Abstract
The reversible conjugation of the small ubiquitin-like modifier (SUMO) to protein substrates occurs as a posttranslational regulatory process in eukaryotic organisms. In Arabidopsis (Arabidopsis thaliana), several stress-responsive SUMO conjugations are mediated mainly by the SUMO E3 ligase SIZ1. In this study, we observed a phenotype of hypersensitivity to excess copper in the siz1-2 and siz1-3 mutants. Excess copper can stimulate the accumulation of SUMO1 conjugates in wild-type plants but not in the siz1 mutant. Copper accumulated to a higher level in the aerial parts of soil-grown plants in the siz1 mutant than in the wild type. A dramatic difference in copper distribution was also observed between siz1 and wild-type Arabidopsis treated with excess copper. As a result, the shoot-to-root ratio of copper concentration in siz1 is nearly twice as high as that in the wild type. We have found that copper-induced Sumoylation is involved in the gene regulation of metal transporters YELLOW STRIPE-LIKE 1 (YSL1) and YSL3, as the siz1 mutant is unable to down-regulate the expression of YSL1 and YSL3 under excess copper stress. The hypersensitivity to excess copper and anomalous distribution of copper observed in the siz1 mutant are greatly diminished in the siz1ysl3 double mutant and slightly in the siz1ysl1 double mutant. These data suggest that SIZ1-mediated sumoylation is involved specifically in copper homeostasis and tolerance in planta.
Copper (Cu) is an essential metal for normal plant growth and development. It is also an important cofactor for many metalloproteins such as plastocyanin, Cu/zinc (Zn) superoxide dismutase, cytochrome c oxidase, laccase, amino oxidase, and polyphenol oxidase in plants (Clarkson and Hanson, 1980; Yruela, 2005, 2009; Pilon et al., 2006; Burkhead et al., 2009). Furthermore, Cu participates in numerous physiological processes, including photosynthesis, respiration, antioxidant activity, cell wall metabolism and lignification, and ethylene perception (Himelblau and Amasino, 2000; Burkhead et al., 2009). Because Cu has a high redox activity, excess Cu in the plant is toxic and easily interferes with numerous biochemical and physiological processes (Luna et al., 1994; Shen et al., 1998; Nielsen et al., 2003; Demirevska-Kepova et al., 2004). Consequently, plants have evolved different strategies and mechanisms to avoid such toxicity, which include the regulation of Cu uptake, chelation, efflux, sequestration, and storage to tightly regulate Cu homeostasis (Clemens, 2001; Puig and Thiele, 2002; Wintz and Vulpe, 2002; Meharg, 2005; Sharma and Dietz, 2006; Palmer and Guerinot, 2009; Pilon et al., 2009; Puig and Peñarrubia, 2009; Yruela, 2009). To date, several mechanisms for Cu tolerance have been identified. Excess Cu can be entrapped by cell wall binding in the apoplastic space (Nishizono et al., 1987; Konno et al., 2005). In the cytosol, Cu can be chelated by small cellular molecules like amino acids, Cu-binding proteins, or phenolic compounds to reduce the toxicity of the free forms of Cu. Sequestration into vacuolar compartments can also block the contact between free Cu ions and cellular components (Palma et al., 1990; Backor et al., 2004; Sharma and Dietz, 2006; Kovacik and Backor, 2007). In addition, Cys-rich metallothionein proteins and glutathione-derived phytochelatins can also chelate Cu to buffer the cellular Cu concentration (Lolkema et al., 1984; Zhou and Goldsbrough, 1994; Cobbett and Goldsbrough, 2002; Guo et al., 2003, 2008).
SUMO (for small ubiquitin-related modifier) proteins, small proteins with a molecular mass of about 12 kD, are ubiquitously expressed throughout the eukaryotic kingdom. Although the overall amino acid sequence identity between SUMO and ubiquitin is less than 18% in Arabidopsis (Arabidopsis thaliana; Kurepa et al., 2003), both proteins share a similar three-dimensional globular structure called the Ub fold that consists of an α-helix and four β-strands (Hay, 2001; Miura and Hasegawa, 2010). A cascade of enzymatic steps is required for sumoylation that is similar to that for ubiquitination. These steps require the participation of an E1-activating enzyme, an E2-conjugating enzyme (UBC9), and an E3 SUMO ligase to facilitate the transfer of SUMO from UBC9 to the acceptor Lys residue(s) in target proteins. Based on annotated databases and recent genetic and biochemical analyses, the components of sumoylation systems are also present in plants including algae (Chlamydomonas reinhardtii), dicots (Arabidopsis), and monocots (rice [Oryza sativa]; Colby et al., 2006; Miura et al., 2007; Nigam et al., 2008; Wang et al., 2008; Park et al., 2010; Shin et al., 2010). Sumoylation is involved in controlling cell growth and development (Miura and Hasegawa, 2010), embryogenesis (Colby et al., 2006; Saracco et al., 2007), and regulation of flowering time (Jin et al., 2008). In addition, sumoylation is involved in actions of both biotic and abiotic stresses (Kurepa et al., 2003) including salicylic acid-dependent pathogen defense (Lee et al., 2007), phosphate starvation responses (Miura et al., 2005), cold tolerance (Agarwal et al., 2006), drought response (Catala et al., 2007), and basal thermotolerance (Yoo et al., 2006).
A previous screening experiment did not detect changes in SUMO conjugation in Arabidopsis seedlings subjected to heavy metal stress (Kurepa et al., 2003). However, recent reports indicated that reactive oxygen species may function as key regulators of the sumoylation-desumoylation equilibrium by influencing the redox states of SUMO cascade enzymes and SUMO protease (Bossis and Melchior, 2006; Xu et al., 2007). Excess Cu can induce reactive oxygen species production through Haber-Weiss and Fenton reactions (Stadtman and Oliver, 1991; Maksymiec et al., 1994; Schützendübel and Polle, 2002). Therefore, we were interested to find out whether excess Cu can induce sumoylation responses and, if yes, whether such responses play a role in the tolerance of excess Cu stress.
Although several SUMO E3 ligases exist in plants, stress-responsive SUMO conjugation is mainly mediated by the SUMO E3 ligase SIZ1 (Ishida et al., 2009; Lois, 2010). In this study, we examined the role of sumoylation in Cu homeostasis and tolerance using the siz1 mutant and an anti-SUMO1 antibody. In summary, we detected Cu hypersensitivity in the siz1 mutant and a SIZ1-dependent accumulation of SUMO1 conjugates under excess Cu treatment. Interestingly, we observed an elevated shoot-to-root Cu concentration ratio in the siz1 mutant. By examining the gene expression patterns and through functional studies of mutants, the involvement of YSL1 and YSL3 regulation in the control of Cu translocation is proposed.
RESULTS
The siz1 Mutant Is Sensitive to Excess Cu
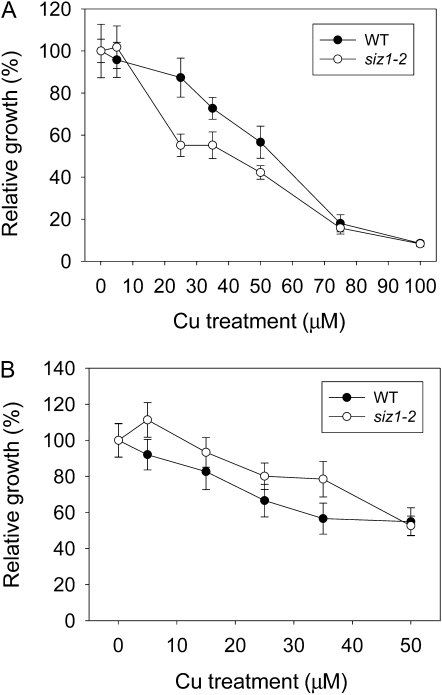
Stress-responsive SUMO conjugation is mediated mainly by the SIZ1 SUMO E3 ligase in Arabidopsis. To investigate whether sumoylation is involved in plant responses to excess Cu, we first examined the phenotype of the siz1-2 mutant at exposure to a wide range of CuSO4 concentrations (0.05–100 μm). We found that the siz1-2 mutants were more sensitive to excess Cu than the wild type within a window of Cu concentration between 25 and 50 μm (Fig. 1A). Root growth experiments showed that the relative growth rate of primary root in the siz1 mutant is slightly higher than that in the wild type in response to 5 to 50 μm Cu treatments (Fig. 1B). To confirm that the Cu-induced effect is specific to siz1, we further examined the phenotype of another mutant, siz1-3, which contains a different mutated allele, upon exposure of seedlings to excess Cu. As shown in Supplemental Figure S1, the sensitivity of both mutants to excess Cu is indistinguishable. The phenotype of siz1 demonstrates the involvement of SIZ1 in the tolerance to Cu stress. Notably, the growth inhibition only occurred in shoots but not in roots (Supplemental Fig. S1).
Figure 1.
Growth of wild-type (WT) and siz1 mutant seedlings under Cu stress. One-week-old wild-type (black circles) and siz1-2 (white circles) seedlings were treated with various Cu concentrations, as indicated, for 10 d before fresh weights (A) or for 3 d (vertical growth) before root lengths (B) were measured. The normalized value against the growth in half-strength MS medium is presented. Means and error bars were calculated from six repeats with 10 plants each.
Excess Cu Induces SIZ1-Dependent SUMO1 Sumoylation
Based on these observations, we hypothesized that sumoylation is involved in the tolerance to excess Cu in Arabidopsis. To investigate whether this posttranslational modification of proteins occurs in plants encountering Cu stress, we performed immunoblotting analyses with anti-SUMO1 antibody to detect sumoylation. The antibody was prepared by immunizing a rabbit with purified recombinant SUMO1 (Supplemental Fig. S2). Before use, the antibody was affinity purified with recombinant SUMO1 protein immobilized on a polyvinylidene difluoride (PVDF) membrane. Furthermore, we used media-grown plant materials in this experiment instead of liquid-cultured seedlings that could be experiencing leaf-submerging stresses.
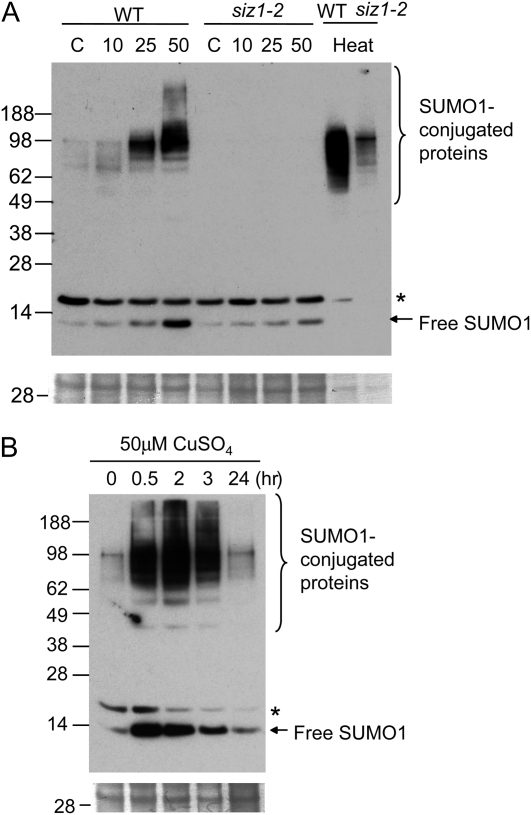
Considering that the root tissue is the uptake organ and could elicit a first response to excess Cu stress, we focused at first on protein sumoylation in roots. Immunoblot analysis with anti-SUMO1-specific antibodies resulted in the detection of SUMO1-protein conjugates in crude extracts of roots. The conjugates were induced by excess Cu in the wild type, while no conjugate formed in siz1 mutants under Cu treatment (Fig. 2A). In the wild type, the induction is dose dependent in the range of tested Cu concentrations (10-50 μm). Compared with heat shock-induced sumoylation, Cu-induced sumoylation gives rise to a different pattern and is completely SIZ1 dependent (Fig. 2A). The extent of sumoylation is much less (about one-fifth) under Cu stress than under heat shock conditions. Time-course analysis revealed that the formation of SUMO1-protein conjugates as well as free SUMO1 increases rapidly upon exposure to excess Cu for about 2 h and then rapidly decreases after treatment for about 3 h (Fig. 2B; Supplemental Fig. S3). Cu-induced SUMO1 sumoylation was also observed at delayed time points in the shoot tissue (Supplemental Fig. S3). These data clearly demonstrate that sumoylation can be induced under excess Cu in Arabidopsis.
Figure 2.
Sumoylation in roots under conditions of excess Cu. Western-blot analysis was performed with homemade anti-SUMO1 antibody. A, Sumoylation in roots of 12-d-old wild-type (WT) and siz1-2 seedlings treated for 1 h with half-strength MS solution (control [C]) or half-strength MS solution containing 10, 25, or 50 μm CuSO4 or subjected to heat shock at 37°C for 1 h. Loadings of heat shock samples are one-fifth of control and excess Cu-treated samples. B, Sumoylation of root proteins isolated from plants treated with 50 μm CuSO4 for the indicated time periods. Asterisks indicate unidentified protein. Bottom panels show a gel portion stained with Coomassie blue.
Cu Distribution Is Abnormal in siz1
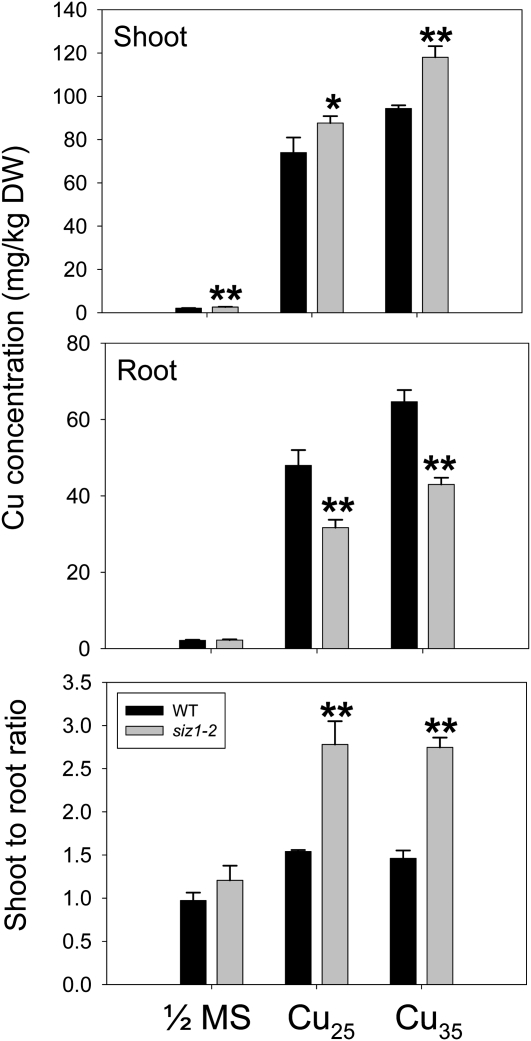
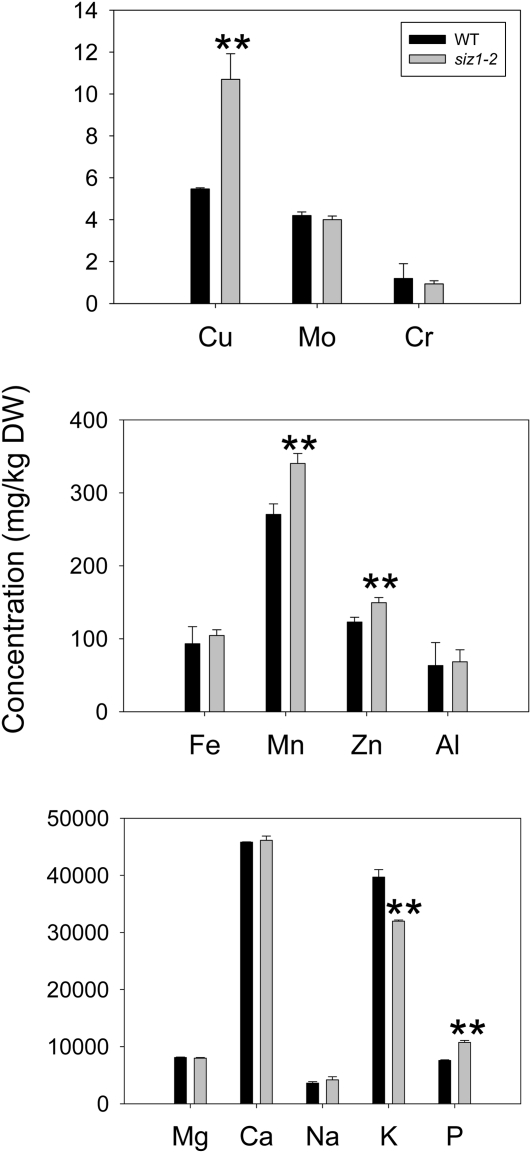
To examine whether siz1 mutants accumulate more Cu than wild-type seedlings, we measured the Cu content in shoots and roots of plants after treatment with excess Cu. Results obtained at treatment of seedlings with 25 or 35 μm CuSO4 were very similar. Interestingly, we found that shoots of siz1 mutant seedlings accumulated higher concentrations of Cu as compared with the wild type. By contrast, the Cu concentration was more elevated in roots of wild-type seedlings. As a result, the siz1 mutant possesses a shoot-to-root ratio for Cu that is about twice as high as that of the wild type (Fig. 3). In order to gain more insight into the accumulation of metals, the elemental profile of shoots of wild-type and siz1 mutant plants, which were grown in soil for 3 weeks under normal conditions, was determined by inductively coupled plasma-optical emission spectroscopy (ICP-OES; Fig. 4). Among all metals inspected, Cu showed the most dramatic difference, with a concentration twice as high in the siz1 mutant as compared with the wild type; by comparison, accumulation of other metals exhibited a small difference (manganese [Mn], Zn, and potassium [K]) or no difference (iron [Fe], aluminum, molybdenum, chromium, magnesium [Mg], calcium, and sodium). These results suggest that the Cu sensitivity of the siz1 shoot is associated with Cu overaccumulation and that the Cu translocation from root to shoot is aberrant in siz1.
Figure 3.
Cu content in shoot and root tissues. One-week-old wild-type (WT) and siz1-2 seedlings were treated with excess Cu (25 or 35 μm CuSO4, Cu25, and Cu35) for 10 d. Cu concentrations in the shoot (top panel) and root (middle panel) were determined by ICP-OES. In the bottom panel, calculated shoot-to-root ratios of Cu concentration are shown. Means and error bars were calculated from three repeats (* P < 0.05, ** P < 0.01). DW, Dry weight.
Figure 4.
Elemental analysis of soil-grown wild-type (WT) and siz1-2 plants. Determination of metal elements was done in the leaves of 3-week-old, soil-grown wild-type and siz1-2 plants by ICP-OES. Means and error bars were calculated from three repeats (** P < 0.01). Al, Aluminum; Ca, calcium; Cr, chromium; DW, dry weight; Mo, molybdenum; Na, sodium; P, phosphorus.
Expression of Cu-Related Transporter Genes in the siz1 Mutant
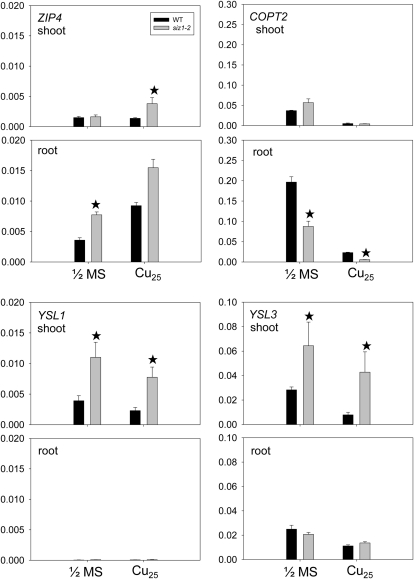
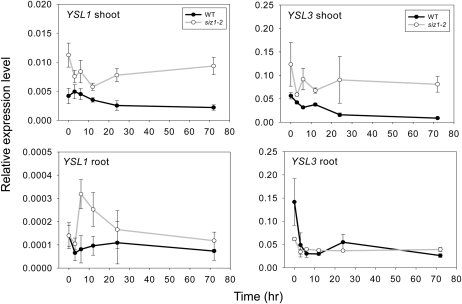
Owing to the atypical distribution of Cu in the siz1 mutant, we hypothesized that sumoylation contributes to the control of certain Cu-related transporters, including members of the ZIP, COPT, P-1b ATPase, and YSL families (Colangelo and Guerinot, 2006; Puig et al., 2007; Yruela, 2009; del Pozo et al., 2010). Thus, we examined the gene expression of these transporters in both wild-type and siz1 plants under excess Cu treatment by quantitative real-time reverse transcription (RT)-PCR (qPCR). The gene expression patterns of some transporters, including ZIP4, COPT2, YSL1, and YSL3, are different between the wild type and the siz1 mutant (Fig. 5), while others are similar (Supplemental Fig. S4). Among these transporters, root COPT2 expression is down-regulated after treatment with excess Cu. COPT2 transcript levels in root were significantly lower in the siz1 mutant than in the wild type under both normal and excess Cu conditions. The opposite trend was observed for the expression of root ZIP4. In addition, we found that ZIP4 is up-regulated in the shoot of siz1 but not in the wild type under excess Cu. Indeed, down-regulation of COPT2 and up-regulation of ZIP4 under excess Cu conditions were also reported previously (Sancenón et al., 2003; Wintz et al., 2003; del Pozo et al., 2010). The trends of regulation are comparable in the siz1 mutant and the wild type. These observations may reflect the regulation by endogenous Cu status. On the other hand, we found that the expression of YSL1 and YSL3 in the shoot of siz1 is roughly 2- to 3-fold higher than that in the wild type under both normal and excess Cu conditions. In the presence of excess Cu, the shoot expression of YSL1 and YSL3 was down-regulated in the wild type but not significantly regulated in the siz1 mutant. This expression pattern was confirmed in a time-course experiment. The high expression of YSL1 and YSL3 in the shoot lasted for the time examined (Fig. 6). These data imply that the action of sumoylation directly or indirectly regulates the mRNA levels of YSL1 and YSL3 through either transcription or mRNA stability under excess Cu conditions and that SIZ1 is required for the control of basal transcription levels of YSL1 and YSL3 under normal conditions.
Figure 5.
Differential expression of Cu-related transporter genes in siz1. Twelve-day-old wild-type (WT) and siz1-2 plants were treated with half-strength MS medium or half-strength MS medium with 25 μm CuSO4 (Cu25) for 1 d. Gene expression of Cu homeostasis-related transporters in the shoot and root tissues was measured by qPCR. The y axis values represent expression relative to ACT2. Means and error bars were calculated from six samples of two biological repeats. Asterisks indicate genes with an expression ratio greater than 2 or lower than 0.5 for siz1:the wild type).
Figure 6.
Time-course expression profiles of YSL1 and YSL3 in shoot and root tissues. Twelve-day-old wild-type (WT) and siz1-2 plants were treated with half-strength MS medium or half-strength MS medium containing 25 μm CuSO4 for 3, 6, 12, 24, and 74 h. Transcript levels of YSL1 and YSL3 were determined by qPCR analysis. The y axis values represent expression relative to ACT2. Means and error bars were calculated from six samples of two biological repeats.
YSL1 and YSL3 Are Downstream of Excess Cu-Induced SIZ1-Dependent Sumoylation and Involved in Cu Translocation
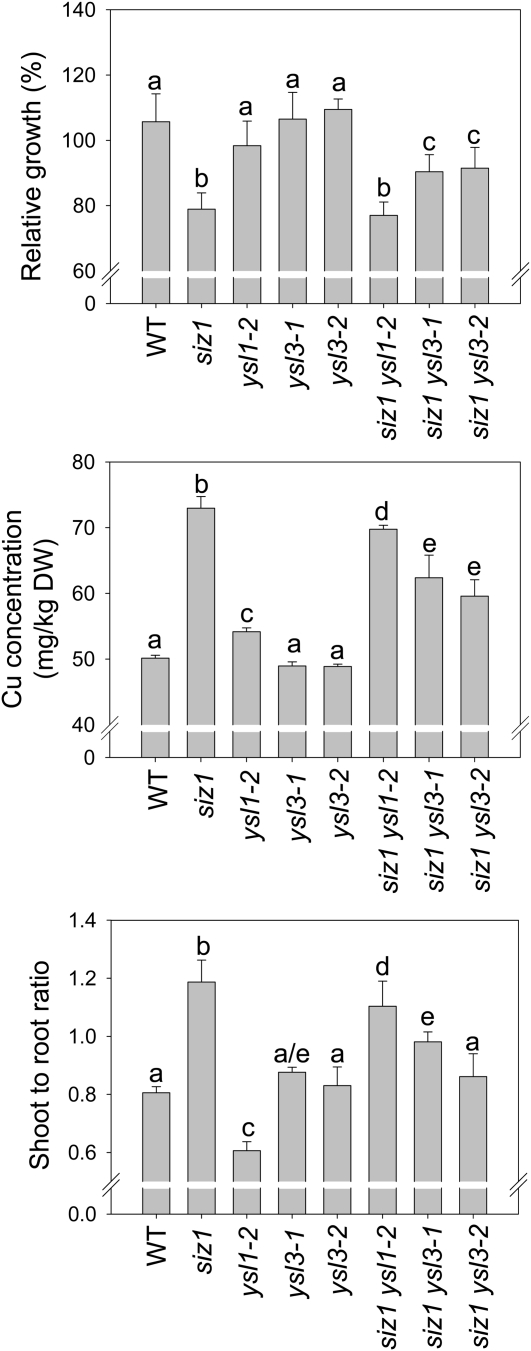
YSL1 and YSL3 were previously reported to possess a similar function with regard to Fe loading into seeds (Chu et al., 2010). In order to examine whether YSL1 and YSL3 are responsible for the high accumulation of Cu in shoot and for the hypersensitivity of the siz1 mutant to Cu, we created siz1-2ysl1-2 and siz1-2ysl3-1 double mutants for further tests. With regard to the overall phenotype, both siz1-2ysl1-2 and siz1-2ysl3-1 double mutants are similar to the siz1 mutant grown on medium or soil (data not shown). However, a major recovery effect of ysl3 on siz1 was associated with Cu tolerance, suggesting that YSL3 is situated downstream of sumoylation in the regulatory network (Fig. 7A). ICP-OES analysis further revealed that the siz1ysl3 double mutant accumulated less Cu in shoot than the siz1 mutant under excess Cu treatment. The reduction of Cu accumulation in the shoot and of the shoot-to-root Cu concentration ratio is smaller in siz1ysl1 than in siz1ysl3 but still statistically significant (Fig. 7, B and C). This result may reflect the lower relative expression of YSL1 as compared with YSL3 (Waters et al., 2006).
Figure 7.
Phenotypes of siz1, ysl1, and ysl3 single mutants as well as siz1ysl1and siz1ysl3 double mutants under excess Cu. One-week-old seedlings (wild type [WT], siz1-2 [siz1], ysl1-2, ysl3-1, ysl3-2, siz1ysl1-2, siz1ysl3-1, and siz1ysl3-2) were transferred and grown vertically for 5 d on half-strength MS, treated with 35 μm CuSO4 for 5 d, and dried before analysis of Cu in shoot and root tissues by ICP-OES. A, Relative growth of shoots normalized to their growth on half-strength MS medium. Data represent means plus sd from six repeats with 20 plants. B, Shoot Cu contents. C, Shoot-to-root ratio of Cu concentration. Means and error bars were calculated from three biological repeats. Letters a through e indicate independent groups of statistical significance. DW, Dry weight.
To confirm the Cu phenotype of siz1-2ysl3-1, experiments in two double mutants with different ysl3 alleles, siz1-2ysl3-1 and siz1-2ysl3-2, were conducted. Their reductions of Cu sensitivity, Cu accumulation, and shoot-to-root Cu concentration ratio are nearly identical (Fig. 7). These data support the downstream role of YSL3 in the Cu-induced SIZ1-dependent sumoylation and the involvement of YSL3 in the Cu translocation from roots to shoots.
DISCUSSION
In this study, we found that siz1 mutants clearly display a shoot growth-retarded phenotype under conditions of excess Cu (Fig. 1; Supplemental Fig. S1). Dose-response experiments were also performed at excess Zn, Fe, or cadmium levels; however, no obvious phenotype different from that of the wild type was observed, with the exception of a slightly more yellowish color of siz1 as compared with wild-type plants under conditions of excess Zn (data not shown). In addition, excess Cu-induced sumoylation was demonstrated to occur in a SIZ1-dependent manner (Fig. 2). These data strongly suggest that sumoylation is involved in the mechanism of Cu tolerance. Although other SUMO E3 ligases have been found (i.e. MMS21 and HIGH PLOIDY2) in Arabidopsis (Ishida et al., 2009; Zhang et al., 2010), Cu-induced sumoylation is SIZ1 dependent. By contrast, SUMO1-conjugated complexes were detected in the siz1 mutant under heat treatment (Fig. 2). Therefore, SUMO E3 ligases other than SIZ1 are involved in the process of heat stress but do not participate in Cu homeostasis and tolerance. These data indicate that the sumoylation response induced by excess Cu is not the same as that induced by heat shock in Arabidopsis. At this point, it remains to be investigated if other SUMO E3 ligases or SUMO paralogues are also involved in regulating plant responses to excess Cu.
The elemental profile of shoots of soil-grown plants revealed that the major difference of metal accumulation in siz1 is Cu (Fig. 4). Moreover, a dramatic difference in the shoot-to-root ratio of Cu concentration between wild-type and siz1 plants was observed under excess Cu treatment (Fig. 3). These data suggest that the mechanism controlling the partition of Cu between root and shoot is dysfunctional in siz1, causing a higher transport of Cu ions to the shoot, and indicate that sumoylation is involved in the regulation of Cu translocation from roots to shoots. In addition, we observed minor but significant increases in the accumulation of Zn and Mn and a decrease in the accumulation of K in the siz1 mutant (Fig. 4). In the Cu-tolerant plant Commelina communis, Cu, Zn, and Mn have similar uptake patterns and are distributed at high levels in the vascular cylinder. This distribution pattern is different from that of Fe, which is located in the epidermis and endodermis (Shi et al., 2011). SIZ1-dependent sumoylation seems to be involved in the regulation of one or more common component(s) related to Cu, Zn, and Mn increases. It suggests that a possible SIZ1-dependent controlling mechanism cannot only regulate Cu partition but also influences the uptake and translocation of Zn and Mn. Interestingly, a reduction in K accumulation was also found in the Cu hyperaccumulator Erica andevalensis (Oliva et al., 2010). Excess Cu can also induce K efflux from roots in many plant species such as Agrostis capillaries, Silene vulgaris, Mimulus guttatus, and wheat (Triticum aestivum; De Vos et al., 1991; Strange and Macnair, 1991; Quartacci et al., 2001). The reduced accumulation of K could be due to the impairment of Cu distribution in the siz1 mutant.
In general, Cu accumulation levels are correlated with Cu hypersensitivity. Overexpression of Cu uptake transporters, COPT1 and COPT3, caused a hypersensitive phenotype to Cu (Andrés-Colás et al., 2010). The hma5 mutant accumulates high Cu levels in the root and shows Cu hypersensitivity in the root (Andrés-Colás et al., 2006). A common feature of Cu toxicity in most plants is inhibition of primary root growth and a reduction in biomass (Lequeux et al., 2010). We observed that the reduction in primary root growth was lower in the siz1 mutant than in the wild type under conditions of excess Cu. On the other hand, the shoot of siz1 is more sensitive to excess Cu (Fig. 1; Supplemental Fig. S1). These phenotypes can be rationalized by the Cu accumulation levels.
We also observed that the Cu distribution was anomalous in the siz1 mutant. The abnormal distribution could be due to differences in either Cu uploading in the root or Cu unloading in the shoot. Hence, we examined the expression patterns of Cu-related transporters (Colangelo and Guerinot, 2006; Puig et al., 2007; Yruela, 2009; del Pozo et al., 2010). Among them, we found that the expression of YSL1 and YSL3 in shoot differed largely between siz1 and the wild type (Figs. 5 and 6). Previous studies suggested that YSL1 and YSL3 function in the remobilization of Cu and Zn from senescing leaves and are required for the formation of pollen and Fe, Zn, and Cu loading in seed development (Himelblau and Amasino, 2001; Curie et al., 2009; Chu et al., 2010). The roles of YSL1 and YSL3 are redundant in Fe transport and transfer of metal micronutrients to or from vascular tissues (Le Jean et al., 2005; Waters et al., 2006; Chu et al., 2010). By creating siz1ysl1 and siz1ysl3 double mutants, we were able to demonstrate that these two genes that are highly expressed in the siz1 mutant play a role downstream of the action of SIZ1, causing the irregular Cu distribution (Fig. 7). Our results further support a functional role of YSL1 and YSL3 in Cu transport.
In yeast experiments, nicotianamine (NA)-metal complexes were demonstrated to constitute substrates of YSL2 transporters (DiDonato et al., 2004). NA possesses affinities to various metal ions, including Cu, Fe, nickel, Mn, cobalt, and Zn. NA-Cu could be the major NA-metal complex in the xylem due to the low pH value in this transport tissue (Rellán-Alvarez et al., 2008; Curie et al., 2009). Indeed, the Cu-NA complex has been found in xylem saps and is likely an intermediate for the translocation of Cu from roots to shoots (Pich et al., 1994; Rellán-Alvarez et al., 2008; Curie et al., 2009). Although there is no evidence that YSL1 and YSL3 possess a direct function on NA-Cu or Cu transport so far, transgenic plants overexpressing YSL3 accumulated more Cu in leaves (Chu et al., 2010). In our study, we found that the Cu concentration in the shoot and the shoot-to-root ratio of Cu concentration were reduced in the siz1ysl3 and siz1ysl1 double mutants as compared with siz1 (Fig. 7). In addition, the siz1ysl3 double mutant is more efficient than siz1ysl1, as siz1ysl3 contains a lower Cu concentration in shoot and has a lower shoot-to-root ratio of Cu concentration, suggesting that YSL3 plays a major role in Cu accumulation in the shoot (Fig. 7). This result supports the conclusion that YSL3 is important for Cu distribution controlled by SIZ1 under excess Cu. Together, the high expression of YSL3 and YSL1 in the siz1 mutant could be responsible for the high accumulation of Cu in shoots.
In conclusion, we have found that sumoylation is involved in excess Cu tolerance and distribution. In siz1, Cu overly translocates to the shoot, and this translocation is accompanied by an anomalously high expression of YSL1 and YSL3. The sumoylation induced by excess Cu is specifically mediated by SIZ1. Therefore, we suggest that the SUMO E3 ligase SIZ1 is required for excess Cu tolerance and distribution. Our data support the notion that a regulatory pathway exists that prevents excessive translocation of Cu from root to shoot under Cu stress by down-regulation of YSL1 and YSL3 that is mediated by SIZ1-dependent sumoylation. Although sumoylation of multiple substrates was observed, similar to sumoylation induced by other environmental stresses, we hypothesize that the control of at least one component involved in Cu distribution requires the regulation of SIZ1-mediated sumoylation; this component is possibly an upstream component in the regulatory mechanism of YSL1 and YSL3 expression.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana ecotype Columbia) and siz1 mutant plants were used. Seeds of the T-DNA insertion lines siz1-2 (SALK_065397), siz1-3 (SALK_034008), ysl1-2 (SALK_034534), ysl3-1 (SALK_064683C), and ysl3-2 (SALK_045218) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Homozygote T-DNA insertion mutants were confirmed using specific primers. To create siz1-2ysl1-2, siz1-2ysl3-1, and siz1-2ysl3-2 double mutants, we crossed ysl1-2, ysl3-1, and ysl3-2 with siz1-2, respectively. The T-DNA insertion and gene expression were confirmed by PCR and RT-PCR, respectively. Primers are listed in Supplemental Tables S2 and S3. Seeds were surface sterilized with 70% ethanol for 5 min and then treated with 1.2% bleach containing 0.02% SDS for 15 min, rinsed five times with sterilized water, and kept in darkness at 4°C for 4 d to break dormancy. Treated seeds were plated on half-strength Murashige and Skoog (MS) medium (half-strength MS salt, pH 5.7, 1% Suc, and 0.35% phytagel) before the excess Cu treatments indicated in the figure legends. Soil-grown plants were obtained by sowing seeds in pots containing a mixture of organic substrate, vermiculite, and mica sheet (9:1:1, v/v/v). In all cases, plants were subjected to a 16-h-light (70 μmol m−2 s−1)/8-h-dark cycle at 22°C.
Plasmid Construction, and Expression and Purification of Recombinant SUMO1
The full-length Arabidopsis SUMO1 (At4g26840) cDNA was generated by RT-PCR from total RNA isolated from Arabidopsis ecotype Columbia as the template. The forward primer 5′-GGTGGTCATATGTCTGCAAACCAGGAG-3′ and reverse primer 5′-GCCACCAGTCTGATGGAGCATCG-3′ were designed to introduce NdeI and SmaI restriction sites at the predicted start codon and the C-terminal diglycine motif, respectively. The RT-PCR product was then cloned into the vector pTYB2 with an in-frame fusion to chitin-binding protein (CBD) at the C terminus. Expression of the CBD-SUMO-1 fusion protein was induced in Escherichia coli BL21 (DE3) culture by the addition of isopropylthio-β-galactoside (0.3 mm). Cells were grown at 28°C for an additional 4 h after induction and lysed using a cell disruptor (Constant Systems). Total soluble protein was applied to a chitin column according to the manufacturer’s recommendations (New England Biolabs). The bound protein was treated with 30 mm dithioerythritol for 48 h at 25°C. Then, the resulting intein cleavage product was eluted with the elution buffer.
Production and Purification of Anti-SUMO1 Antibody
The SUMO1-containing elution was boiled in 2× SDS sample buffer for 10 min and analyzed on a NuPAGE 4% to 12% Bis-Tris gel (Invitrogen). After Commassie Brilliant Blue staining, the SUMO1-containing gel band was excised and used as the antigen by injecting it directly into a rabbit. The antibody raised against full-length recombinant Arabidopsis SUMO1 was further purified using the PVDF method. The recombinant SUMO1 protein was run on a SDS-PAGE gel and then transferred to a PVDF membrane. The membrane was stained with Ponceau S and rinsed with distilled, deionized water to destain. The band corresponding to the region of the correct molecular mass was excised and collected. The membrane strip was blocked with 5% dry milk in 1× phosphate-buffered saline (PBS; pH 7.4) buffer for 1 h at room temperature and then washed three times for 5 min in 1× PBST (PBS with 0.1% Tween 20). The membrane strips containing recombinant SUMO1 were then incubated with 1 mL of antiserum in 5 mL of 1× PBS buffer for 1 h at room temperature. After incubation, the depleted antiserum was removed and the membrane was washed three times for 15 min in 1× PBS buffer. To strip the antibody off the membrane, the membrane was incubated with 1.5 mL of ImmunoPure IgG elution buffer, pH 2.8 (Pierce), for 5 min at room temperature. The eluted antibody was transferred to a new microcentrifuge tube containing 100 μL of 1 m Tris (pH 9.5). After neutralization, the antibody was dialyzed against 1× PBS buffer with a centrifuged filtrate tube (Vivaspin 500, VS0111; Sartorius Stedium Biotech). The purified antibody is able to recognize subnanogram amounts of recombinant SUMO1 (Supplemental Fig. S2).
Plant Protein Extraction and Immunoblot Analysis of SUMO1 Conjugates
Samples were extracted with extraction buffer (2× SDS sample buffer containing 20 mm N-ethylmaleimide, 100 mm Na2S2O5, and one tablet of protease inhibitor cocktail [Roche Applied Science] per 50 mL). Samples were centrifuged at 12,000g for 10 min, and the protein concentration was determined using the BCA Protein Assay Kit (Thermo Scientific). Total proteins (200 μg) were separated on a NuPAGE 4% to 12% Bis-Tris gel (Invitrogen) and transferred to a PVDF membrane (Immobilon-P; Millipore). For immunodetection, the membrane was blotted with 5% fat-free milk and PBST for 1 h, incubated with 1:10,000-diluted purified anti-SUMO1 antibody, washed with PBST, and incubated for 1 h with 1:10,000-diluted secondary antibody (peroxidase-conjugated goat anti-rabbit IgG; Millipore). The membrane was washed five times for 10 min each with PBST solution before development. Specific protein bands were visualized using the Immobilon Western Chemiluminescent horseradish peroxidase substrate (Millipore).
Elemental Analysis
Elemental analysis was conducted according to the procedure described previously (Lin et al., 2009). Harvested plant samples were washed with CaCl2 and water and dried for 3 d before digestion. Microwave-digested samples were analyzed by ICP-OES (OPTIMA 5300; Perkin-Elmer).
RNA Isolation and qPCR
Frozen shoot and root tissues (approximately 100 mg) were ground in liquid nitrogen using a tissue homogenizer (SH-48, J&H Technology) to which 1 mL of TRIzol reagent was immediately added. Samples were mixed briefly and incubated for 30 min at room temperature. Subsequently, chloroform (200 μL) was added to the sample and the mixture was vigorously shaken for 30 s. The samples were centrifuged at 15,000g at 4°C for 15 min, and the upper aqueous phase was carefully transferred to a new tube. RNA was precipitated by the addition of 0.5 mL of isopropanol and incubation at −80°C for 30 min. Following centrifugation at 15,000g at 4°C for 15 min, the resulting pellet was washed twice with 75% ethanol. The residual ethanol was evaporated in a chemical fume hood for 10 min. RNA was redissolved in 30 μL of diethyl pyrocarbonate-treated water. The concentration of the RNA was determined at 260 nm on a NanoDrop ND-1000 Spectrophotometer (Isogen Life Science).
Subsequently, 2 μg of RNA was treated with RQ1 RNase-Free DNase (Promega), and the reaction buffer was replaced with 5× First-Strand RT buffer (Invitrogen). The cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen). qPCR analyses were conducted with SYBR Green I dye (ABI). The expression of ACTIN2 (ACT2) was used as the internal control for all tested genes. The sequences of primers used for qPCR are listed in Supplemental Table S1. The efficiency of primers was tested based on the manufacturer’s instructions. Relative expression was calculated using a previously described method (Livak and Schmittgen, 2001).
Statistical Analysis
Assessments of statistical difference between controls and treatments were made based on two-sample t tests and methods of multiple comparisons (Montgomery, 2009).
To control overall levels of confidence in multiple comparisons, Fisher’s protected lsd method was used throughout (Fig. 7), as it is one of the most commonly used multiple testing procedures. All possible pairwise comparisons were performed once the F test rejected the hypothesis that all group means are equal in the one-way ANOVA. In this study, the outcome of Fisher’s lsd coincides with that of Duncan’s multiple range tests. As a result, statistical significance in the pairwise differences between samples was used to determine the group membership of an individual sample.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes of two siz1 mutants grown on excess Cu.
Supplemental Figure S2. Antibody production.
Supplemental Figure S3. Sumoylation of shoot and root proteins under Cu stress.
Supplemental Figure S4. Expression of Cu-related transporter genes in the wild type and siz1 under excess Cu conditions.
Supplemental Table S1. List of Cu-related transporter families and primers used for real-time PCR in Arabidopsis.
Supplemental Table S2. Primers used to confirm T-DNA insertion lines.
Supplemental Table S3. Primers used in RT-PCR.
Acknowledgments
We thank Varanavasiappan Shanmugam for plant crossing. We are grateful to Drs. Yee-yung Charng and Shu-Hsing Wu for valuable discussion and thank Dr. Heiko Kuhn for manuscript editing.
References
- Agarwal M, Hao YJ, Kapoor A, Dong CH, Fujii H, Zheng XW, Zhu JK. (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281: 37636–37645 [DOI] [PubMed] [Google Scholar]
- Andrés-Colás N, Perea-García A, Puig S, Peñarrubia L. (2010) Deregulated copper transport affects Arabidopsis development especially in the absence of environmental cycles. Plant Physiol 153: 170–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Colás N, Sancenón V, Rodríguez-Navarro S, Mayo S, Thiele DJ, Ecker JR, Puig S, Peñarrubia L. (2006) The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J 45: 225–236 [DOI] [PubMed] [Google Scholar]
- Backor M, Fahselt D, Wu CT. (2004) Free proline content is positively correlated with copper tolerance of the lichen photobiont Trebouxia erici (Chlorophyta). Plant Sci 167: 151–157 [Google Scholar]
- Bossis G, Melchior F. (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell 21: 349–357 [DOI] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KAG, Abdel-Ghany SE, Cohu CM, Pilon M. (2009) Copper homeostasis. New Phytol 182: 799–816 [DOI] [PubMed] [Google Scholar]
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH. (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HH, Chiecko J, Punshon T, Lanzirotti A, Lahner B, Salt DE, Walker EL. (2010) Successful reproduction requires the function of Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson DT, Hanson JB. (1980) The mineral nutrition of higher plants. Annu Rev Plant Physiol 31: 239–298 [Google Scholar]
- Clemens S. (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212: 475–486 [DOI] [PubMed] [Google Scholar]
- Cobbett C, Goldsbrough P. (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53: 159–182 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. (2006) Put the metal to the petal: metal uptake and transport throughout plants. Curr Opin Plant Biol 9: 322–330 [DOI] [PubMed] [Google Scholar]
- Colby T, Matthäi A, Boeckelmann A, Stuible HP. (2006) SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142: 318–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Cassin G, Couch D, Divol F, Higuchi K, Le Jean M, Misson J, Schikora A, Czernic P, Mari S. (2009) Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot (Lond) 103: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo T, Cambiazo V, González M. (2010) Gene expression profiling analysis of copper homeostasis in Arabidopsis thaliana. Biochem Biophys Res Commun 393: 248–252 [DOI] [PubMed] [Google Scholar]
- Demirevska-Kepova K, Simova-Stoilova L, Stoyanova Z, Holzer R, Feller U. (2004) Biochemical changes in barley plants after excessive supply of copper and manganese. Environ Exp Bot 52: 253–266 [Google Scholar]
- De Vos CHR, Schat H, Dewaal MAM, Vooijs R, Ernst WHO. (1991) Increased resistance to copper-induced damage of the root cell plasmalemma in copper tolerant Silene cucubalus. Physiol Plant 82: 523–528 [Google Scholar]
- DiDonato RJ, Jr, Roberts LA, Sanderson T, Eisley RB, Walker EL. (2004) Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J 39: 403–414 [DOI] [PubMed] [Google Scholar]
- Guo WJ, Bundithya W, Goldsbrough PB. (2003) Characterization of the Arabidopsis metallothionein gene family: tissue-specific expression and induction during senescence and in response to copper. New Phytol 159: 369–381 [DOI] [PubMed] [Google Scholar]
- Guo WJ, Meetam M, Goldsbrough PB. (2008) Examining the specific contributions of individual Arabidopsis metallothioneins to copper distribution and metal tolerance. Plant Physiol 146: 1697–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT. (2001) Protein modification by SUMO. Trends Biochem Sci 26: 332–333 [DOI] [PubMed] [Google Scholar]
- Himelblau E, Amasino RM. (2000) Delivering copper within plant cells. Curr Opin Plant Biol 3: 205–210 [PubMed] [Google Scholar]
- Himelblau E, Amasino RM. (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158: 1317–1323 [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K. (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, et al. (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 53: 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H, Nakato T, Nakashima S, Katoh K. (2005) Lygodium japonicum fern accumulates copper in the cell wall pectin. J Exp Bot 56: 1923–1931 [DOI] [PubMed] [Google Scholar]
- Kovacik J, Backor M. (2007) Phenylalanine ammonia-lyase and phenolic compounds in chamomile tolerance to cadmium and copper excess. Water Air Soil Pollut 185: 185–193 [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD. (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis: accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278: 6862–6872 [DOI] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- Le Jean M, Schikora A, Mari S, Briat JF, Curie C. (2005) A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J 44: 769–782 [DOI] [PubMed] [Google Scholar]
- Lequeux H, Hermans C, Lutts S, Verbruggen N. (2010) Response to copper excess in Arabidopsis thaliana: impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol Biochem 48: 673–682 [DOI] [PubMed] [Google Scholar]
- Lin YF, Liang HM, Yang SY, Boch A, Clemens S, Chen CC, Wu JF, Huang JL, Yeh KC. (2009) Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol 182: 392–404 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lois LM. (2010) Diversity of the SUMOylation machinery in plants. Biochem Soc Trans 38: 60–64 [DOI] [PubMed] [Google Scholar]
- Lolkema PC, Donker MH, Schouten AJ, Ernst WHO. (1984) The possible role of metallothioneins in copper tolerance of Silene cucubalus. Planta 162: 174–179 [DOI] [PubMed] [Google Scholar]
- Luna CM, Gonzalez CA, Trippi VS. (1994) Oxidative damage caused by an excess of copper in oat leaves. Plant Cell Physiol 35: 11–15 [Google Scholar]
- Maksymiec W, Russa R, Urbaniksypniewska T, Baszynski T. (1994) Effect of excess Cu on the photosynthetic apparatus of runner bean-leaves treated at 2 different growth-stages. Physiol Plant 91: 715–721 [Google Scholar]
- Meharg AA. (2005) Mechanisms of plant resistance to metal and metalloid ions and potential biotechnological applications. Plant Soil 274: 163–174 [Google Scholar]
- Miura K, Hasegawa PM. (2010) Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol 20: 223–232 [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Hasegawa PM. (2007) Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 10: 495–502 [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA.et al (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery D. editor (2009) Design and Analysis of Experiments. John Wiley & Sons, New York [Google Scholar]
- Nielsen HD, Brownlee C, Coelho SM, Brown MT. (2003) Inter-population differences in inherited copper tolerance involve photosynthetic adaptation and exclusion mechanisms in Fucus serratus. New Phytol 160: 157–165 [DOI] [PubMed] [Google Scholar]
- Nigam N, Singh A, Sahi C, Chandramouli A, Grover A. (2008) SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa L.) genes: genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol Genet Genomics 279: 371–383 [DOI] [PubMed] [Google Scholar]
- Nishizono H, Ichikawa H, Suziki S, Ishii F. (1987) The role of the root cell-wall in the heavy-metal tolerance of Athyrium yokoscense. Plant Soil 101: 15–20 [Google Scholar]
- Oliva SR, Mingorance MD, Valdes B, Leidi EO. (2010) Uptake, localisation and physiological changes in response to copper excess in Erica andevalensis. Plant Soil 328: 411–420 [Google Scholar]
- Palma JM, Yanez J, Gomez M, Delrio LA. (1990) Copper-binding proteins and copper tolerance in Pisum sativum L: characterization of low-molecular-weight metalloproteins from plants with different sensitivity to copper. Planta 181: 487–495 [DOI] [PubMed] [Google Scholar]
- Palmer CM, Guerinot ML. (2009) Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat Chem Biol 5: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Kim H, Koo SC, Park HJ, Cheong MS, Hong MJ, Baek D, Chung WS, Kim DH, Bressan RA, et al. (2010) Functional characterization of the SIZ/PIAS-type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice. Plant Cell Environ 33: 1923–1934 [DOI] [PubMed] [Google Scholar]
- Pich A, Scholz G, Stephan UW. (1994) Iron-dependent changes of heavy-metals, nicotianamine, and citrate in different plant organs and in the xylem exudate of 2 tomato genotypes: nicotianamine as possible copper translocator. Plant Soil 165: 189–196 [Google Scholar]
- Pilon M, Abdel-Ghany SE, Cohu CM, Gogolin KA, Ye H. (2006) Copper cofactor delivery in plant cells. Curr Opin Plant Biol 9: 256–263 [DOI] [PubMed] [Google Scholar]
- Pilon M, Cohu CM, Ravet K, Abdel-Ghany SE, Gaymard F. (2009) Essential transition metal homeostasis in plants. Curr Opin Plant Biol 12: 347–357 [DOI] [PubMed] [Google Scholar]
- Puig S, Andrés-Colás N, García-Molina A, Peñarrubia L. (2007) Copper and iron homeostasis in Arabidopsis: responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ 30: 271–290 [DOI] [PubMed] [Google Scholar]
- Puig S, Peñarrubia L. (2009) Placing metal micronutrients in context: transport and distribution in plants. Curr Opin Plant Biol 12: 299–306 [DOI] [PubMed] [Google Scholar]
- Puig S, Thiele DJ. (2002) Molecular mechanisms of copper uptake and distribution. Curr Opin Chem Biol 6: 171–180 [DOI] [PubMed] [Google Scholar]
- Quartacci MF, Cosi E, Navari-Izzo F. (2001) Lipids and NADPH-dependent superoxide production in plasma membrane vesicles from roots of wheat grown under copper deficiency or excess. J Exp Bot 52: 77–84 [PubMed] [Google Scholar]
- Rellán-Alvarez R, Abadía J, Alvarez-Fernández A. (2008) Formation of metal-nicotianamine complexes as affected by pH, ligand exchange with citrate and metal exchange: a study by electrospray ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 22: 1553–1562 [DOI] [PubMed] [Google Scholar]
- Sancenón V, Puig S, Mira H, Thiele DJ, Peñarrubia L. (2003) Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol Biol 51: 577–587 [DOI] [PubMed] [Google Scholar]
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD. (2007) Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145: 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schützendübel A, Polle A. (2002) Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J Exp Bot 53: 1351–1365 [PubMed] [Google Scholar]
- Sharma SS, Dietz KJ. (2006) The significance of amino acids and amino acid-derived molecules in plant responses and adaptation to heavy metal stress. J Exp Bot 57: 711–726 [DOI] [PubMed] [Google Scholar]
- Shen ZG, Zhang FQ, Zhang FS. (1998) Toxicity of copper and zinc in seedlings of mung bean and inducing accumulation of polyamine. J Plant Nutr 21: 1153–1162 [Google Scholar]
- Shi J, Yuan X, Chen X, Wu B, Huang Y, Chen Y. (2011) Copper uptake and its effect on metal distribution in root growth zones of Commelina communis revealed by SRXRF. Biol Trace Elem Res 141: 294–304 [DOI] [PubMed] [Google Scholar]
- Shin YC, Liu BY, Tsai JY, Wu JT, Chang LK, Chang SC. (2010) Biochemical characterization of the small ubiquitin-like modifiers of Chlamydomonas reinhardtii. Planta 232: 649–662 [DOI] [PubMed] [Google Scholar]
- Stadtman ER, Oliver CN. (1991) Metal-catalyzed oxidation of proteins: physiological consequences. J Biol Chem 266: 2005–2008 [PubMed] [Google Scholar]
- Strange J, Macnair MR. (1991) Evidence for a role for the cell-membrane in copper tolerance of Mimulus guttatus Fischer Ex Dc. New Phytol 119: 383–388 [Google Scholar]
- Wang Y, Ladunga I, Miller AR, Horken KM, Plucinak T, Weeks DP, Bailey CP. (2008) The small ubiquitin-like modifier (SUMO) and SUMO-conjugating system of Chlamydomonas reinhardtii. Genetics 179: 177–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, Chu HH, Didonato RJ, Roberts LA, Eisley RB, Lahner B, Salt DE, Walker EL. (2006) Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol 141: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintz H, Fox T, Wu YY, Feng V, Chen WQ, Chang HS, Zhu T, Vulpe C. (2003) Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem 278: 47644–47653 [DOI] [PubMed] [Google Scholar]
- Wintz H, Vulpe C. (2002) Plant copper chaperones. Biochem Soc Trans 30: 732–735 [DOI] [PubMed] [Google Scholar]
- Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I. (2007) NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19: 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM. (2006) SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 142: 1548–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yruela I. (2005) Copper in plants. Braz J Plant Physiol 17: 145–146 [Google Scholar]
- Yruela I. (2009) Copper in plants: acquisition, transport and interactions. Funct Plant Biol 36: 409–430 [DOI] [PubMed] [Google Scholar]
- Zhang ZK, Liu SQ, Hao SQ, Liu SH. (2010) Grafting increases the copper tolerance of cucumber seedlings by improvement of polyamine contents and enhancement of antioxidant enzymes activity. Agric Sci China 9: 985–994 [Google Scholar]
- Zhou JM, Goldsbrough PB. (1994) Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell 6: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]