs were involved in the synthesis of this cellulose, mucilage released from the seeds of mutants affected in the 10 different CESA genes was examined using ruthenium red staining. Direct staining of seed mucilage found no major difference between mutants and the wild type, although there appeared to be more diffuse mucilage around cesa5-1 mutant seeds (Fig. 1, A and B). After imbibition and shaking in water, however, seeds from the cesa5-1 mutant had a very different staining pattern from those of the wild type (Fig. 1, C and D). The amount of adherent mucilage attached to the seed coat of mutant seeds appeared to be reduced. To confirm that the ruthenium red-staining phenotype observed in the mutant was due to the T-DNA insertion in the CESA5 gene, homozygous lines for two additional T-DNA mutants were isolated and called cesa5-2 and cesa5-3 (Fig. 1E). The adherent seed mucilage released from both lines appeared reduced, like that of cesa5-1. Furthermore, backcrossed seeds from all three mutants still exhibited the phenotype, as did seeds of progeny from crosses between cesa5-1 and cesa5-2 or cesa5-3. The presence of normal mucilage released from F1 seed coats of F2 seeds from these backcrosses indicated that all three mutant alleles were recessive and maternally inherited.
Figure 1.
CESA5/MUM3 is involved in the formation of adherent seed mucilage. A to D, Visualization of seed mucilage released from the Arabidopsis seed coat by ruthenium red staining. Wild-type (WT) seeds and cesa5-1 seeds were stained directly (A and B) or after imbibition and shaking in water (C and D). E, Schematic representation of the structure of the CESA5 gene as annotated by The Arabidopsis Information Resource (http://www.arabidopsis.org/index.jsp) indicating the sites and nature of the cesa5 mutants and the positions of primers used in F. Red bars indicate primers for CESA5-5′, and green bars indicate primers for CESA5-3′ PCR amplifications. LB, Left border. F, Effect of cesa5-2 and cesa5-3 mutations on CESA5 expression compared with the wild type. Reverse transcription-PCR analysis was performed with CESA5-specific primer pairs at the 5′ or 3′ end of the CESA5 gene (CESA5-5′ or CESA5-3′, respectively). A control amplification was carried out with primers for the EF1α4 gene. G and H, mum3-1 seed (G) and F1 seed coat/F2 seed from a cesa5-1 × mum3-1 cross (H) stained after imbibition and shaking in water. Bars = 150 μm (A–D) and 100 μm (G and H).
Analysis of CESA5 expression in the two new mutant alleles was determined by reverse transcription-PCR on RNA extracts from developing seeds using CESA5-specific primers situated 5′ of the insertions or at the 3′ end of the gene (Fig. 1E). Amplification was observed with the 5′ primer pair in the wild type and the two mutants, with similar differences in intensity between sample amplifications to those observed for the EF1α4 control (Fig. 1F). With the 3′ primer pair, no fragment was amplified for cesa5-3, indicating that the mutant is a knockout, whereas a faintly observed fragment in cesa5-2 suggested that some transcripts are produced in this mutant (Fig. 1F).
mum3-1 Is a CESA5 Mutant Allele
A previous forward genetic screen for modified seed mucilage had identified two mutants, mum3-1 and mum5-1, that released mucilage with modified ruthenium red staining properties only after shaking in water prior to staining (Western et al., 2001). Ruthenium red-stained adherent mucilage from cesa5-1 more closely resembled that of the mum3-1 mutant than that of mum5-1, where the adherent layer thickness was more significantly reduced (Fig. 1G; Macquet et al., 2007a). Progeny from reciprocal crosses between the three cesa5 mutant alleles and mum3-1 were examined for mucilage staining in F1 and F2 seed coats. In all cases, there was noncomplementation of the adherent seed mucilage phenotype, which was less intensely stained with ruthenium red and appeared thinner than that of the wild type (Fig. 1H). Sequencing of the CESA5 gene in the mum3-1 mutant identified two G-to-A polymorphisms compared with the wild type. The first, at amino acid 399, changes GAG to GAA, which are synonymous codons, whereas the second, at amino acid 997, changes a Trp to a stop codon. Although there is evidence that synonymous codon changes are not always silent and can modify gene expression (Hurst, 2011), the introduction of a stop codon and the resulting truncation of the CESA5 protein at the C terminus would be expected to have a more significant effect on protein function in the mum3-1 mutant.
The cesa5 Mucilage Phenotype Is Not Due to Defects in Seed Coat Differentiation
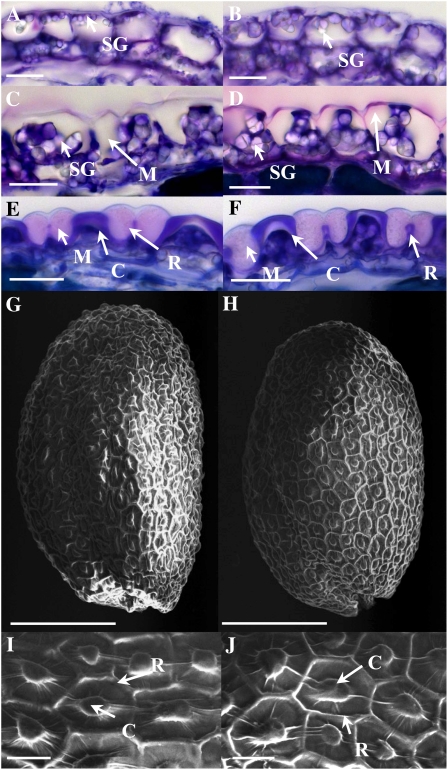
Developing seed sections were examined to determine whether CESA5 mutation also affected seed coat morphology or whether mucilage accumulation was modified during seed coat differentiation. Comparison of mutant and wild-type seed coats at identical stages found no obvious difference in the structure or timing of epidermal cell differentiation or in the aspect of accumulated mucilage (Fig. 2, A–F). The form of epidermal cells and the height of the columella on mature seeds were also unchanged (Fig. 2, G–J; Supplemental Fig. S1; Supplemental Movies S1 and S2).
Figure 2.
Seed coat epidermal cell differentiation in the cesa5 mutant. A to F, Sections of developing seeds stained with toluidine blue O. G to J, Scanning electron micrographs of the surface of mature dry seeds. A and B show seeds with linear cotyledon stage embryos/9 DAP, C and D show seeds with embryos at bent cotyledon stage/12 DAP, and E and F show seeds with mature embryos at midseed development/15 DAP. A, C, E, G, and I show wild-type seeds, and B, D, F, H, and J show cesa5-2 seeds. C, Columella; M, mucilage; R, radial cell wall; SG, starch granule. Bars = 20 μm (A–F, I, and J) and 150 μm (G and H).
CESA5 Is Expressed in Epidermal Cells of the Developing Seed Coat
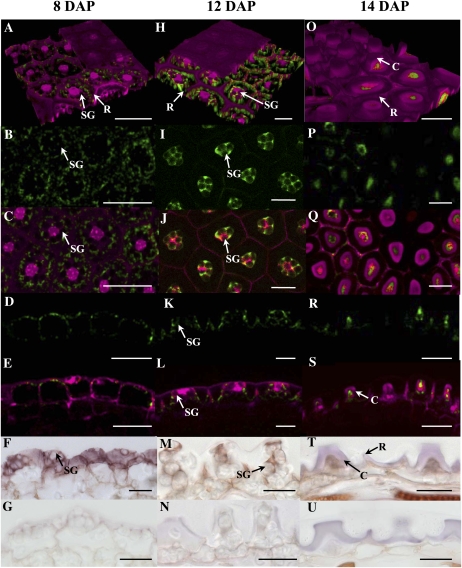
CESA5 transcripts have previously been shown to accumulate in young plants, flowers, and stems of mature plants and embryos (Beeckman et al., 2002). In addition, data available from transcriptome analyses using RNA extracted from laser-dissected seed coat tissues (Le et al., 2010; http://seedgenenetwork.net/arabidopsis) indicate that CESA5 is expressed at low levels in the seed coat during early embryogenesis and that levels rise from the linear cotyledon stage onward (Supplemental Fig. S2). In order to confirm the latter data and to determine precisely which seed coat cells expressed CESA5, developing seeds were analyzed from wild-type plants expressing reporter genes under the control of the CESA5 promoter. A transcriptional fusion with GUS and a translational fusion with GFP were used. To facilitate visualization, cell walls were counterstained with propidium iodide. The GFP-CESA5 protein produced with the translation fusion construct was functional, as when it was used to transform the cesa5-1 mutant, complementation of the adherent mucilage phenotype was observed (Supplemental Fig. S3, B versus A). In agreement with the low levels of expression indicated by the transcriptome data, reporter gene expression was only observed once seed embryo development was at the linear cotyledon stage (Fig. 3, A–E; Supplemental Fig. S4, A and B). Based on the laser gain used to generate fluorescence signals, the expression appeared to be highest during the bent cotyledon to mature green embryo stages (Fig. 3, H–L and O–S; Supplemental Fig. S4, C–F). In the seed coat, expression was limited to the epidermal cell layer, and at 8 d after pollination (DAP), it could be seen in the cytoplasm surrounding the large central vacuole and the starch granules that had accumulated (Fig. 3, A–F; Supplemental Fig. S4, A and B). At 12 DAP, the cytoplasm of the epidermal cells is present in a central column and at the base of the seed coat epidermal cells, and as well as labeling around the starch granules, GFP-CESA5 delimited the cytoplasmic column bordering the apoplasmic space where mucilage polysaccharides accumulate (Fig. 3, K and L). At the end of the seed coat epidermal cell differentiation process, the cytoplasm is reduced with the accumulation of secondary cell wall material in the columella and programmed cell death is initiated. Reporter gene expression was restricted to this reduced cytoplasm (Fig. 3, O–S; Supplemental Fig. S4, C–F), and the apparent reduction in reporter gene signal after the mature green embryo stage is in agreement with the process of programmed cell death. In situ hybridization was also carried out at different stages of seed development using a CESA5-specific probe. An identical pattern of CESA5 expression was detected to that observed using reporter genes, with labeling in the seed coat restricted to the cytoplasm of the epidermal cells (Fig. 3, F, M, and T); this was absent when using the sense control (Fig. 3, G, N, and U).
Figure 3.
Expression of CESA5 in the developing seed coat. A to E, H to M, and O to S show GFP-CESA5 fluorescence in the wild type (green), A, C, E, H, J, L, O, Q, and S show composite images with propidium iodide labeling (magenta), F, M, and T show in situ hybridization using a CESA5-specific antisense probe, and G, N, and U show in situ hybridization using a CESA5-specific sense probe in the seed coat of developing seeds (brown staining). A to G, Seeds with linear cotyledon stage embryos/8 DAP. H to N, Seeds with embryos at bent cotyledon stage/12 DAP. O to U, Seeds with mature embryos at midseed development/14 DAP. A, H, and O are three-dimensional reconstructions of seed coat epidermal cells, B, C, I, J, P, and Q are transverse optical sections through the basal region of the seed coat epidermal cells, and D to G, K to N, and R to U are longitudinal sections through the seed coat. B, D, I, K, P, and R show the same images as in C, E, J, L, Q, and S, respectively, with CESA5 fluorescence alone. C, Columella; R, radial cell wall; SG, starch granule. Bars = 50 μm (A–E), 20 μm (F–N, T, and U), and 25 μm (O–S).
CESA5 Mutation Induces Major Changes in the Structure of Adherent Mucilage
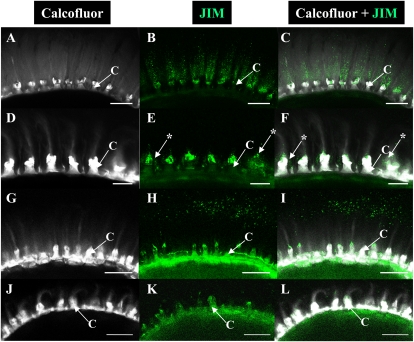
The adherent mucilage layer of wild-type Col-0 has previously been shown to be composed of two domains that differ in their composition and structure, cellulose being restricted to the innermost domain (Macquet et al., 2007a). To determine whether the structure and composition of adherent mucilage are modified in the reduced layer observed in cesa5, whole mount immunolabeling or staining of polysaccharides was carried out on mature dry seeds. Labeling of cellulose was carried out with Calcofluor, a fluorescent probe for β-glycans, and the carbohydrate-binding module (CBM) protein CBM3a, which binds to the planar surface of crystalline cellulose (Dagel et al., 2011); both have previously been shown to label adherent layers of seed mucilage (Willats et al., 2001; Blake et al., 2006; Macquet et al., 2007a). Calcofluor labeling of wild-type mucilage showed the expected combination of intense rays originating from the tops of the columella and diffuse labeling (Figs. 4, A and D, and 5, A and G; Supplemental Fig. S5A). In the mutant, Calcofluor labeling was very different, as although the intense rays associated with the tops of the columella were still observed, the diffuse staining was no longer evident (Figs. 4, G and J, and 5, D and J; Supplemental Fig. S5D). CBM3a labeled wild-type adherent mucilage in a similar manner to Calcofluor, with stronger labeling of the rays emanating from the tops of the columella and punctate labeling within the mucilage (Fig. 4, B, C, E, and F; Supplemental Fig. S3, C and D). In the mutant, CBM3a labeling of the rays radiating from the top of the columella was particularly prominent (Fig. 4, H, I, K, and L). Interestingly, composite images of Calcofluor and CBM3a labeling did not completely overlap, with CBM3a marking the peripheral ends of cellulose rays more strongly; this was most noticeable for cesa5 mucilage (Fig. 4, C, F, I, and L). Adherent seed mucilage was also examined for the cesa5-1 mutant expressing GFP-CESA5 under the control of the CESA5 promoter. Calcofluor and CBM3a labeling were found to be similar to that of the wild type, although the width of the cellulose domain appeared slightly reduced (Supplemental Fig. S3, F–H), suggesting partial complementation of cellulose synthesis.
Figure 4.
Cellulose labeling of adherent mucilage released from wild-type and cesa5-2 seeds. Confocal microscopy optical sections through mature, imbibed seeds and mucilage are shown. A, D, G, and J, Staining of β-glycans with Calcofluor. B, E, H, and K, Indirect immunofluorescence detection of His-tagged CBM3a binding to crystalline cellulose. C, F, I, and L, Composite images of double labeling with Calcofluor and CBM3a. D to F and J to L correspond to the magnification of regions in A to C and G to I, respectively. A to F show the wild type and G to L show cesa5-2. C, Columella; M, mucilage. Bars = 100 μm (A–I) and 50 μm (J–K). [See online article for color version of this figure.]
Figure 5.
Labeling of methyl esterified homogalacturonan in adherent mucilage released from wild-type and cesa5-2 seeds. Confocal microscopy optical sections of adherent mucilage released from mature, imbibed seeds are shown. A, D, G, and J show staining of β-glycans with Calcofluor, with immunofluorescence corresponding to B and E (JIM5) or H and K (JIM7) antibody labeling; C and F or I and L show composite images of double labeling with Calcofluor and JIM5 or JIM7, respectively. A to C and G to I, The wild type. D to F and J to L, cesa5-2. C, Columella. Asterisks indicate JIM5 labeling of mucilage around the columella. Bars = 50 μm (A–C and G–L) and 20 μm (D–F).
HG is present as a minor component of the wild-type adherent mucilage layer and was shown to be highly or moderately methyl esterified in the outer or inner domain of adherent mucilage, respectively (Macquet et al., 2007a; Figure 5, B, C, H, and I). In cesa5-2, moderately methyl esterifed HG, labeled with the JIM5 antibody, was only detectable in mucilage around the columella (Fig. 5, E and F), whose cell wall material is also labeled with this antibody (Macquet et al., 2007a). Labeling of highly methyl esterified HG in adherent mucilage with the JIM7 antibody was no longer observed in the adherent mucilage and was only detectable for columella and cell wall debris in a similar manner to the wild type (Fig. 5, K and L). The major component of adherent mucilage is RG I (Macquet et al., 2007a). In accordance, wild-type mucilage was strongly labeled with the INRA-RU2 antibody (Supplemental Fig. S5, B and C); this antibody was raised against RG I from Arabidopsis soluble mucilage (Ralet et al., 2010). The INRA-RU2 antibody only labeled a small part of the peripheral mucilage, probably because the large quantities of RG I epitope present had depleted all available antibody. Strong labeling was also observed against cesa5-2 adherent mucilage, and labeling extended further into the mucilage layer, suggesting that the mucilage was less dense than in wild-type RG I (Supplemental Fig. S5, E and F). Furthermore, the mucilage surface appeared uneven, suggesting that cells released variable quantities of mucilage. In agreement with ruthenium red staining (Fig. 1D), the adherent mucilage layer thickness was reduced compared with the wild type.
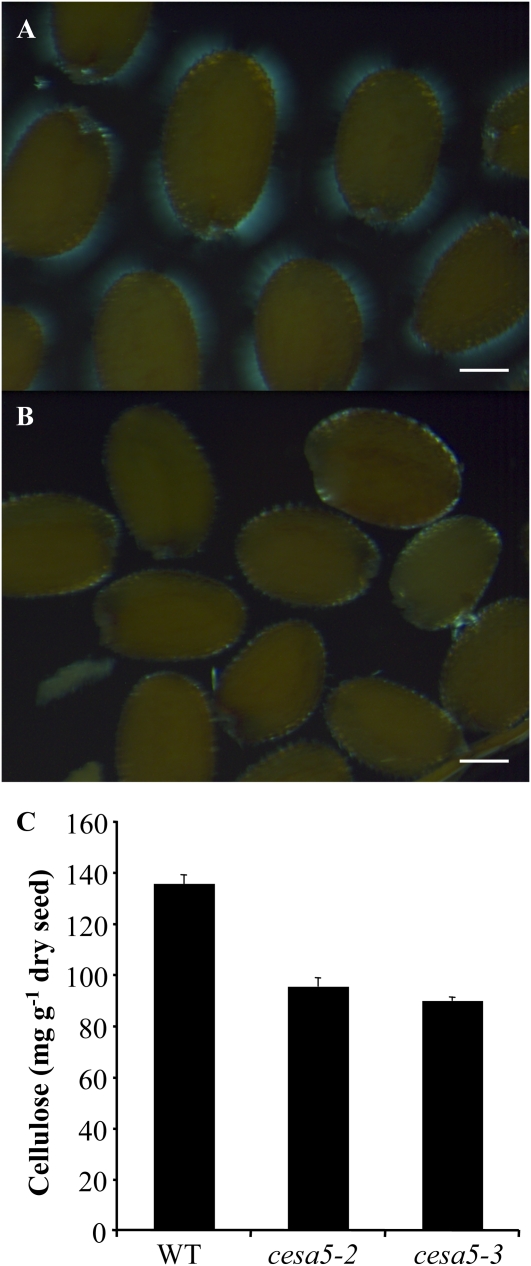
Crystalline Cellulose Is Reduced in cesa5 Mucilage and Seeds
Aligned cellulose microfibrils in crystalline cellulose produce birefringence of polarized light, and mutants with defects in trichome cellulose content have previously been identified based on their altered birefringence (Potikha and Delmer, 1995). The adherent mucilage of wild-type seeds was observed for birefringence by any crystalline cellulose present therein. Wild-type seeds showed bright regions with visible rays of crystalline cellulose within the adherent mucilage (Fig. 6A). In contrast, cesa5-2 and cesa5-3 seeds only exhibited bright spots on the edges of seeds under polarized light, indicating that the crystalline cellulose observed in the wild type was absent (Fig. 6B). Furthermore, quantification of crystalline cellulose contents in whole mature seeds showed that cesa5 mutants contained around 30% less crystalline cellulose than the wild type (Fig. 6C). During desiccation, the five cell layers of the seed coat are compressed to essentially the tannins accumulated in the endothelium and the epidermal cell layer containing mucilage polysaccharides and secondary cell wall material (Haughn and Chaudhury, 2005). Previous studies have shown that approximately 2% (w/w) of the weight of an Arabidopsis Col-0 seed is seed coat-derived tannins (Routaboul et al., 2006) and a further 3% (w/w) is seed coat-derived mucilage polysaccharides (Fig. 8; Macquet et al., 2007a). We estimate, therefore, that including the secondary cell wall material accumulated, the seed coat represents 7% of the mature dry seed weight. The observed reduction in crystalline cellulose represents around 4% of wild-type seed weight and more than half of the dry seed coat. This suggests that crystalline cellulose production is probably affected in other seed tissues in addition to the seed coat, in agreement with the expression of CESA5 in embryos (Beeckman et al., 2002; http://seedgenenetwork.net/arabidopsis).
Figure 6.
Crystalline cellulose is reduced in cesa5 mutants. A and B, Observation of birefringence of polarized light by crystalline cellulose in adherent mucilage released from mature, imbibed wild-type (A) and cesa5-2 (B) seeds. Bars = 150 μm. C, Crystalline cellulose contents determined by the Updegraff (1969) protocol for whole mature seeds of the wild type (WT), cesa5-2, and cesa5-3. Error bars represent se (n = 4). Similar results were obtained in two biological repeats. [See online article for color version of this figure.]
Figure 8.
The allocation of monosaccharides between adherent and water-soluble mucilage is altered in cesa5 mutants. Amounts of sugars are shown in water-soluble mucilage (black bars) or adherent mucilage (white bars) for wild-type (WT), cesa5-2 (c5-2), and cesa5-3 (c5-3) plants. Values are means of two independently extracted samples from three biological repeats. For sample sd, see Supplemental Table S1.
Water-Soluble Mucilage Amount and Macromolecular Properties Are Modified in cesa5 Mutants
Extracts of water-soluble mucilage were obtained from the wild type and cesa5 mutants and analyzed for their sugar composition. In agreement with previous studies (Macquet et al., 2007a, 2007b), water-soluble mucilage from wild-type Col-0 seeds was mainly composed of sugars (20 mg g−1 intact seeds), as shown in Table I. In wild-type Col-0, the majority of neutral sugars were previously shown to be Rha, in a molar ratio to GalA close to 1, typical of RG I (Macquet et al., 2007a, 2007b). Calculation of the molar ratio of GalA/neutral sugars measured in this study yielded a similar value of 1.13 ± 0.055. In cesa5-1 water-soluble mucilage, an increase was observed in the amount of total sugars, compared with that of wild-type Col-0. The amount of both GalA and neutral sugars increased, yet their molar ratio remained unchanged (1.16 ± 0.092). An average increase of 22% in the amount of water-soluble mucilage was calculated for cesa5-1 on the basis of sugar analyses.
Table I. Sugar composition and macromolecular characteristics of wild-type, cesa5-1, and mum5-1 water-extracted mucilage.
Values in parentheses are sd of four independently extracted samples from two biological repeats. Mn, Number-average molar mass; Mp, molar mass at peak maximum; Mw, weight-average molar mass; I, polydispersity index; [η], average intrinsic viscosity; a, Mark-Houwink coefficient; –, not determined.
| Measurement | Wild-Type Total Water-Extracted Mucilage | Wild-Type Polymeric Fraction 1 | Wild-Type Polymeric Fraction 2 | cesa5-1 Total Water-Extracted Mucilage | mum5-1 Total Water-Extracted Mucilage |
| Composition (mg g−1 intact seeds) | |||||
| GalA | 10.6 (1.07) | – | – | 13.1 (1.53) | 14.7 (1.27) |
| Neutral sugars | 9.4 (0.58) | – | – | 11.3 (0.44) | 12.4 (0.62) |
| GalA-neutral sugars | 1.13 (0.055) | – | – | 1.16 (0.092) | 1.18 (0.067) |
| Total sugars | 20.0 (1.63) | – | – | 24.3 (1.97) | 27.1 (1.79) |
| Total polymeric materiala | 22.0 (2.75) | 1.6 (0.10) | 20.4 (2.71) | 27.2 (3.58) | 30.6 (3.08) |
| Macromolecular characteristics | |||||
| Mn (kD) | 815 (95.1) | 29,953 (2623) | 754 (81.0) | 448 (45.9) | 422 (50.9) |
| Mp (kD) | 659 (90.9) | 32,650 (3519) | 659 (90.9) | 453 (12.7) | 450 (7.5) |
| Mw (kD) | 3,180 (587.8) | 30,593 (2763) | 911 (98.1) | 667 (167.7) | 650 (202.1) |
| I (Mw/Mn) | 3.9 (0.32) | 1.0 (0.00) | 1.2 (0.04) | 1.5 (0.68) | 1.5 (0.31) |
| [η] (mL g−1) | 601 (8.3) | 808 (10.7) | 583 (10.4) | 594 (12.3) | 633 (16.9) |
| a | – | – | 0.70 (0.166) | 0.69 (0.120) | 0.75 (0.145) |
From HP-SEC refractometric signal, using dn/dc = 0.146.
The macromolecular parameters of wild-type and cesa5-1 water-soluble mucilage were determined using high-performance size-exclusion chromatography (HP-SEC) coupled with refractive index, dual laser light scattering, and viscosimetry detection (Table I). As previously observed (Macquet et al., 2007a), wild-type water-extracted mucilage had an elution profile that separated two polymeric populations (Fig. 7). The first one, representing 7% ± 1% of the extracted mucilage, eluted at the void volume and was of extremely high average molar mass (Mn approximately 30,000 kD). The intrinsic viscosity of polymeric fraction 1 was moderate ([η] approximately 800 mL g−1) considering its very high molar mass. The second polymeric population was of high average molar mass (Mn approximately 750 kD) and exhibited a high intrinsic viscosity ([η] approximately 600 mL g−1). Added together, the two polymeric populations accounted for 22.0 ± 2.75 mg g−1 intact seeds, in good agreement with values calculated from sugar analyses. All values obtained were in excellent agreement with previously published data (Macquet et al., 2007a) and indicated the presence of small amounts of entangled, collapsed, or aggregated macromolecules together with large amounts of a homogeneous population of “coiled” macromolecules of high molar mass (Mn approximately 750 kD).
Figure 7.
Separation of the different macromolecular populations present in wild-type, cesa5-1, and mum5-1 water-extracted mucilage by HP-SEC. RI, Normalized refractive index signal voltage. Wild type, black circles; cesa5-1, white circles; mum5-1, white triangles. Each point represents the mean of four independent extractions from two biological repeats.
The macromolecular characteristics of the cesa5-1 water-soluble mucilage exhibited major differences from those of the wild type. The high molar mass population (polymeric fraction 1) was totally absent, while the amount of the second population (polymeric fraction 2) was notably increased (Fig. 7). The unique polymeric population of cesa5-1 water-soluble mucilage accounted for 27.2 ± 3.58 mg g−1 intact seeds, in good agreement with values calculated from sugar analyses. Altogether, using the refractive index data, an average increase of 23% in the water mucilage amount was calculated for cesa5-1 compared with the wild type. Comparison of different molar mass characteristics for the polymeric population 2 indicated a moderate decrease in average Mn, weight-average molar mass (Mw), and molar mass at peak maximum (Mp) for cesa5-1 water-soluble mucilage compared with the wild type (−40%, −27%, and −31%, respectively). Intrinsic viscosity and Mark-Houwink coefficient values, however, were unaltered. This revealed that there were no major conformational differences between wild-type and cesa5-1 water-soluble mucilage in polymeric population 2.
The modifications observed for cesa5-1 water-soluble mucilage were similar to those previously reported for mum5-1 (Macquet et al., 2007a). To determine the level of similarity in water-soluble mucilage characteristics between cesa5-1 and mum5-1, extracts of the latter were analyzed in parallel (Table I). The increase in the amount of total sugars in water-soluble mucilage was even higher in mum5-1 than in cesa5-1, with an average increase compared with the wild type calculated as 45% based on sugar analyses. Nevertheless, as with cesa5-1 mucilage and as observed previously (Macquet et al., 2007a), this was due to increases in both GalA and neutral sugar amounts, and the molar ratio was unchanged (1.18 ± 0.067). The macromolecular characteristics of the mum5-1 mucilage were similar, with the same loss of polymeric population 1 and increase in polymeric population 2, although this increase was higher for mum5-1, corresponding to 30.6 ± 3.08 mg g−1 intact seeds, in agreement with the increased amounts of sugars measured from water-soluble mucilage (Fig. 7). Previously, the polymeric population 2 was resolved into two peaks for mum5-1, probably because HP-SEC was carried out with two columns in series compared with only one in our study (Macquet et al., 2007a). The average increase in the amount of mum5-1 water-soluble mucilage, calculated using refractive index data, was 39%. The analyses of molar mass characteristics, as detailed above for cesa5-1, indicated the same 30% decrease for mum5-1, with no effect on intrinsic viscosity and Mark-Houwink coefficient values. Together, these results show that cesa5-1 and mum5-1 had similar effects on the macromolecular characteristics and quantity of water-soluble mucilage. Nevertheless, a greater increase was observed in the amount of mum5-1 water-soluble mucilage.
The Distribution of Monosaccharides between Water-Soluble and Adherent Mucilage Is Modified in cesa5
To recover and analyze the water-soluble and adherent mucilage layers separately, a sequential extraction method (Macquet et al., 2007a) was used on the wild type, cesa5-2, and cesa5-3. As for cesa5-1, the amounts of both acidic and neutral sugars present in water-soluble mucilage were higher in cesa5-2 and cesa5-3 compared with those of the wild type (Fig. 8; Supplemental Table S1). Average increases of 17% and 19% in the amounts of water-soluble mucilage were calculated for cesa5-2 and cesa5-3, respectively, on the basis of sugar analyses. This correlated with decreased amounts of both acidic and neutral sugars in cesa5-2 and cesa5-3 adherent mucilage compared with the wild type (Fig. 8; Supplemental Table S1). Average decreases of 52% and 56% in the amounts of adherent mucilage were calculated for cesa5-2 and cesa5-3, respectively, on the basis of sugar analyses. Altogether, a clear redistribution of adherent to water-soluble mucilage was observed, as the total amount of each sugar or total sugars in mucilage (i.e. water soluble plus adherent) was unaltered (Fig. 8; Supplemental Table S1). In the wild type, the proportion of extruded mucilage that was water soluble was calculated to be 69% ± 3%, whereas in cesa5, this represented 85% ± 5%.
DISCUSSION
Cellulose is a major component of plant cell walls, where it is intimately associated with pectin and hemicellulose and plays a critical role in plant growth, in particular during cell expansion (Cosgrove, 2005). In seed mucilage, cellulose represents between 12% and 19% of the adherent mucilage layer (Macquet et al., 2007a), and its function remains to be elucidated. An important first step is the identification of the CESAs involved in cellulose synthesis in adherent mucilage. A reverse genetic approach using mutants in all 10 CESA genes showed that mutation of CESA5 affected the mucilage released from seeds on imbibition (Fig. 1, A–D), and the width of the inner layer of adherent mucilage was dramatically reduced. The effects observed were unambiguously shown to be caused by CESA5 mutation, as the phenotypes were consistently observed after backcrossing to the wild type in three independent alleles and were complemented by transformation with a translational fusion of GFP-CESA5 under the control of the CESA5 promoter (Supplemental Fig. S3). Furthermore, the mum3-1 mutant was found to be an allele of cesa5, with a point mutation in the coding sequence that introduces a premature stop codon (Fig. 1E); this mutant had previously been identified in a forward genetic screen for modified mucilage staining with ruthenium red (Western et al., 2001).
Although no obvious seed mucilage phenotype was observed on ruthenium red staining of mutants other than cesa5, the mutants examined for cesa1 and cesa3 are leaky, as knockout alleles are lethal. It remains possible, therefore, that CESA1 and CESA3 participate in cellulose production in mucilage. In vegetative tissues, cellulose synthesis has been shown to require complexes containing at least three different CESA isoforms and other associated proteins (Taylor et al., 2003; Desprez et al., 2007; Gu et al., 2010). It will be important to determine whether similar heteromeric complexes synthesize the cellulose present in adherent mucilage. Nevertheless, the strong phenotype for cesa5 indicates a major and nonredundant contribution of the CESA5 gene to the production of this cellulose. This contrasts with CESA5 mutation in vegetative tissues or dark-grown hypocotyls, where it is only in combination with cesa2 or cesa6 that phenotypes are observed; notably, cesa5 cesa6 is seedling lethal (Desprez et al., 2007).
Morphological defects in seed coat epidermal cells have previously been reported for cesa9 mutants due to modified secondary cell wall synthesis (Stork et al., 2010). Detailed observation of cesa5 seed coat epidermal cell morphology and differentiation during seed development and in mature seeds found no difference from the wild type (Fig. 2; Supplemental Fig. S1). Nonetheless, it is possible that CESA5 plays a redundant role in the production of cellulose in the walls of these cells, which could be determined by functional analysis of multiple cesa mutants. Studies of such mutants could also highlight redundant roles for other CESAs in complexes synthesizing mucilage cellulose, and as transcriptome data obtained using seed coat RNA (Le et al., 2010; http://seedgenenetwork.net/arabidopsis) also indicate the expression of CESA1, CESA2, CESA3, CESA6, and CESA10 in the seed coat, any or all of these could be involved in cellulose production in the epidermal cells.
CESA5 Is Specifically Expressed in the Epidermal Cells of the Seed Coat
In situ hybridization and the expression of reporter genes from the CESA5 promoter found that expression in the seed coat was specific to the epidermal cells that accumulate mucilage (Fig. 3; Supplemental Fig. S4). Furthermore, the timing and apparent peak of expression coincided with the period associated with mucilage accumulation during seed development, in accordance with a requirement for CESA5 for the production of normal seed mucilage. As both transcriptional and translational reporter gene fusions yielded identical results, this indicates that RNA and protein abundance are correlated and that posttranscriptional regulation of CESA5 expression was not significant during seed coat differentiation. In root hairs, CESA5 expression is directly controlled by binding of the transcription factor GL2 to an L1 box in the CESA5 promoter (Tominaga-Wada et al., 2009). GL2 controls the formation of root hairs, an epidermal tissue, as well as the differentiation of the epidermal cells of the seed coat, and it is possible that in the latter, this also involves the control of CESA5 expression.
Cellulose present in mucilage must be synthesized into the apoplasm surrounding the central cytoplasmic column prior to the deposition of secondary cell wall components. CESAs are embedded in the plasma membrane (Cosgrove, 2005), and the observation of GFP-CESA in the cytoplasm delimiting this apoplast (Fig. 3, K and L) is in accord with a role for CESA5 in the production of this cellulose. Further studies will be required to confirm that CESA5 is indeed present at the plasma membrane in seed coat epidermal cells.
CESA5 Synthesizes Crystalline Cellulose Present in Seed Mucilage
Comparison of cellulose in the mucilage of cesa5 mutants with the fluorescent probe Calcofluor showed that there was a clear reduction in labeled cellulose within adherent seed mucilage (Fig. 4, A, D, G, and H). Cellulose microfibrils are insoluble crystalline β-1,4-glucan produced from the self-association of cellulose chains by van de Waals or hydrogen bonds after their synthesis. The microfibrils can be organized in highly ordered crystalline, semiordered paracrystalline, and disordered noncrystalline or amorphous states (Blake et al., 2006). Crystalline fibrils can cause double refraction, or birefringence, of a ray of polarized light, and the brightness resulting from light refraction by cellulose microfibrils was clearly visible in wild-type adherent mucilage; very little light refraction was observed for cesa5 seeds, indicating an absence of crystalline microfibrils in their mucilage (Fig. 6, A and B). Furthermore, total seed crystalline cellulose contents were reduced by 30% in cesa5 mutants (Fig. 6C). These results corroborate that CESA5 functions as an essential catalytic subunit of the CESA complex in developing epidermal cells of the seed coat.
A number of CBMs have been characterized that target different states of microfibrils, and CBM3a binds to the planar surface of crystalline cellulose (Dagel et al., 2011). In contrast to the observations above, labeling of the cellulose radiating from the columella tops into the adherent mucilage was stronger with CBM3a in cesa5, indicating increased amounts of crystalline cellulose in the mutant. This apparent paradox could arise because in the wild type, the accessibility of the CBM3a protein to crystalline cellulose in the adherent mucilage is blocked by high-molecular-mass pectin components in the outer domain of the adherent mucilage, whereas in the mutant, a reduction in pectin components facilitates the access to remaining crystalline cellulose. In effect, CBM3a labeling of rays was strongest at the outer edge of the mucilage, and labeling of mucilage with INRA-RU2 was observed deeper into the residual mucilage of cesa5 than in wild-type adherent mucilage (Supplemental Fig. S5). The presence of CBM3a and Calcofluor binding in the mutant highlights that some cellulose was still synthesized and indicates that other CESAs could contribute to the production of mucilage cellulose, in particular that in the rays observed from the top of the columella.
Cellulose Synthesis by CESA5 Anchors the Pectin Components of Mucilage to the Seed
In addition to increased CBM3a binding, ruthenium red staining in cesa5 indicated that there was less adherent mucilage as its width was reduced (Fig. 1D). Immunolabeling with anti-pectin antibodies also highlighted the reduced width of the adherent layer in the mutant (Fig. 5, B, E, H, and K; Supplemental Fig. S5). Nevertheless, for mum2, it was previously shown that although the width of the adherent mucilage was reduced, the amount of polysaccharide-derived sugars was higher than in the wild type (Macquet et al., 2007b). Analyses of water-soluble and adherent mucilage extracts from cesa5 seeds established that although the total amounts of GalA, neutral sugars, and total sugars present were approximately the same, the distribution between water-soluble and adherent mucilage layers was modified, with a reduction in adherent mucilage (Fig. 8). Interestingly, as well as being more abundant, the macromolecular characteristics of cesa5 water-soluble mucilage were altered, with the loss of a high-molecular-mass polymeric fraction that corresponds to aggregated, collapsed, or entangled macromolecules (Fig. 7; Table I). These results demonstrate that the crystalline cellulose present in the adherent mucilage is important for the attachment of the pectin components in the adherent layer to the seed coat, as hypothesized previously by Macquet et al. (2007a), but does not influence their basic composition or the total amount produced. Furthermore, cellulose in mucilage is required for the aggregation of pectin polymers in water-soluble mucilage. Interestingly, cesa5 has similar modifications in water-soluble mucilage amounts and macromolecular characteristics as mum5-1 (Fig. 8; Table I; Macquet et al., 2007a). Nevertheless, mum5-1 adherent mucilage was clearly different from that of cesa5, as Calcofluor labeled diffuse cellulose within the residual mum5 adherent mucilage (Supplemental Fig. S6; Macquet et al., 2007a). In addition, pectin methyl esterification of the adherent mucilage appeared to be different, as mum5-1 was previously shown to have a small amount of JIM7 and no JIM5 labeling (Macquet et al., 2007a), whereas the inverse was observed in cesa5 (Fig. 5, E, F, K, and L). These differences suggest that CESA5 and MUM5 produce different structural components of the adherent mucilage that both contribute to the tethering of adherent mucilage to the seed coat.
Precisely how cellulose participates in the attachment of the adherent mucilage remains to be determined, particularly as Calcofluor staining indicates that it is only present in the inner domain of the adherent mucilage. It is possible that the RG I in adherent mucilage is highly entangled with itself and cellulose microfibrils. Alternatively, another component could link the RG I to cellulose. Interestingly, in wild-type adherent mucilage, pectin methyl esterification appears to be reduced in this domain (Fig. 5, B, C, H, and I; Macquet et al., 2007a), and an increased availability of carboxyl groups on HG could participate in providing this link. In addition, cell wall cellulose is deposited parallel to the surface of the plasma membrane, probably due to the presence of existing cell wall components (Cosgrove, 2005), whereas the cellulose in mucilage is embedded in the pectin components of the adherent mucilage, which suggests that it is synthesized out into the apoplasm and that synthesis is organized differently from that in the cell wall. Further studies will be required to clarify these points, and the future cloning of MUM5 will probably contribute toward a complete understanding of the establishment of mucilage structure. Nonetheless, it is clear that the identification of the role of CESA5 in the synthesis of mucilage cellulose is an important step.
MATERIALS AND METHODS
Plant Material
The Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines SALK_099008, SALK_070473, SALK_046453, and SALK_650405 (Col-0 accession), corresponding to cesa5-2, cesa5-3, cesa9-3, and cesa10-1, were identified in the SIGnAL database (Alonso et al., 2003; http://signal.salk.edu). Seeds from these lines and the mutants mum3-1 and mum5-1 (Col-2 accession) were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info). Homozygous lines were identified by PCR using the primers indicated in Supplemental Table S2 on genomic DNA extracts; an amplicon was only obtained for the T-DNA left border and not with primers flanking the insertion. The mutants cesa1 (rsw1-10), cesa2 (N557274), cesa3je5, cesa5-1 (N625535), and cesa6prc1-1 are in the Col-0 accession (Desnos et al., 1996; Fagard et al., 2000a, 2000b; Desprez et al., 2007), and cesa4irx5-1, cesa irx3-1, and cesa irx1-1 are in the Landsberg erecta accession (Turner and Somerville, 1997; Taylor et al., 1999, 2000). CESA5 promoter:GUS fusion transformants (Persson et al., 2007) were a gift of Monica Doblin, and the generation of wild-type and cesa5-1 plants expressing the CESA5 promoter:GFP:CESA5 translational fusion has been described by V. Bischoff, T. Desprez, G. Mouille, S. Vernhettes, M. Gonneau, and H. Höfte (unpublished data). Seed production was carried out in either a growth chamber (photoperiod of 16 h of light at 21°C, 8 h of dark at 18°C, 65% relative humidity, and 170 μmol m−2 s−1) or a glasshouse (18°C–28°C) with a minimum photoperiod of 13 h assured when required by supplementary lighting. Plants were grown in compost (Tref Substrates) and watered with Plan-Prod nutritive solution (Fertil; http://www.plantprodsolutions.com/). Two or three successive backcrosses to wild-type Col-0 were performed on the mutants cesa5-1 or cesa5-2 and cesa5-3, respectively. Reciprocal crosses were performed between homozygous cesa5-1, cesa5-2, cesa5-3, and mum3-1 mutants, and F1 and F2 seed mucilage phenotypes were determined using F2 and F3 seed, respectively. In all subsequent analyses, seeds used for comparisons were from mutant and wild-type plants that had been simultaneously cultivated and harvested.
Expression Analysis
Extraction of total RNA and reverse transcription were carried out as described by Macquet et al. (2007b). For amplification of PCR products (Fig. 1, E and F) from single-stranded cDNA in wild-type, cesa5-2, and cesa5-3 plants, the following exon primers were used: CESA5-5′, forward primer 5′-ACTTTCTCTCGCCGCAACT-3′ and reverse primer 5′-CTCCATCATGTTTAACCACC-3′; CESA5-3′, forward primer 5′-TGCAAGTATCCTCTTCATG-3′ and reverse primer 5′-AGTAACAAACAAATGTGTGA-3′; EF1α4 primers were as described by North et al. (2007). PCR products for CESA5 and EF1α4 amplifications were examined on 1.5% and 3% (w/v) agarose gels, respectively.
In situ hybridization was performed on paraffin-embedded developing seed sections. Tissues were first fixed with 4% (w/v) paraformaldehyde and 0.1% (v/v) Triton X-100 under vacuum for 1 h at 4°C, then overnight at 4°C. Samples were then rinsed with water and dehydrated in ethanol. Then, samples were incubated in Histo-clear II (National Diagnostics; http://www.nationaldiagnostics.com/) before embedding in paraffin (Paraplast plus; Leica Microsystems; http://www.leica-microsystems.com/) as described by Jackson (1991). Tissue sections (8–10 μm) were cut and placed on ProbeOn Plus slides (Fisher Scientific; http://www.fishersci.com/), and paraffin was removed from samples with Histo-clear followed by rehydration. A CESA5-specific probe was PCR amplified using forward primer 5′-GAACTCAAGAGTTGATATGA-3′ and reverse primer 5′-CTAATACGACTCACTATAGGGCTCAAAATATTTCAATGAAAG-3′ with a CESA5 cDNA clone (clone APZL64d06; Kazusa DNA Research Institute; http://www.kazusa.or.jp/) as a template. Transcription was performed using Riboprobe combination system SP6-T7 RNA polymerase (Promega; http://www.promega.com/) followed by prehybridization, hybridization, and washes essentially as described by Jackson (1991) with the following modifications. After dehydration, slides were rinsed in phosphate-buffered saline (PBS) and incubated for 10 min at 37°C in a solution (100 mm Tris-HCl, pH 7.5, 50 mm EDTA) containing 1 μg mL−1 proteinase K; the incubation in 4% (v/v) formaldehyde was omitted. Hybridization was performed at 42°C overnight, and slides were then washed with 0.1× SSC, 0.5% SDS for 30 min at 42°C, followed by 90 min in 50% (v/v) formamide, 2× SSC at 42°C, and then 0.5 m NaCl, 10 mm Tris-HCl, pH 8.0, 1 mm EDTA (NTE) for 5 min at 42°C. Nonhybridized RNAs were removed by incubation in 10 mg mL−1 RNase in NTE for 10 min at 37°C. Slides were then rinsed with NTE prior to incubation in 50% (v/v) formamide, 2× SSC at 42°C. After rinsing with 0.1× SSC at 42°C, then in PBS, hybridized transcripts were detected using anti-digoxigenin antibodies conjugated with alkaline phosphatase (Roche Applied Science; http://www.roche-applied-science.com/).
Cytochemical Staining and Immunolabeling Procedures
Mucilage released from mature dry seeds was either stained directly with 500 μg mL−1 ruthenium red or after imbibition in water for 10 min and observed with a light microscope (Axioplan 2; Zeiss; http://www.zeiss.fr/). Developing seed sections were obtained from siliques staged by tagging flowers on the day of pollination and sampling at the specified number of days afterward. Siliques were fixed and paraffin embedded as above before staining with 0.1% (w/v) toluidine blue O as described previously (Macquet et al., 2007b). Three primary monoclonal antibodies, JIM5, JIM7, and INRA-RU2, and the carbohydrate-binding module CBM3a containing a His tag were used for immunolabeling (Plant Probes; http://www.plantprobes.net/); these bind low-ester HG, high-ester HG, RG I, and crystalline cellulose, respectively (Knox et al., 1990; McCartney et al., 2004; Ralet et al., 2010). Immunolabeling and observation with an inverted TCS-SP2-AOBS spectral confocal laser microscope (Leica Microsystems) were carried out as described by Macquet et al. (2007a). For CBM3a binding and detection, a three-stage immunolabeling technique was used employing a mouse anti-His antibody (Sigma-Aldrich; http://www.sigmaaldrich.com/) and a rat anti-mouse IgG secondary antibody conjugated to Alexa Fluor 488 (Invitrogen; http://www.invitrogen.com/; McCartney et al., 2004); double labeling was then carried out with Calcofluor (Macquet et al., 2007a). Laser gain values were fixed within a given experiment to facilitate image comparison.
Scanning Electron Microscopy and Reporter Gene Analyses
Dry seeds were mounted onto a Peltier cooling stage using adhesive discs (Deben; http://www.deben.co.uk/) and observed with a SH-1500 tabletop scanning electron microscope (Hirox; http://www.hirox-europe.com/). Histochemical detection of GUS activity was performed as described by Jefferson et al. (1987) on seeds dissected from siliques staged as above using 0.5 or 2 mm potassium ferricyanide/potassium ferrocyanide. Seeds were then stained using the modified pseudo-Schiff propidium iodide protocol described by Truernit et al. (2008). Seeds were also dissected from siliques of plants expressing the CESA5 promoter:GFP:CESA5 construct and stained for 10 min with 1 mg mL−1 propidium iodide, then rinsed twice with PBS. Reporter gene expression was observed with a TCS-SP2-AOBS spectral confocal laser scanning microscope (Leica Microsystems) with excitation wavelengths and emission or reflection signal collection as described by Truernit et al. (2008) for pseudo-Schiff propidium iodide and GUS. For GFP and propidium iodide fluorescence, an excitation wavelength of 488 nm was used with emission signal collection at 497 to 526 nm and 595 to 640 nm, respectively. Three-dimensional reconstructions were obtained from RGB stacks of confocal images as described previously (Truernit et al., 2008).
DNA Sequencing
Genomic DNA was extracted from flower buds of wild-type Col-2 and the mum3-1 mutant as described by Doyle and Doyle (1990). DNA fragments for the CESA5 gene were then amplified by PCR, gel purified using the Wizard PCR clean-up kit according to the manufacturer’s instructions (Promega), and sequenced (GenoScreen; http://www.genoscreen.fr/).
Crystalline Cellulose Observation and Content Determination
Mucilage was released from seeds by imbibition in water for 10 min. Seeds were then mounted in water on a glass slide and observed with an SMZ 1500 binocular microscope equipped with a MNN4092 polarizing attachment (Nikon; http://www.nikon.fr/). For determination of crystalline cellulose contents, 100 mg of mature dry seeds was ground in liquid N2 and suspended in 3 mL of Updegraff reagent (acetic acid:nitric acid:water, 8:1:2 [v/v]) before boiling for 30 min in a water bath. Remaining crystalline cellulose was pelleted by centrifugation (2,500g) and dissolved in 3 mL of 67% (v/v) sulfuric acid. Crystalline cellulose amounts were quantified colorimetrically at 620 nm in a spectrophotometer using the anthrone reagent (Scott and Melvin, 1953; Updegraff, 1969).
Mucilage Extraction
Water-soluble mucilage was extracted from 200 mg of intact seeds by mixing head over tail for 3 h in distilled water (4 mL) at 20°C. The resulting suspension was centrifuged (8,000g, 3 min). Supernatants were carefully recovered, and seeds were rinsed twice with 4 mL of distilled water. Supernatants and washes were pooled and filtered through a disposable glass microfiber filter (13 mm diameter, 2.7 μm pore size; Whatman; http://www.whatman.com/index.aspx). For digestion and analysis of the inner, adherent layer of mucilage, hydrated seeds remaining after water-soluble mucilage extraction were adjusted with 100 mm sodium acetate, pH 4.5, to a final molarity of 50 mm in a final volume of 4 mL. Rhamnogalacturonan hydrolase (Swiss-Prot Q00018) provided by Novozymes (http://www.novozymes.com/) was added (0.1 nkat), and samples were incubated for 16 h at 40°C. A control reaction without enzyme and a blank reaction without seeds were also carried out. After centrifugation (8,000g, 3 min), supernatants were removed for analysis.
For analysis of the macromolecular characteristics of water-soluble mucilage, mucilage was extracted from samples of 100 mg of intact seeds by mixing head over tail for 3 h in distilled water (2 mL) at 20°C. One milliliter of supernatant was carefully recovered after centrifugation (8,000g, 3 min), filtered through a disposable glass microfiber filter, boiled for 5 min, and filtered through a polyvinylidene difluoride filter (13 mm diameter, 0.45 μm pore size; Whatman) prior to HP-SEC injection.
HP-SEC
HP-SEC was performed at room temperature on a system comprising a Shodex OH SB-G precolumn followed by a Shodex OH-Pack SB-805 HQ column (http://www.shodex.de/). Elution was carried out with 50 mm sodium nitrate buffer at a constant flow rate of 60 mL h−1. Measurements were performed using a differential refractometer (VE 3580 RI Detector) and a Viscotek 270 Dual Detector (dual laser light scattering, λ = 670 nm, 90° and 7°, combined with a differential pressure viscometer; Malvern Instruments; http://www.malvern.com/). All detectors were calibrated with a pullulan standard having narrow molecular mass distribution (Mw = 145,618 D, Mn = 139,180 D, [η] = 54.0 mL g−1 at 30°C in 0.1 m sodium nitrate, dn/dc = 0.147 mL g−1; Malvern Instruments). Samples were injected automatically through a 50-μL loop. Data analyses were performed using OmniSec version 4.5 software (Malvern Instruments).
Analytical
The uronic acid (as GalA) and total neutral sugar (as Rha) contents were determined by the automated m-hydroxybiphenyl and orcinol methods, respectively (Thibault, 1979; Tollier and Robin, 1979).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JN222801.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Columella height is not affected in cesa5.
Supplemental Figure S2. CESA5 transcript levels in the developing seed coat.
Supplemental Figure S3. Complementation of cesa5-1 adherent mucilage phenotypes by transformation with CESA5 promoter:GFP:CESA5.
Supplemental Figure S4. GUS expression from the CESA5 promoter in the developing seed coat.
Supplemental Figure S5. The reduced width of cesa5 adherent mucilage is highlighted with an anti-RG I antibody.
Supplemental Figure S6. Comparison of Calcofluor labeling in the adherent mucilage of cesa5mum3-1 and mum5-1.
Supplemental Table S1. Sugar composition of wild-type, cesa5-2, and cesa5-3 water-extracted, adherent and total mucilage.
Supplemental Table S2. Sequences of primers used for the identification of homozygous mutants.
Supplemental Movie S1. Z-scan through wild-type seed coat epidermal cells and mucilage labeled with Calcofluor.
Supplemental Movie S2. Z-scan through cesa5-2 seed coat epidermal cells and mucilage labeled with Calcofluor.
Acknowledgments
We are grateful to Marie-Jeanne Crépeau, Damien Poulain, Maxime Magne, and David Viterbo for technical aid. We are also indebted to Herman Höfte for stimulating scientific exchanges and critical reading of the manuscript, to Bertrand Dubreucq for advice on acquiring GUS/propidium iodide images, and to Michaël Anjuère and Fabrice Petitpas for assistance with plant culture.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL. (2009) AtBXL1 encodes a bifunctional β-d-xylosidase/α-l-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, De Rycke R, Viane R, Inzé D. (2000) Histological study of seed coat development in Arabidopsis thaliana. J Plant Res 113: 139–148 [Google Scholar]
- Beeckman T, Przemeck GKH, Stamatiou G, Lau R, Terryn N, De Rycke R, Inzé D, Berleth T. (2002) Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiol 130: 1883–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP. (2006) Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem 281: 29321–29329 [DOI] [PubMed] [Google Scholar]
- Bui M, Lim N, Sijacic P, Liu Z. (2011) LEUNIG_HOMOLOG and LEUNIG regulate seed mucilage extrusion in Arabidopsis. J Integr Plant Biol 53: 399–408 [DOI] [PubMed] [Google Scholar]
- Burn JE, Hocart CH, Birch RJ, Cork AC, Williamson RE. (2002) Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol 129: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall KH, Pattathil S, Phillips SE, Hahn MG, Mohnen D. (2009) Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol Plant 2: 1000–1014 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Dagel DJ, Liu Y-S, Zhong L, Luo Y, Himmel ME, Xu Q, Zeng Y, Ding S-Y, Smith S. (2011) In situ imaging of single carbohydrate-binding modules on cellulose microfibrils. J Phys Chem B 115: 635–641 [DOI] [PubMed] [Google Scholar]
- Dean GH, Zheng H, Tewari J, Huang J, Young DS, Hwang YT, Western TL, Carpita NC, McCann MC, Mansfield SD, et al. (2007) The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos T, Orbović V, Bellini C, Kronenberger J, Caboche M, Traas J, Höfte H. (1996) Procuste1 mutants identify two distinct genetic pathways controlling hypocotyl cell elongation, respectively in dark- and light-grown Arabidopsis seedlings. Development 122: 683–693 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissues. Focus 12: 13–15 [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. (2000a) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Höfte H, Vernhettes S. (2000b) Cell wall mutants. Plant Physiol Biochem 38: 15–25 [Google Scholar]
- Gonzalez A, Mendenhall J, Huo Y, Lloyd A. (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev Biol 325: 412–421 [DOI] [PubMed] [Google Scholar]
- Goto N. (1985) A mucilage polysaccharide secreted from testa of Arabidopsis thaliana. Arabidopsis Inf Serv 22: 143–145 [Google Scholar]
- Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin TI, Persson S, Somerville CR. (2010) Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci USA 107: 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterman Y, Shem-Tov S. (1996) Structure and function of the mucilaginous seed coats of Plantago coronopus inhabiting the Negev Desert of Israel. Isr J Plant Sci 44: 125–133 [Google Scholar]
- Haughn G, Chaudhury A. (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Hurst LD. (2011) Molecular genetics: the sound of silence. Nature 471: 582–583 [DOI] [PubMed] [Google Scholar]
- Jackson D. (1991) In situ hybridization in plants. Bowles DJ, Gurr SJ, McPherson M, , Molecular Plant Pathology: A Practical Approach. Oxford University Press, Oxford, pp 163–174 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, King J, Cooper C, Roberts K. (1990) Pectin esterification is spatially regulated both within cell walls and between developing tissues or root apices. Planta 181: 512–521 [DOI] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW. (2009) The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21: 72–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A, Ralet M-C, Kronenberger J, Marion-Poll A, North HM. (2007a) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet M-C, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM. (2007b) A naturally occurring mutation in an Arabidopsis accession affects a β-d-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAbee JM, Hill TA, Skinner DJ, Izhaki A, Hauser BA, Meister RJ, Venugopala Reddy G, Meyerowitz EM, Bowman JL, Gasser CS. (2006) ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J 46: 522–531 [DOI] [PubMed] [Google Scholar]
- McCartney L, Gilbert HJ, Bolam DN, Boraston AB, Knox JP. (2004) Glycoside hydrolase carbohydrate-binding modules as molecular probes for the analysis of plant cell wall polymers. Anal Biochem 326: 49–54 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North HM, De Almeida A, Boutin J-P, Frey A, To A, Botran L, Sotta B, Marion-Poll A. (2007) The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J 50: 810–824 [DOI] [PubMed] [Google Scholar]
- Oka T, Nemoto T, Jigami Y. (2007) Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J Biol Chem 282: 5389–5403 [DOI] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potikha T, Delmer DP. (1995) A mutant of Arabidopsis thaliana displaying altered patterns of cellulose deposition. Plant J 7: 453–460 [Google Scholar]
- Ralet M-C, Tranquet O, Poulain D, Moïse A, Guillon F. (2010) Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 231: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T. (2008) A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J 54: 466–480 [DOI] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L. (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224: 96–107 [DOI] [PubMed] [Google Scholar]
- Scott TA, Melvin EH. (1953) Determination of dextran with anthrone. Anal Chem 25: 1656–1661 [Google Scholar]
- Stork J, Harris D, Griffiths J, Williams B, Beisson F, Li-Beisson Y, Mendu V, Haughn G, Debolt S. (2010) CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol 153: 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR. (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault J-F. (1979) Automatisation du dosage des substances pectiques par la méthode au métahydroxydiphényle. Lebensm Wiss Technol 12: 247–251 [Google Scholar]
- Tollier MT, Robin JP. (1979) Adaptation de la méthode à l’orcinol sulfurique au dosage automatique des glucides neutres totaux: conditions d’application aux extraits d’origine végétale. Ann Technol Agric 28: 1–15 [Google Scholar]
- Tominaga-Wada R, Iwata M, Sugiyama J, Kotake T, Ishida T, Yokoyama R, Nishitani K, Okada K, Wada T. (2009) The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J 60: 564–574 [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui J-C. (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Usadel B, Kuschinsky AM, Rosso MG, Eckermann N, Pauly M. (2004) RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol 134: 286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M, Tehseen M, Doblin MS, Pettolino FA, Wilson SM, Bacic A, Golz JF. (2011) The transcriptional regulator LEUNIG_HOMOLOG regulates mucilage release from the Arabidopsis testa. Plant Physiol 156: 46–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW. (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW. (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW. (2004) MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Knox JP. (2001) In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213: 37–44 [DOI] [PubMed] [Google Scholar]
- Williamson RE, Burn JE, Birch R, Baskin TI, Arioli T, Betzner AS, Cork A. (2001) Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215: 116–127 [DOI] [PubMed] [Google Scholar]
- Windsor JB, Symonds VV, Mendenhall J, Lloyd AM. (2000) Arabidopsis seed coat development: morphological differentiation of the outer integument. Plant J 22: 483–493 [DOI] [PubMed] [Google Scholar]
- Witztum A, Gutterman Y, Evenari M. (1969) Integumentary mucilage as an oxygen barrier during germination of Blepharis persica (Burm) Kuntze. Bot Gaz 130: 238–241 [Google Scholar]
- Young JA, Evans RA. (1973) Mucilaginous seed coats. Weed Sci 21: 52–54 [Google Scholar]
- Young RE, McFarlane HE, Hahn MG, Western TL, Haughn GW, Samuels AL. (2008) Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]