Abstract
In tomato (Solanum lycopersicum) fruit, the number of locules (cavities containing seeds that are derived from carpels) varies from two to up to 10 or more. Locule number affects fruit shape and size and is controlled by several quantitative trait loci (QTLs). The large majority of the phenotypic variation is explained by two of these QTLs, fasciated (fas) and locule number (lc), that interact epistatically with one another. FAS has been cloned, and mutations in the gene are described as key factors leading to the increase in fruit size in modern varieties. Here, we report the map-based cloning of lc. The lc QTL includes a 1,600-bp region that is located 1,080 bp from the 3′ end of WUSCHEL, which encodes a homeodomain protein that regulates stem cell fate in plants. The molecular evolution of lc showed a reduction of diversity in cultivated accessions with the exception of two single-nucleotide polymorphisms. These two single-nucleotide polymorphisms were shown to be responsible for the increase in locule number. An evolutionary model of locule number is proposed herein, suggesting that the fas mutation appeared after the mutation in the lc locus to confer the extreme high-locule-number phenotype.
The domestication of wild plant species has altered particular sets of traits, such as the size and the diversity of the consumed parts (Frary and Doganlar, 2003). During this process, genetic diversity decreased, not only in those genes controlling domestication traits as a result of artificial selection but also at the whole genome level as a result of genetic bottlenecks. Improvements of cultivars and production systems have increased crops yield. At the genetic level, the yield of domesticated plants has increased considerably compared with their wild ancestors (Buckler et al., 2001), particularly over the last 40 years (http://faostat.fao.org). However, this increase will probably reach its limit due to the increase of the world human population and the decrease in arable lands. The loss of genetic diversity at important loci that accompanied the yield increase could limit improvements in the future. The identification of new alleles or new allelic combinations from wild species or tomato (Solanum lycopersicum) accessions that did not experience an erosion of their genetic diversity is essential but requires an understanding of crop domestication. Linking the evolution of genotypes and phenotypes to understand domestication is one of the main challenges of modern genetics.
The tomato clade, Solanum section Lycopersicum, is composed of several wild species and the cultivated tomato S. lycopersicum. The wild species are distributed along the Andean coast from Ecuador to Chile, with two species (S. cheesmaniae and S. galapagense) endemic to the Galapagos Islands. The cultivated tomato is a self-pollinating species that is thought to be derived from its closest wild ancestor S. pimpinellifolium (Nesbitt and Tanksley, 2002). Cherry tomato accessions (S. lycopersicum var cerasiforme) have an intermediate position between these two species, as their genomes are mosaics of those from S. lycopersicum and S. pimpinellifolium (Ranc et al., 2008).
Compared with wild species, domesticated accessions show a range of morphological diversity, particularly with respect to fruit shape and size (Paran and van der Knaap, 2007; Supplemental Fig. S1), despite the strong loss of molecular diversity throughout the whole genome (Van Deynze et al., 2007).
A number of genes controlling tomato fruit morphology have been cloned: FW2.2 (Frary et al., 2000) controls fruit weight; SUN (Xiao et al., 2008) and OVATE (Liu et al., 2002) control elongated fruit shape; and FASCIATED (FAS; Cong et al., 2008) controls locule number, fruit size, and flat fruit shape. fw2.2 controls up to 30% of fruit weight variation and was the first gene underlying a quantitative trait locus (QTL) that was identified by a positional cloning approach (Frary et al., 2000). The gene is differentially expressed in the carpel, resulting in increased cell number in large fruit. FAS encodes a YABBY-like transcription factor that is expressed early in the development of stamen and carpels. The characterization of both FW2.2 and FAS illustrates the link between fruit size and genes involved in developmental processes.
In tomato, locule number influences fruit shape and size. The locules are directly derived from the carpels in the flower. In addition to fas, several QTLs controlling locule number have been mapped, and a candidate gene approach has been used to map genes regulating floral meristem development that might colocalize with known QTLs for locule number (Barrero et al., 2006). Although none of the genes responsible for these QTLs were successfully identified, the results of this study were used to develop several interesting hypotheses.
Two QTLs, fas and locule number (lc; also named lcn2.1), have major effects on the phenotype. FAS, which could be considered a major gene, has the strongest effect by increasing the number of locules from two to more than six. lc has a weaker effect by increasing the number of locules from two to three or four. An epistatic interaction between the two QTLs influences the phenotype, with both lc and fas synergistically contributing to extremely high locule number (Lippman and Tanksley, 2001; Barrero and Tanksley, 2004). WUSCHEL, a gene controlling stem cell fate in the apical meristem, was mapped to the same region as lc (Barrero et al., 2006). The lack of polymorphisms in the gene and its promoter and the similar expression level of the gene in both wild-type and mutant accessions led the authors to conclude that the DNA region that has been analyzed did not correspond to lc. The gene responsible for lc and its role during domestication thus remains unknown.
We report here the map-based cloning and identification of two single-nucleotide polymorphisms (SNPs) immediately downstream of WUSCHEL that control the trait. The lc locus shows a remarkable pattern of diversity. In addition, we demonstrate that the selection of this locus has been necessary to increase locule number during the domestication of tomato.
RESULTS
Physical Mapping of lc
The lc locus, which was first described in 1937 (Yeager, 1937), was mapped close to the ovate region (Butler, 1952). This QTL is a major locus known to modify the number of locules in tomato fruits, with a phenotypic effect that varies according to the genetic background. The QTL location on chromosome 2 was more accurately defined using molecular mapping with polymorphic markers (Lippman and Tanksley, 2001; van der Knaap and Tanksley, 2003).
Our study exploited a recombinant inbred line population obtained from an intraspecific cross between two cultivated tomato accessions (Causse et al., 2002), Cervil, a cherry tomato line, and Levovil, a classical French fresh market line carrying round fruits with many locules weighing approximately 100 g. The QTL was initially mapped to the TG463 region (Lecomte et al., 2004) using two near-isogenic lines (CF12-C and CF13-L) that were genetically identical except for a 30-centiMorgan (cM) region containing lc. CF12-C contains the low-locule-number allele from Cervil, and CF13-L contains the high-locule-number allele from Levovil (Fig. 1A).
Figure 1.
Phenotypic analysis and mendelization of the locule number QTL (lc). A, Fruit morphology of lines used in the study. The parental lines Cervil and Levovil were crossed to obtain the F1 hybrid and the near-isogenic recombinant F8 lines CF12-C and CF13-L. The L2 line was derived from Levovil, and its lc region of chromosome 2 containing the Cervil allele has been introgressed by marker-assisted selection. B, Phenotypic effect of the QTL. The average locule number of F2 heterozygous plants (H) or F3 plants homozygous for either the Cervil (C) or the Levovil (L) allele was determined. The plants were selected based on genotyping data, and heterozygous plants were selected by analyzing genotypic and phenotypic segregation in their corresponding F3 progeny. Each histogram, with its sd, is partitioned according to the proportion of fruits with two locules (white), three locules (black), and four or more locules (gray). A Student’s t test indicated that the means are significantly different between C and H (P < 0.001), between H and L (P < 0.001), and between H and C (P < 0.01). [See online article for color version of this figure.]
An F2 segregating population (2,688 plants) derived from the cross between CF12-C and CF13-L was used to identify 215 plants with a recombination between the markers T1555 and TG191 surrounding lc in a 4.8-cM region. The segregation of the phenotype was tested in the progeny of each recombinant line (Supplemental Table S1), allowing the mendelization of the QTL (Fig. 1B). Heterozygous plants have an intermediate phenotype (2.7 ± 0.50 locules) that was between homozygous plants with low locule number (2.4 ± 0.51 locules) and homozygous plants with high locule number (3.5 ± 0.75 locules). This indicated that the mutant allele was partially recessive to the wild-type allele. Homozygous plants carrying both the Cervil wild-type alleles produced fruits with mostly two locules. In contrast, homozygous plants carrying both the Levovil mutant alleles produced fruits mostly containing three or more locules. Given the semidominant nature of the QTL and the limited phenotypic difference between the two extreme values of the two alleles, heterozygous plants could not be differentiated from either homozygous plant unless the recombinant plant was subjected to progeny testing.
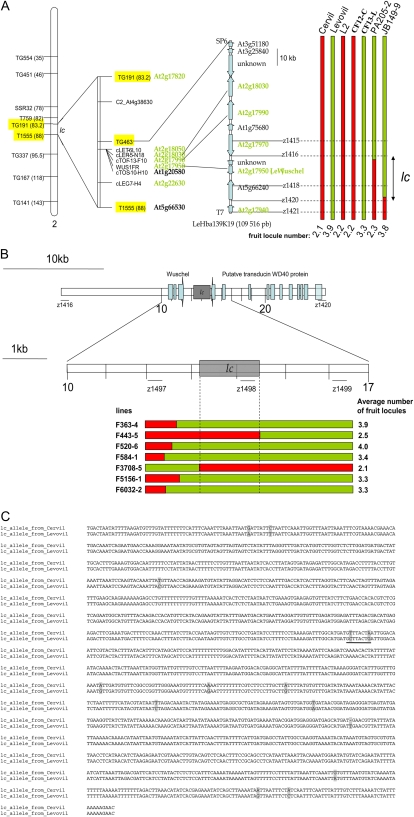
Sequence analysis of the region around the closest linked marker, TG463, indicated that the lc region was partially syntenic to a region in Arabidopsis (Arabidopsis thaliana) surrounding At2g18000 (Fig. 2A). Sequence analysis of TG463 showed homology with the bacterial artificial chromosome (BAC) end of the tomato clone Le_HBa0139K19. The mapping of an insertion/deletion marker found at the other BAC end confirmed that this clone originated from the lc region. The ctof-13-f10 marker sequence was present on this BAC, as confirmed by PCR amplification. Two recombinant plants, PA205-2 and JB149-9, showed a recombination event in the Le_HBa0139K19 BAC. The PA205-2 line produced fruits with few locules (2.3). The JB149-9 line produced fruits with a higher number of locules (3.8). Taken together, these observations indicated that the Le_HBa0139K19 BAC covered the lc QTL.
Figure 2.
Fine mapping (A), ultra-high-resolution mapping (B), and polymorphism (C) of lc QTL. A, All sequences highlighted in green represent tomato genes syntenic with the At2g18000 region in Arabidopsis. Markers highlighted in yellow were used to screen for recombinant plants. The PA205-2 and JB149-9 recombinant lines allowed the identification of the BAC clone containing the QTL. This BAC, LeHBa139K19, contains 11 putative ORFs, with some showing homology to known genes or expressed sequences. lc is located in the region between z1416 and z1420. B, The z1416 and z1420 markers were used to screen for recombinant plants. Newly designed markers finally localized lc between z1497 and z1499. Seven plants with recombination events between these two markers were identified. The molecular characterization of these seven lines restricted lc to a 1,608-bp noncoding DNA region. The orientation (from ATG to stop codon) and the exons (in blue) of the cDNAs coding for WUSCHEL and the WD40 protein are shown. C, Sequence alignment of the two alleles of lc from Cervil and Levovil. The 14 polymorphic sites are highlighted in gray. The two underlined SNPs are responsible for lc (Fig. 5). [See online article for color version of this figure.]
Sequence analysis of the BAC clone Le_HBa0139K19 (109.5 kb) showed that it contained 11 putative open reading frames (ORFs). Polymorphisms found in the BAC sequence were used to identify the recombinant breakpoints in PA205-2 and JB149-9, which allowed us to narrow down the locus to 26.6 kb (Fig. 2A). This region contained three ORFs encoding a transducin WD40 repeat regulatory protein (similar to At5g66240), an unknown protein, and tomato WUSCHEL.
WUSCHEL (Laux et al., 1996; Reinhardt et al., 2003) was a likely candidate for the lc QTL because its function is associated with floral organ number (Mayer et al., 1998) and meristem size via the regulation of another key regulator of apical meristem development, Clavata3 (Brand et al., 2000; Schoof et al., 2000). The WD40 repeat protein was also a likely candidate because these proteins are linked to several important functions in eukaryotes (Smith et al., 1999), such as cell division (Feldman et al., 1997) and chromatin remodeling (Vitaliano-Prunier et al., 2008).
To further refine the recombination map, an additional 6,768 F2 plants from the same cross between CF12-C and CF13-L were genotyped with the z1416 and z1420 markers that flanked the lc locus. This experiment led to an additional 52 recombinant lines. Newly designed markers, located every 2 kb within this region, restricted lc to a 3-kb region (Fig. 2B). Final sequence analysis of the region restricted lc to a 1,608-bp region that showed 14 polymorphic sites (13 SNPs and a 1-bp insertion/deletion) between the two alleles (Fig. 2C).
Functional Analysis of lc
The 1,608-bp region to which lc was fine mapped corresponded to a noncoding region located 1,080 bp downstream of the stop codon of WUSCHEL. None of the in silico analyses of the region provided any information about its putative function. In addition, this region did not show any homology with known microRNAs, nor did its sequence have a clear secondary structure that would predict a novel microRNA. Using northern blots and reverse transcription (RT)-PCR did not reveal any possible expression of this locus (data not shown).
We also examined the effect of the lc locus on the expression of the two adjacent genes (Fig. 3). WUSCHEL expression was restricted to flower buds, whereas the WD40 repeat protein was expressed in all tissues examined. There was no significant expression difference between the wild-type and mutant alleles of lc. Therefore, these results could not determine conclusively which of the two candidate genes underlies lc.
Figure 3.
Expression analysis of the two candidate genes. RT-PCR was performed on total RNA extracted from floral buds, leaves, or fruits. Primers were designed on WUSCHEL (accession no. AJ538329) and the WD40 repeat protein (tomato Unigene no. SGN-U585584). eIF4A2 (tomato Unigene no. SGN-U593757) was used as a reference gene. DAA, Days after anthesis.
Given that WUSCHEL expression was restricted to floral buds, the effect of lc on floral development was evaluated by determining floral organ number in several representative near-isogenic lines (Fig. 4). The increase in locule number was positively correlated with an increase in petal number but was not associated with an increase in flower size.
Figure 4.
Effect of lc on locule number (A), flower size (B), and petal number (C). Cervil and Levovil are the parental lines. L2 and L4 have been obtained by marker-assisted selection from Levovil; they are both identical to Levovil except for a region of chromosome 4 from Cervil (L4) and a region of chromosome 2 from Cervil that contains lc (L2). L2 and L4 must be compared with Levovil. CF12-C and CF13-L are a pair of identical isogenic lines except for a 30-cM region that contains lc. JB1538 and JB1546 are identical to CF12-C and CF13-L, respectively, but differ for less than 30 cM. PA205-2 and JB149-9, on the same principle, differ from one another by a 28-kb region surrounding lc. The lines are all homozygous at the lc locus and contain either the alleles from Cervil, which produce fruits with low locule number, or from Levovil, which produce fruits with high locule number. The genetic background is either Cervil (C) or Levovil (L); it is recombinant (R) between Cervil and Levovil for the isogenic lines. The mean of each line has been compared with Cervil. Statistical significance of the t test is indicated as ns (not significant) or *** (P < 0.001).
Sequence Polymorphisms among Different Tomato Accessions Validated lc
To further characterize lc, the 1,608-bp region was sequenced in a set of 88 accessions composed of 16 S. lycopersicum, 62 S. lycopersicum var cerasiforme, and 10 S. pimpinellifolium (Supplemental File S1). This panel of varieties was chosen to represent a large spectrum of tomato diversity (Ranc et al., 2008). Sequence analysis revealed 21 new polymorphic sites, with the majority present in the wild species. Two SNPs were found to be associated with locule number (P < 1 × 10−6), and these two SNPs were in almost complete linkage disequilibrium, as shown by the existence of only one accession (LA2402) that showed a recombination event between the two SNPs. Together, the two SNPs were thus considered a unique haplotype. The correlation between the number of locules and the lc haplotypes was consistent except for three lines. The Pescio, Muchamiel, and Stupicke Polni Rane cultivars produced fruits with high locules (3.8, 5.5, and 4.2, respectively) and contained the low-locule-number haplotypes, but the mutant genotype of these three lines at the fas locus explained their phenotype.
To validate the functional effect of the two SNPs linked to lc by association genetics, a 235-bp region containing the two SNPs was sequenced in 92 additional lines (Supplemental Table S2). The statistical association between the genotypes at the two SNPs and the number of locules was highly significant (P < 1.30 × 10−12), demonstrating that the two SNPs located in the noncoding sequence were responsible for lc and thus could be considered quantitative trait nucleotides (Fridman et al., 2004). None of the other SNPs in the region was associated with fruit locule number. The potential effect of lc on fruit weight was also examined. The two SNPs explained more than 12% of fruit weight variation (association P < 3.5 × 10−6) in the core collection of 88 accessions. In the same set, one SNP in the 5′ untranslated region of fw2.2 explained the same range of variation in fruit weight but with a weaker significance level (P < 3 × 10−4) and no association with locule number. The mutations responsible for fw2.2 remain unknown; thus, another SNP may be more significantly associated with fruit weight at this locus.
Molecular Evolution of lc during Domestication
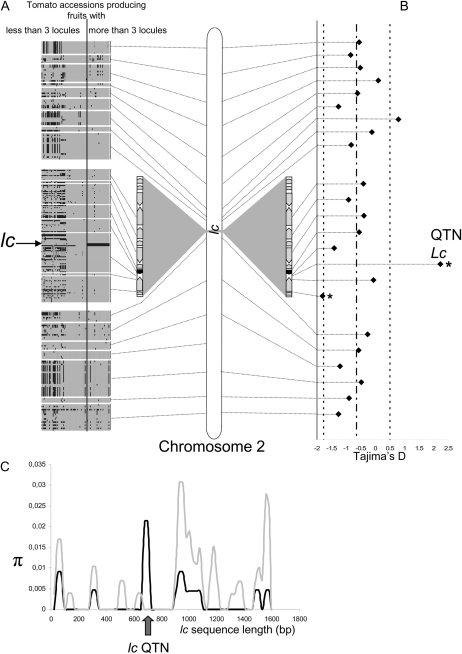
Detailed sequence analysis revealed that the diversity of the locus was drastically reduced in the cultivated species, with the exception of the two SNPs responsible for lc (Fig. 5, A and C). This observation was particularly evident for a 311-bp window surrounding the two SNPs, in which no sequence differences were observed in the cultivated accessions. These results indicate that lc is under high selective pressure, probably due to the functional importance of the locus. An analysis of eight additional loci located on the Le_HBa0139K19 BAC and 16 molecular markers located along the entire length of chromosome 2 indicated that the two SNPs responsible for lc (positive Tajima’s D) evolved differently from other loci on the chromosome (negative Tajima’s D for the 23 other loci). The negative Tajima’s D values for the 23 loci located along the entire chromosome 2 indicated an increase of the population size, in contrast to lc with a positive value. The two SNPs were under balancing selection (Fig. 5B; Tajima’s D = 2.227, P < 0.05), indicating that both alleles were selected during the domestication process, in contrast to other loci selected during domestication, such as tb1 in maize (Zea mays; Wang et al., 1999) or fw2.2 in tomato (Nesbitt and Tanksley, 2002), that were under positive selection.
Figure 5.
Molecular diversity of the lc locus. A, Tomato haplotypic structure of lc and other loci on chromosome 2. Each block corresponds to one of the 24 amplicons (average length of 500 bp). Columns and rows represent individuals and SNPs, respectively. With Heinz1706 used as a reference, polymorphisms are indicated either in gray (identical allele) or in black (different allele). B, Tajima test over the whole chromosome and within lc. Stars indicate significant departures from the neutrality hypothesis. A significant positive value of Tajima’s D for lc indicated either a balancing selection of the locus or a population decrease. An analysis of the entire chromosome showed that lc evolved differently from the entire chromosome: lc underwent a balancing selection, whereas chromosome 2 evolved following a population expansion. Dashed lines represent mean and se for the whole chromosome. C, Molecular diversity of lc in wild and cultivated lines. The diversity (π) of the locus was drastically reduced in cultivated lines (black lines) except for two SNPs (arrow) in comparison with wild accessions (gray lines). A total of 17 cultivated and 11 wild lines were used in this analysis.
The tomato accessions studied for association mapping were not selected based on fruit locule number but rather for their molecular diversity at neutral markers. They were mostly composed of S. lycopersicum var cerasiforme accessions producing fruits with low locules. The fasciated phenotype was underrepresented in all tested accessions (5.6%). Because lc and fas are known to interact epistatically, another collection of accessions was used to determine whether lc was necessary for the phenotypic expression of the fasciated phenotype. Thus, a complementary set of 87 modern cultivars producing fruits with more than six locules and considered fasciated were analyzed. Among these 87 cultivars, only three carried the low-locule-number allele of lc (Supplemental Table S3). Approximately 97% of the cultivars carried the allele of lc producing a high locule number. These results suggested that lc is necessary for the expression of a clear fasciated phenotype. Surprisingly, only 39 cultivars (44%) among the 87 had the mutant allele of fas, confirming that fas is not the only locus that produces fasciated fruits.
DISCUSSION
An Intraspecific Cross between Two Cultivated Tomato Lines Allowed a Map-Based Cloning of the lc QTL
Almost all wild tomato species produce bilocular fruits. During domestication, the number of locules in fruits has increased together with fruit weight, which ranges from a few grams in wild species to up to 1,000 g in some cultivated varieties. Compared with other plant species, tomato fruit size is an excellent model for understanding the molecular basis underlying the domestication of fruit-bearing crops. The molecular characterization of the loci that control fruit size also provides insight into fleshy fruit development. fw2.2, the first QTL controlling fruit weight to be cloned, was identified via a map-based cloning strategy using an interspecific cross. The same strategy was employed to clone the gene responsible for the fas QTL, which determines the multilocular phenotype in beefsteak-type tomatoes. In both examples, the wild-type allele came from S. pennellii. This distant wild relative was used because it had high polymorphism levels compared with S. lycopersicum varieties. The population of introgression lines from S. pennellii (Eshed et al., 1992) could not be used in this study because both parents have the same haplotype at the two SNPs and the same low-locule-number phenotype. Although lc is described as a major QTL, its effect on the number of fruit locules is weak compared with that of fas. Thus, all genetic background effects needed to be overcome to determine which phenotypic variations could be attributed to lc alone, without any segregation of other minor QTLs. The best method to ensure this requirement was the use of near-isogenic lines. An intraspecific cross between two cultivated accessions was selected to clone the lc QTL, which had been previously mapped in our segregating population (Lecomte et al., 2004). The two near-isogenic lines only differed from each other in the region containing lc. A 1,608-bp region responsible for the phenotype was identified based on the genotyping of 9,456 F2 plants from this cross and the phenotyping of 267 recombinant sub-near-isogenic lines. The success of a positional cloning approach is based on reliable phenotyping. Because lc is semidominant, heterozygous plants have an intermediate phenotype between those associated with the two homozygous alleles. The difference between the two extreme values of the two alleles was too close to distinguish the heterozygous plants from either homozygous plant. Thus, the segregation of the phenotype was systematically studied by self-pollinating the F2 recombinant plants and measuring the phenotype in the resulting homozygous F3 plants.
Association Mapping Permitted the Identification of Two SNPs Responsible for lc But Alone Would Not Have Identified the QTL
Linkage disequilibrium mapping allows the identification of the genes responsible for complex traits (Long and Langley, 1999). This approach was first developed to overcome the impossibility of obtaining the large segregating populations necessary for QTL mapping in humans (Spielman et al., 1993). The principle is simple, involving the comparison of two groups of genetically unlinked individuals that differ for a particular trait. The two groups are then genotyped with molecular markers, and statistical associations between the phenotype and the genotype at the markers are determined.
Association mapping is based on linkage disequilibrium, which varies significantly between species. In maize, linkage disequilibrium is low within distances ranging from 200 to 1,500 bp (Remington et al., 2001). In tomato, it remains high within distances of 20 cM (van Berloo et al., 2008). Because high linkage disequilibrium increases the risk of detecting false-positive associations, this study used a combination of map-based cloning to identify the locus region and association mapping to refine its molecular characterization. The map-based cloning step identified a 1,608-bp region. For the association mapping step, this region was sequenced in a core collection composed of 88 accessions selected from a larger population that was designed to maximize the molecular diversity. In addition to the 14 polymorphic sites found between both parental lines, 21 polymorphisms were identified from this core collection. As expected, more than 81% of the new polymorphic sites were found in S. pimpinellifolium wild accessions. Association mapping identified two SNPs that had an almost perfect association with the locule-number phenotype. This association was then validated using a larger set of tomato accessions. The same result was obtained, confirming that the two SNPs were responsible for the phenotype.
An additional 24 DNA fragments located on chromosome 2 were sequenced in the core collection (N. Ranc, unpublished data). The two SNPs responsible for lc, however, were not in linkage disequilibrium with any other polymorphic site adjacent to the locus or distantly located on chromosome 2. Given this observation, the identification of the molecular region corresponding to lc using association mapping alone would have been impossible except by sequencing the region by chance. New high-throughput sequencing tools allow genomes to be resequenced more quickly and cheaply. Because the tomato genome is now sequenced (http://solgenomics.net/genomes/Solanum_lycopersicum/index.pl; Mueller et al., 2009), such an approach involving the comparison of a large number of full genomes could be used in the near future to identify an increasing number of genes for complex traits.
A Model for Locule Number Evolution in Tomato Fruits
Using simple sequence repeat markers, the genome of cherry tomato accessions (S. lycopersicum var cerasiforme) has been shown to be a mosaic of the genomes from S. pimpinellifolium and S. lycopersicum (Ranc et al., 2008). Cherry tomatoes are thus a useful tool for association mapping because of their intermediate molecular diversity level and admixture genome structure. The 180 accessions used for association mapping were mostly composed of S. lycopersicum var cerasiforme tomatoes. Wild tomato species, however, mainly produce fruits with a low locule number. Among the 180 accessions, 81.1% produced fruits with two to four locules, and 18.9% produced fruits with more than four locules. This collection allowed the identification of the two SNPs responsible for lc but could not be used to determine the evolutionary history of locule number during tomato domestication. The mechanism by which fas could have evolved compared with lc was studied using a third set of tomato accessions that only included accessions producing fruits with more than six locules. Interestingly, only 44% of the accessions carried the fas high-locule-number allele, but 97% carried the high-locule-number allele of lc. This result indicates that the high-locule-number allele of lc is necessary to express the fasciated phenotype in combination with the fas locus and that it is also required to produce the beefsteak tomato phenotype in combination with one or more other loci in addition to fas.
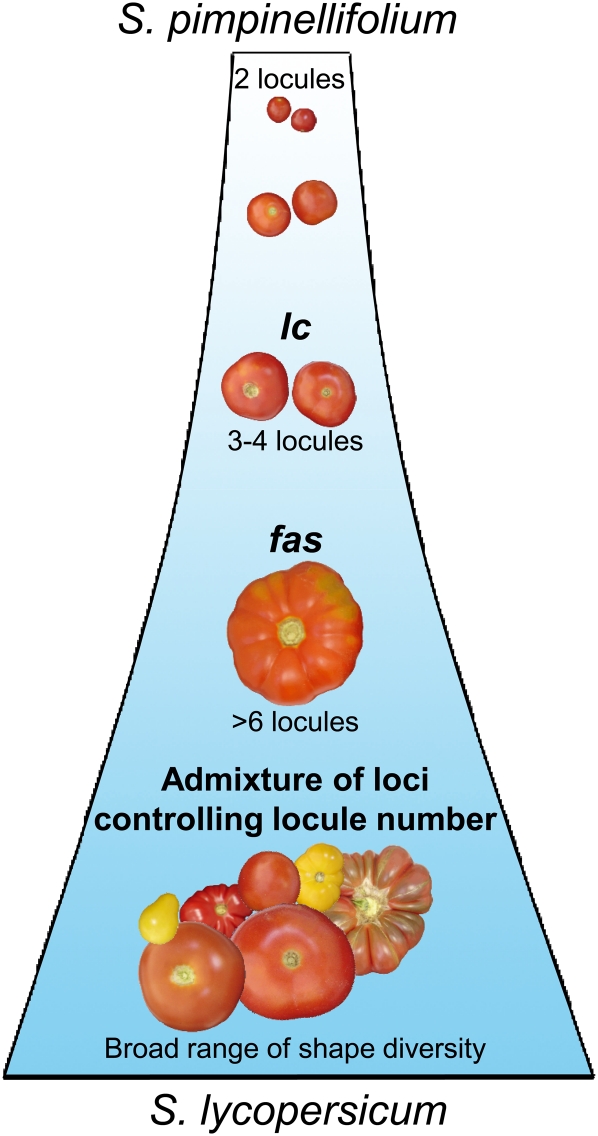
Additionally, all wild species tested contained the lc haplotypes that produce fruits with a low number of locules, suggesting that the two SNPs responsible for the increase of locule number appeared during tomato domestication in a S. lycopersicum var cerasiforme cultivar and then spread among these cultivars. fas appeared later, and the combination of the two loci produced the fasciated phenotype exhibited by the first cultivars introduced in Europe (Daunay et al., 2007). A model is thus proposed to explain the evolution of locule number during tomato domestication and breeding (Fig. 6).
Figure 6.
Model of locule number evolution in tomato fruits during domestication. S. pimpinellifolium is considered the wild ancestor of the cultivated tomato S. lycopersicum. Based on the analysis of 267 tomato accessions, we propose a model that could explain the history of locule number evolution during tomato domestication. In our study, only 4.6% of the high-locule-number accessions (i.e. those with more than three locules) had the low-locule allele of lc, and 96.9% of the fasciated accessions (i.e. those with more than six locules) had the high-locule allele of lc, but only 49.4% of them had the fas allele. These results indicate that lc was required for the increase in locule number in tomato fruits during domestication. The lc locus could have appeared before the fas locus. These two QTLs are the major loci controlling locule number. Modern breeding has used other loci to expand phenotypic diversity. [See online article for color version of this figure.]
The average value of Tajima’s D for the entire chromosome 2 is −0.633, suggesting a recent demographic expansion that followed the bottleneck of domestication (Fig. 5B). The two SNPs responsible for lc exhibit a significantly different pattern. The evolutionary history of lc is more likely explained by a particular pattern of evolution than by the demographic evolution of the sample used to validate the QTL. The positive Tajima’s D value for the two SNPs indicates a balancing selection. This type of selection is explained by the maintenance of the two alleles at the locus, with an equilibrated frequency.
In the proposed model, lc existed as a polymorphic locus in the wild species, and all alleles resulted in the production of fruits with two locules. Then, the two SNPs responsible for the increase in the number of locules appeared in a particular allele of lc, which has subsequently expanded in S. lycopersicum accessions because it conferred larger fruits. Later, the fas mutation allowed a further increase in locule number when expressed in the lc background. In our hypothesis, we proposed that lc appeared prior to other loci responsible for the increase of locule number. The wild type allele of lc has been recently reintroduced in S. lycopersicum by modern breeding to diversify fruit shape.
How to Characterize the Molecular Function of lc?
Interestingly, the genomic region of lc is located in a noncoding region between two putative candidate genes, WUSCHEL and a gene encoding a WD40 repeat protein. WUSCHEL, which was previously proposed as a candidate gene for lc (Barrero et al., 2006), has a central role in apical meristem development, being responsible for stem cell fate and affecting meristem size. No differences in the expression of the two genes could be detected in a set of sub-near-isogenic lines. To determine whether the lc region could be expressed, quantitative RT-PCR assays using several primer pairs inside and surrounding the locus were performed. These assays did not reveal any expression of this region. Instead of using transformation to try to validate the QTL, we preferred to use a combination of fine mapping and association mapping. This method successfully identified the two SNPs responsible for lc. Although transgenic plants are used to help understand the function of a gene, this method would have been difficult for lc due to the unclear limits of the functional region necessary to express the phenotype. Indeed, the locus did not show any homology with a known coding sequence. Even though we concluded that the 1,608-bp region contains the polymorphic sites responsible for the phenotype, we cannot exclude that the functional region could be larger than 1,608 bp.
lc had previously been shown to be necessary to obtain a fas phenotype (Lippman and Tanksley, 2001). The results presented here further the understanding of the interaction between lc and fas. First, fas was not the only locus producing tomato fruits with a high number of locules. Second, a high-locule-number allele at lc was necessary for the fasciated phenotype. At the molecular level, fas encodes a YABBY-like transcription factor (Cong et al., 2008). Interestingly, a YABBY transcription factor acts on the partitioning of shoot apical meristem in Arabidopsis (Goldshmidt et al., 2008). This observation could favor WUSCHEL as the best candidate interacting with fas. WUSCHEL expression is regulated by several factors (Dodsworth, 2009). Given that the lc SNPs are located 1,080 bp from the stop codon of WUSCHEL, they could act as posttranscriptional regulators.
WD40 repeat proteins play important roles in eukaryotes. Interestingly, the WD40 protein near lc is orthologous to At5g66240 in Arabidopsis. At5g66240 is itself homologous to the At5g66430 gene that encodes the FAS2 protein (Kaya et al., 2001). FAS1 and FAS2 are two subunits of the chromatin-remodeling factor 1 complex. The FAS genes are described as regulating the maintenance of the expression state of WUSCHEL in the shoot apical meristem. Mutations in fas1 and fas2 were also described as causing stem fasciation together with altered floral development (Leyser and Furner, 1992). Stem fasciation is often linked to fruit fasciation in tomato, which is due to the high increase of locule number.
Altogether, these observations do not allow discrimination between WUSCHEL and FAS2-like as the gene that is modified by the two SNPs and thus is responsible for the phenotype. lc could act on shoot apical meristem development by regulating both genes or either gene in a complex way. The two SNPs could also act on the regulation of one or several genes elsewhere in the genome.
CONCLUSION
Although we were unable to identify the function of the two polymorphic nucleotides responsible for lc, we can assume that they play an important role in meristem development. Although the lc region is not expressed, the two SNPs have a pleiotropic effect on locule number, floral organ number, and fruit weight and might act by regulating meristem size. These quantitative trait nucleotides are flanked by two genes that could affect tomato fruit development. Thus, these two SNPs may regulate either or both of these two genes, but at a very specific stage.
The two QTLs fas and lc make tomato an excellent model to study floral meristem and fleshy fruit development. Here, two SNPs responsible for the increase of locule number were identified and used to develop a model of the evolutionary history of tomato domestication. An understanding of the mechanism by which this noncoding region affects locule number in the fruits and floral organ number in the flowers still remains of great interest and will require additional experiments. Although this study did not reveal any evidence for the expression of the lc region, it might be expressed in specific cells or at a precise time of development. In situ hybridization experiments should be performed to determine whether the lc region is expressed at the cellular level in meristems, where it should act. Similar experiments using WUSCHEL and the FAS2-like protein as probes may also reveal differential expression between lines not identified by RT-PCR on floral buds.
MATERIALS AND METHODS
Plant Material and Phenotyping
All tomato (Solanum lycopersicum) accessions are maintained in the Génétique et Amélioration des Fruits et Légumes research unit (INRA, Avignon, France) and are available upon request (helene.burck@avignon.inra.fr, mathilde.causse@avignon.inra.fr).
All plants were grown in greenhouses in Montfavet (southern France) between 2004 and 2008.
Fine mapping and ultra-high-resolution mapping were performed on an F2 population derived from a cross between the two near-isogenic lines CF12-C and CF13-L (described as F8-V-C and F8-V-L, respectively, by Lecomte et al., 2004). Plants were sown on 96-well plates and transferred to 3-L containers after genotype selection. The locule number of each plant was determined by phenotyping 20 mature fruits (10 from the second truss and five each from the third and fourth trusses). The petal number and flower diameter of each plant were determined by analyzing 20 flowers.
For the diversity analysis, 10 fruits per accession were phenotyped.
Genotyping
The z100_CAPS and z274 primers were used to amplify the T1555 and TG191 markers, and the resulting PCR products were digested with BamHI and NdeI, respectively, and mapped on the F6 recombinant inbred line population derived from the cross between Cervil and Levovil (Causse et al., 2002). Taqman markers developed from the associated SNPs were used to screen 2,688 F2 plants for the fine-mapping analysis. Those plants with different genotypes at T1555 and TG191 (i.e. recombination between the two markers) were selected for fine mapping. The same markers were also used to screen eight F3 plants from the self-progeny of each F2 recombinant plant. Two plants homozygous for the segregating markers were then selected for phenotyping.
Polymorphisms in the z1416 and z1420 markers were used to develop Taqman markers that were used to screen 6,768 F2 plants for the ultra-high-resolution mapping analysis. As above, these markers were also used to genotype eight F3 plants from the self-progeny of each recombinant F2 plant.
Alleles and BAC Sequencing
All sequencing reactions were performed either manually on the ABI3730XL (Philippe et al., 2010) or by Genome Express, GATC (http://www.gatc-biotech.com), or MWG (http://www.eurofinsdna.com). The TG463 sequence was obtained by sequencing the plasmid. All other sequencing reactions were performed using PCR products. All primers used for PCR amplification are described in Supplemental Table S4.
The LeHBa0139K19 BAC sequencing was performed by Genome Express.
Sequence and Polymorphism Analysis
Sequence alignment and SNP detection were performed manually using the Genalys software (Takahashi et al., 2003), which is available at http://software.cng.fr.
Expression Analysis
Samples from leaves, flower buds, or fruits at 7 or 14 d after anthesis were randomly harvested (four plants per line) and immediately frozen in liquid nitrogen. Total RNA were extracted from three different pools with TRI Reagent Solution (Ambion) following the procedure described by the manufacturer.
Quantitative RT-PCR was performed and analyzed as described by Prudent et al. (2010). The genes of interest coding for WUSCHEL (AJ538329), the WD40 protein (SGN-U585584), and the eIF-4A-2 gene used as an internal control (SGN-U593757) were amplified; a specific PCR product was obtained with corresponding efficiency of 100.8%, 108.1%, and 110.8%, respectively.
RT-PCR was performed in the same conditions with the same samples but amplified without SYBR Green on a classic thermal cycler.
Northern-blot analysis was performed using the lc locus as a probe on the same RNA samples as described by Muños et al. (2004).
Diversity Analysis
A total of 180 accessions (37 S. lycopersicum, 128 S. lycopersicum var cerasiforme, 15 S. pimpinellifolium) were phenotyped during the summers of 2007 and 2008. Part of this sample represents a core collection of 88 individuals selected to maximize the diversity of the whole collection (Ranc et al., 2008). A total of 24 genomic 500-bp fragments (eight on the BAC identified by positional cloning and 16 on the rest of chromosome 2) were used to sequence the plants in this core collection. A mixed linear model (Yu et al., 2006) implemented using Tassel software was used for the association tests of the core collection. The genetic structure of each sample was taken into account and calculated using Structure2.0 software (http://pritch.bsd.uchicago.edu/structure.html) based on microsatellite genotyping information (Ranc et al., 2008). The genetic background interaction was also taken into account using an estimation of kinship matrix following the recommendations of Yu et al. (2006). To validate any associations, fruit weight was used as a covariate in the model. A 200-bp window surrounding the two SNPs was sequenced on an additional set of 92 accessions, and the association between the two SNPs and fruit locule number was then validated.
DNAsp version 4 software was used to compare the molecular diversity between S. lycopersicum (17 accessions) and S. pimpinellifolium (11 accessions) using a sliding-window analysis method. Sequences obtained from the BAC clone and whole chromosome 2 were used to estimate Tajima’s D and to test for evidence of selection pressures applied to the lc locus or to the entire chromosome for cultivated accessions (65 accessions). The lc fragment was restricted to a 500-bp region surrounding the SNPs to maintain the same length as the other amplicons.
All sequences have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank) with the Sequin version 10.3 software (http://www.ncbi.nlm.nih.gov/projects/Sequin). The accession numbers are listed in Supplemental Table S5.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Photograph of fruits from some varieties used in this study.
Supplemental Table S1. Genotypes and phenotypes of the 215 recombinant F2 plants and their F3 progeny used for lc fine mapping.
Supplemental Table S2. Validation of the association by genotyping the two SNPs in 92 tomato accessions.
Supplemental Table S3. Analysis of lc and fas loci in tomato accessions producing fasciated fruits with more than six locules.
Supplemental Table S4. Primer sequences used in the study.
Supplemental Table S5. Accession numbers of the sequences.
Supplemental File S1. Locule number phenotypes and genotypes of fas and lc loci for association studies in 88 tomato accessions.
Acknowledgments
We thank the master students who helped in the experiments: Abir Youssef, David Tricon, Xavier Titeca, and Claire-Emmanuelle Modin. Aurélie Chauveau and Rémi Bounon helped in the allele sequencing experiments. Florent Leydet and Laure David participated in the fine-mapping experiment and in the association mapping, respectively. Hélène Burck manages and characterizes all the accessions of the tomato collection at INRA in Avignon and provided the seeds. We thank the greenhouse experimental team of the Génétique et Amélioration des Fruits et Légumes research unit for growing all plants. Jean-Paul Bouchet helped on primer design. We gratefully thank Esther Van Der Knaap for critical review of the manuscript and for providing one marker to confirm some data for fas genotyping. We thank Rebecca Stevens for editing the manuscript.
References
- Barrero LS, Cong B, Wu F, Tanksley SD. (2006) Developmental characterization of the fasciated locus and mapping of Arabidopsis candidate genes involved in the control of floral meristem size and carpel number in tomato. Genome 49: 991–1006 [DOI] [PubMed] [Google Scholar]
- Barrero LS, Tanksley SD. (2004) Evaluating the genetic basis of multiple-locule fruit in a broad cross section of tomato cultivars. Theor Appl Genet 109: 669–679 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Buckler ES, IV, Thornsberry JM, Kresovich S. (2001) Molecular diversity, structure and domestication of grasses. Genet Res 77: 213–218 [DOI] [PubMed] [Google Scholar]
- Butler L. (1952) The linkage map of the tomato. J Hered 43: 25–36 [Google Scholar]
- Causse M, Saliba-Colombani V, Lecomte L, Duffé P, Rousselle P, Buret M. (2002) QTL analysis of fruit quality in fresh market tomato: a few chromosome regions control the variation of sensory and instrumental traits. J Exp Bot 53: 2089–2098 [DOI] [PubMed] [Google Scholar]
- Cong B, Barrero LS, Tanksley SD. (2008) Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat Genet 40: 800–804 [DOI] [PubMed] [Google Scholar]
- Daunay M-C, Laterrot H, Janick J. (2007) Iconography and history of Solanaceae: antiquity to the 17th century. Hortic Rev 34: 1–119 [Google Scholar]
- Dodsworth S. (2009) A diverse and intricate signalling network regulates stem cell fate in the shoot apical meristem. Dev Biol 336: 1–9 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Abu-Abied M, Saranga Y, Zamir D. (1992) Lycopersicon esculentum lines containing small overlapping introgressions from L. pennellii. Theor Appl Genet 83: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. (1997) A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91: 221–230 [DOI] [PubMed] [Google Scholar]
- Frary A, Doganlar S. (2003) Comparative genetics of crop plant domestication and evolution. Turk J Agric For 27: 59–69 [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88 [DOI] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D. (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y. (2008) Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 20: 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. (2001) FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142 [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G. (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lecomte L, Saliba-Colombani V, Gautier A, Gomez-Jimenez MC, Duffé P, Buret M, Causse M. (2004) Fine mapping of QTLs of chromosome 2 affecting the fruit architecture and composition of tomato. Mol Breed 13: 1–14 [Google Scholar]
- Leyser HMO, Furner IJ. (1992) Characterization of 3 shoot apical meristem mutants of Arabidopsis thaliana. Development 116: 397–403 [Google Scholar]
- Lippman Z, Tanksley M. (2001) Dissecting the genetic pathway to extreme fruit size in tomato using a cross between the small-fruited wild species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom. Genetics 158: 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Van Eck J, Cong B, Tanksley SD. (2002) A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci USA 99: 13302–13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AD, Langley CH. (1999) The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res 9: 720–731 [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Lankhorst RK, Tanksley SD, Giovannoni JJ, White R, Vrebalov J, Zhangjun F, van Eck J, Buels R, Mills AA, et al. (2009) A snapshot of the emerging tomato genome sequence. Plant Genome 2: 78–92 [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16: 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt TC, Tanksley SD. (2002) Comparative sequencing in the genus Lycopersicon: implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics 162: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran I, van der Knaap E. (2007) Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J Exp Bot 58: 3841–3852 [DOI] [PubMed] [Google Scholar]
- Philippe R, Courtois B, McNally KL, Mournet P, El-Malki R, Le Paslier MC, Fabre D, Billot C, Brunel D, Glaszmann JC, et al. (2010) Structure, allelic diversity and selection of Asr genes, candidate for drought tolerance, in Oryza sativa L. and wild relatives. Theor Appl Genet 121: 769–787 [DOI] [PubMed] [Google Scholar]
- Prudent M, Bertin N, Génard M, Muños S, Rolland S, Garcia V, Petit J, Baldet P, Rothan C, Causse M. (2010) Genotype-dependent response to carbon availability in growing tomato fruit. Plant Cell Environ 33: 1186–1204 [DOI] [PubMed] [Google Scholar]
- Ranc N, Muños S, Santoni S, Causse M. (2008) A clarified position for Solanum lycopersicum var. cerasiforme in the evolutionary history of tomatoes (Solanaceae). BMC Plant Biol 8: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Frenz M, Mandel T, Kuhlemeier C. (2003) Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development 130: 4073–4083 [DOI] [PubMed] [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES., IV (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98: 11479–11484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 24: 181–185 [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52: 506–516 [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Matsuda F, Margetic N, Lathrop M. (2003) Automated identification of single nucleotide polymorphisms from sequencing data. J Bioinform Comput Biol 1: 253–265 [DOI] [PubMed] [Google Scholar]
- van Berloo R, Zhu A, Ursem R, Verbakel H, Gort G, van Eeuwijk FA. (2008) Diversity and linkage disequilibrium analysis within a selected set of cultivated tomatoes. Theor Appl Genet 117: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E, Tanksley SD. (2003) The making of a bell pepper-shaped tomato fruit: identification of loci controlling fruit morphology in Yellow Stuffer tomato. Theor Appl Genet 107: 139–147 [DOI] [PubMed] [Google Scholar]
- Van Deynze A, Stoffel K, Buell CR, Kozik A, Liu J, van der Knaap E, Francis D. (2007) Diversity in conserved genes in tomato. BMC Genomics 8: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Menant A, Hobeika M, Géli V, Gwizdek C, Dargemont C. (2008) Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol 10: 1365–1371 [DOI] [PubMed] [Google Scholar]
- Wang RL, Stec A, Hey J, Lukens L, Doebley J. (1999) The limits of selection during maize domestication. Nature 398: 236–239 [DOI] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. (2008) A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527–1530 [DOI] [PubMed] [Google Scholar]
- Yeager A. (1937) Studies on the inheritance and development of fruit size and shape in the tomato. J Agric Res 55: 141–152 [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, et al. (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208 [DOI] [PubMed] [Google Scholar]