Abstract
The plant hormone cytokinin is perceived by membrane-located sensor histidine kinases. Arabidopsis (Arabidopsis thaliana) possesses three cytokinin receptors: ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and CYTOKININ RESPONSE1/AHK4. The current model predicts perception of the cytokinin signal at the plasma membrane. However, cytokinin-binding studies with membrane fractions separated by two-phase partitioning showed that in the wild type, as well as in mutants retaining only single cytokinin receptors, the major part of specific cytokinin binding was associated with endomembranes. Leaf epidermal cells of tobacco (Nicotiana benthamiana) expressing receptor-green fluorescent protein fusion proteins and bimolecular fluorescence complementation analysis showed strong fluorescence of the endoplasmic reticulum (ER) network for all three receptors. Furthermore, separation of the microsomal fraction of Arabidopsis plants expressing Myc-tagged AHK2 and AHK3 receptors by sucrose gradient centrifugation followed by immunoblotting displayed the Mg2+-dependent density shift typical of ER membrane proteins. Cytokinin-binding assays, fluorescent fusion proteins, and biochemical fractionation all showed that the large majority of cytokinin receptors are localized to the ER, suggesting a central role of this compartment in cytokinin signaling. A modified model for cytokinin signaling is proposed.
Cytokinins are a class of phytohormones regulating such diverse processes as root and shoot growth, leaf senescence, and chloroplast development. They also function in mediating the plant response to environmental signals, such as biotic and abiotic stresses (Argueso et al., 2009; Werner and Schmülling, 2009). The cytokinin signal is perceived by membrane-bound sensor His kinases and further transduced by a multistep variant of the two-component signaling system (Heyl and Schmülling, 2003; Kakimoto, 2003; Bishopp et al., 2006). Cytokinin receptors consist of an extracytosolic cyclase, His kinase-associated sensory extracellular (CHASE) domain flanked by two transmembrane domains at the N terminus, followed toward the C terminus by a His kinase and a response regulator domain in the predicted cytosolic part. Arabidopsis (Arabidopsis thaliana) encodes three cytokinin receptors, ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and CYTOKININ RESPONSE1 (CRE1)/AHK4 (Inoue et al., 2001; Suzuki et al., 2001a; Ueguchi et al., 2001; Yamada et al., 2001), which differ in their biological functions and biochemical properties (for review, see Heyl et al., 2011). The cytoplasmic part of all three receptors interact redundantly with His phosphotransfer proteins (AHP), which transmit the signal to the nucleus (Hwang and Sheen, 2001; Punwani et al., 2010), where type B response regulators, a class of Myb transcription factors, are activated. These mediate the majority, if not all, of the transcriptional responses to cytokinin (Argyros et al., 2008; Heyl et al., 2008; Ishida et al., 2008).
Genetic studies have shown that many of the receptor activities are redundant, but specific activities for single receptors have been reported as well (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). The peculiarities of receptor functions might be due to differences in their expression (Ueguchi et al., 2001; Higuchi et al., 2004; Nishimura et al., 2004; Stolz et al., 2011), differences in the recognition of different cytokinin compounds (Yamada et al., 2001; Spíchal et al., 2004; Romanov et al., 2006; Stolz et al., 2011), specific interactions with other proteins (Dortay et al., 2008), or differences in their subcellular localization. Current cytokinin signaling models predict that the cytokinin receptors are localized to the plasma membrane (PM), which was initially based on bioinformatic analysis of the protein sequence and analogy with sensor His kinase localization in bacteria and yeast (Inoue et al., 2001; Ueguchi et al., 2001). This assignment was experimentally supported by the localization of an overexpressed AHK3-GFP fusion protein at the PM of Arabidopsis protoplasts (Kim et al., 2006). However, additional experiments were not carried out, and localization to endomembranes was not excluded. Instead, biochemical studies of cytokinin receptors revealed that their maximal hormone-binding activity is close to a neutral or weak alkaline pH (Romanov et al., 2006), which is characteristic for the cytoplasm and the endoplasmic reticulum (ER; Tian et al., 1995) but not the apoplast, leading to the suggestion that cytokinin receptors may function inside the cell (Romanov et al., 2006). We here investigate in more detail the subcellular localization of the three cytokinin receptors of Arabidopsis and present biochemical and cell biological evidence indicating that the cytokinin receptors of Arabidopsis are predominantly localized to the ER.
RESULTS
Endomembranes Show Higher Cytokinin Binding Than the PM
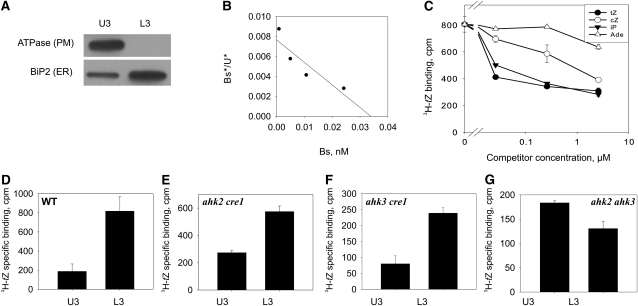
To gain insight into the cellular compartment harboring the cytokinin receptors, isolated membranes of 6-d-old Arabidopsis seedlings were separated, using the two-phase partitioning method, into a PM fraction and an inner membrane (endomembrane) fraction. Microsomes of each fraction retain integral proteins and also proteins associated with these membranes (Larsson et al., 1987). The upper fraction (U3) was strongly enriched with a PM marker (H+-ATPase), while the lower fraction (L3) showed enrichment of the ER marker Binding Protein2 (BiP2; Fig. 1A). The portion of endomembranes was 15 to 20 times higher than that of PMs as measured by their protein content. An in vitro cytokinin-binding assay with microsomes showed [3H]trans-zeatin-specific binding (i.e. the difference between total and nonspecific binding) in both membrane fractions (Fig. 1, B–G). Quantitative analysis of this binding in wild-type plants revealed a high-affinity interaction with an equilibrium dissociation constant (KD) in the low nanomolar range (Fig. 1B). This is similar to the KD for trans-zeatin-receptor complexes of AHK3 and CRE1/AHK4 (Yamada et al., 2001; Romanov et al., 2005, 2006) and the AHK2-CHASE-TM domain (Stolz et al., 2011) expressed in bacteria. The ligand specificity of binding of the isolated membranes (Fig. 1C) also corresponded well with the ligand specificity of cloned Arabidopsis cytokinin receptors (Romanov et al., 2005, 2006; Stolz et al., 2011). These results support the idea that the cytokinin binding of isolated membranes of Arabidopsis is due to these cytokinin receptors. In addition, tobacco (Nicotiana benthamiana) leaves overexpressing a AHK3-GFP fusion gene showed a strong increase of cytokinin binding in their membrane fractions (Supplemental Fig. S1). In microsomes isolated from wild-type plants, the total specific cytokinin binding was approximately four times higher in the endomembranes than in the PM (Fig. 1D). Considering the higher portion of endomembrane proteins, this means that more than 98% of the cytokinin binding sites are located in endomembranes.
Figure 1.
Cytokinin binding to different membrane fractions obtained by aqueous two-phase partitioning. Six-day-old Arabidopsis seedlings of the wild type and cytokinin receptor double mutants were used for membrane isolation. A, Detection of the membrane-specific markers H+-ATPase (PM) and BiP2 (ER) by immunoblot analysis in the upper (U3) and lower (L3) phases. B, Scatchard plot of [3H]trans-zeatin binding to endomembranes of wild-type plants. Bs*, Specifically bound [3H]trans-zeatin; U*, unbound [3H]trans-zeatin; Bs, total bound trans-zeatin (sum of labeled and unlabeled). The deduced KD is 4.7 nm. C, Competition between [3H]trans-zeatin and unlabeled cytokinins for binding to endomembranes of wild-type plants. Data represent mean values ± se (n = 2). tZ, trans-Zeatin; cZ, cis-zeatin; iP, isopentenyladenine; Ade, adenine. D to G, [3H]trans-Zeatin binding to membrane fractions from the wild type (WT) and double receptor mutants as indicated. U3, Upper phase enriched for PM; L3, lower phase enriched for endomembranes. Data represent mean values ± se (n = 2) of specifically bound [3H]trans-zeatin in cpm per 100 μg of membrane protein.
In order to investigate whether this distribution of binding activity is typical for individual cytokinin receptors of Arabidopsis, the experiment was repeated with all three double receptor mutants, each retaining only a single cytokinin receptor (Riefler et al., 2006). The cytokinin binding of microsomes from each of the double mutants was lower than that from the wild type (Fig. 1. D–G), which supports further a crucial role of the three known cytokinin receptors to mediate this binding activity. Mutants harboring solely the AHK3 receptor retained the highest binding activity (Fig. 1E) and showed a clear prevalence of cytokinin binding to endomembranes, similar to the wild type (Fig. 1, D and E). A similar pattern was also revealed in the ahk3 cre1 mutant expressing AHK2 as a single cytokinin receptor (Fig. 1F). In contrast, ahk2 ahk3 mutant plants expressing CRE1/AHK4 displayed a somewhat higher specific binding in the PM fraction than in the fraction enriched with endomembranes (Fig. 1G). However, because the vast majority of membrane proteins are located in endomembranes (see above), in this case, also more than 90% of the cytokinin-binding sites are located in endomembranes. Nevertheless, this result should be taken with caution because of the overall very low binding activity of microsomes isolated from this double mutant. Interestingly, the sum of cytokinin binding of membranes from all three different double receptor mutants corresponds roughly to the binding activity measured in the wild type (Fig. 1, D–G). Taken together, the distribution of cytokinin-binding activity in mutants retaining single receptors indicates that all three cytokinin receptors of Arabidopsis localize mostly to endomembranes.
Fluorescence-Labeled Cytokinin Receptors Show a Predominant Localization to the ER
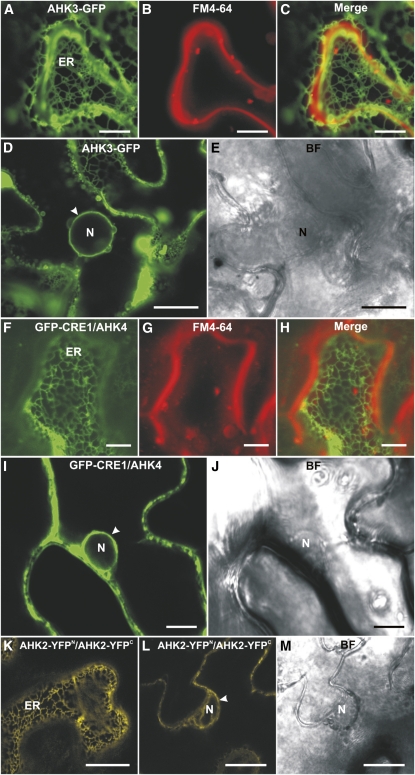
Two-phase partitioning had indicated a predominating location of cytokinin-binding sites in endomembranes but did not reveal which type of intracellular membranes harbors cytokinin receptors. To refine the analysis of their subcellular distribution, we fused all three cytokinin receptors with the GFP (Mathur, 2007). These fusion proteins were transiently expressed under the control of a 35S promoter in the leaf epidermal cells of tobacco and analyzed by laser-scanning confocal microscopy.
The subcellular localization pattern of AHK3-GFP (Fig. 2, A and C) was similar to the pattern known for ER-localized proteins (Boevink et al., 1996, 1998; Batoko et al., 2000; Nelson et al., 2007). This was further corroborated by a strong perinuclear signal for AHK3-GFP, which is a hallmark of ER localization (Fig. 2, D and E). Overlay analysis of the AHK3-GFP signal with the FM4-64 dye staining the PM showed only a limited overlap of the signals, indicating that most of the AHK3-GFP fusion protein does not localize to the PM but to the ER (Fig. 2, A–C).
Figure 2.
Localization of fluorescent cytokinin receptor fusion proteins in epidermal cells of tobacco leaves. A, ER labeled with AHK3-GFP. B, Staining of the PM with FM4-64 (50 μm). C, Merged image of A and B. D, Localization of AHK3-GFP in the perinuclear space (arrowhead). E, Bright-field (BF) image of D. F, ER labeled with GFP-CRE1/AHK4. G, Staining of the PM with FM4-64 (50 μm). H, Merged image of F and G. I, Localization of GFP-CRE1/AHK4 in the perinuclear space (arrowhead). J, Bright-field image of I. K, BiFC analysis reveals the formation of AHK2-YFPN/AHK2-YFPC homodimers in the ER. L, Homodimerization of AHK2-YFPN and AHK2-YFPC in the perinuclear space (arrowhead). M, Bright-field image of L. N, Nucleus. Bars = 7.5 μm in F to H; 10 μm in A to C, I, and J; and 25 μm in D, E, L, and M.
A GFP-CRE1/AHK4 fusion protein showed a similar subcellular distribution in tobacco leaf cells to AHK3-GFP. An ER-like network was decorated by the GFP-CRE1/AHK4 fusion protein (Fig. 2F), and a perinuclear signal was detected as well (Fig. 2, I and J). Staining the cells with the FM4-64 PM marker showed little overlap with the GFP-CRE1/AHK4 signal (Fig. 2, G and H), supporting its predominant localization in the ER membrane.
In the case of AHK2, neither N- nor C-terminal fusion with GFP led to a detectable signal in planta. In contrast, bimolecular fluorescence complementation (BiFC) analysis (Walter et al., 2004) for AHK2 resulted in a clear signal, and these data can be used to analyze the subcellular localization of the AHK2 receptor. The AHK2-Split-yellow fluorescent protein (YFP) dimer decorated an ER-like intracellular network (Fig. 2K), and perinuclear localization was detected for the AHK2-Split-YFP dimer as well (Fig. 2, L and M).
In summary, the subcellular localization of transiently expressed fluorescent fusion proteins supported a predominant localization to the ER for all three cytokinin receptors. Attempts to generate transgenic Arabidopsis lines stably expressing the receptor-GFP fusion genes under the control of the 35S promoter have not been successful. None of the transgenic lines that were obtained showed detectable GFP signal, indicating selection against high expression levels. This finding is in accordance with our inability to obtain transgenic lines constitutively expressing cytokinin receptor genes, most likely due to cellular toxicity of the gene products (data not shown).
Biochemical Fractionation Confirms the ER Localization of Cytokinin Receptors
GFP-based localization experiments do not provide sufficient evidence for determining the true subcellular localization of a given protein, especially of endomembrane-localized proteins (Moore and Murphy, 2009). Other methods need to be applied to validate results obtained by fluorescent protein fusions; therefore, a subcellular localization of Myc-tagged cytokinin receptors following fractionation of cell membranes (microsome fraction) by Suc gradient centrifugation was conducted. The functionality of AHK2-Myc and AHK3-Myc fusion proteins was tested by transforming ahk2 ahk3 mutant plants, which show a distinct phenotype, including a small leaf rosette size, depending on the simultaneous mutation of both receptor genes (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). Transformed plants expressing chimeric PAHK3:AHK3-Myc or PAHK2:AHK2-Myc showed complementation of the ahk2 ahk3 mutant phenotype (Supplemental Fig. S2), demonstrating the functionality of both AHK-Myc fusion proteins. Two independent transgenic lines expressing the fusion genes were analyzed further. The expression of PCRE1:CRE1/AHK4-Myc cannot be verified by this complementation test. In addition, the level of expression of a PCRE1:CRE1/AHK4-Myc gene was too low to reliably detect the tagged receptor protein, which is in accordance with the low cytokinin-binding activity of microsomes from the ahk2 ahk3 mutant (Fig. 1G; data not shown). Therefore, PCRE1:CRE1/AHK4-Myc transgenic plants were not included in this study.
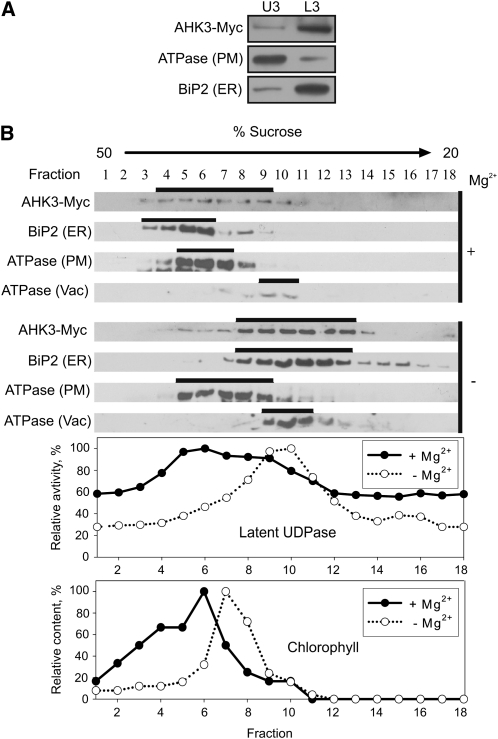
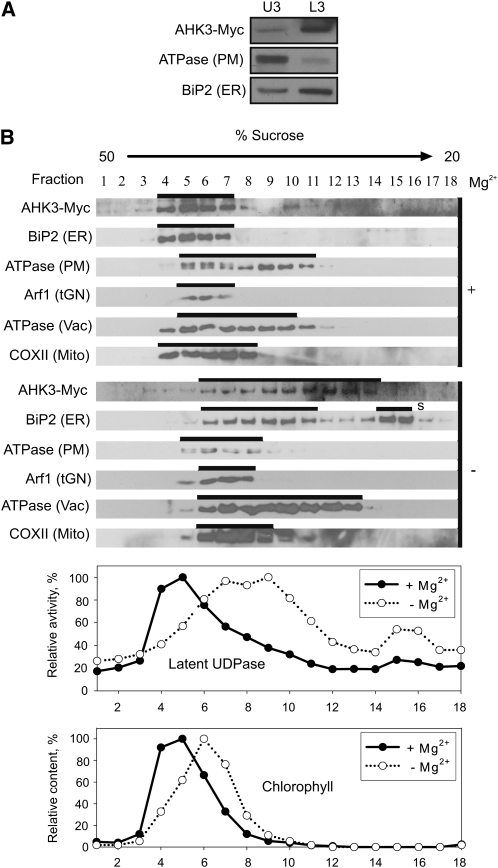
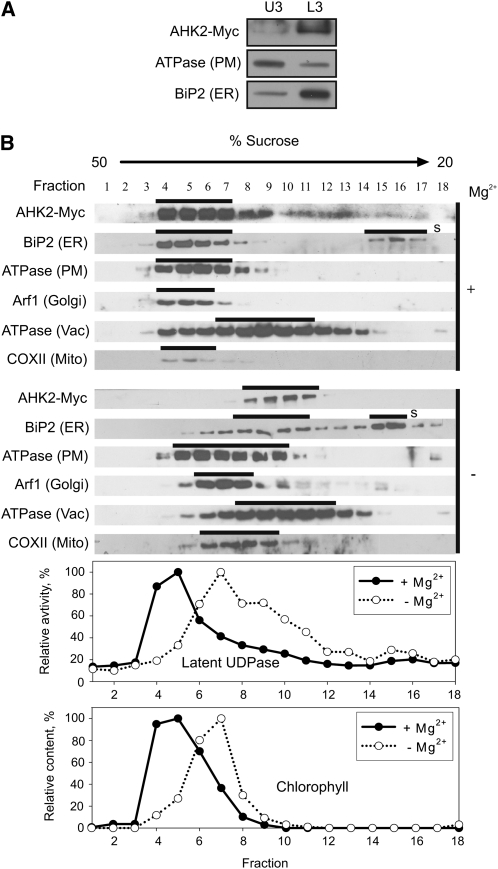
First, membranes of 6- and 18-d-old PAHK3:AHK3-Myc plants and 6-d-old PAHK2:AHK2-Myc transgenic plants were isolated and fractionated using the two-phase partitioning method. Immunoblots showed the expected distribution of the PM-bound H+-ATPase and the ER marker BiP2 to the U3 and L3 fractions, respectively (Figs. 3A,4A, and 5A). Large majorities of both tagged receptors, AHK3-Myc and AHK2-Myc, were detected in the L3 fraction (Figs. 3A, 4A, and 5A), corroborating an endomembrane localization for these two cytokinin receptors.
Figure 3.
Localization of AHK3-Myc in fractionated membranes from 6-d-old Arabidopsis seedlings expressing PAHK3:AHK3-Myc. A, Aqueous two-phase partitioning of microsomes. Equal amounts of protein from upper (U3) and lower (L3) phases were separated by SDS-PAGE and subjected to immunoblot analysis with antibodies specific for Myc, H+-ATPase (PM), and BiP2 (ER). B, Microsomal membranes were fractionated on linear 20% to 50% (w/w) Suc gradients in the presence of Mg2+ (+) to stabilize ribosomes at the ER or in the absence of Mg2+ (−) to dissociate ribosomes from the ER. Samples (50–100 μL) of each fraction were analyzed by immunoblot using antibodies specific for Myc, H+-ATPase, BiP2, and V-ATPase (Vac, vacuole membrane marker). Latent UDPase activity was used as an enzymatic marker for the Golgi apparatus, and chlorophyll was used to localize thylakoid membranes. In both cases, the highest measured enzyme activity (A650) and chlorophyll content (A652) were set at 100%.
Figure 4.
Localization of AHK3-Myc in fractionated membranes from shoots of 18-d-old Arabidopsis plants expressing PAHK3:AHK3-Myc. A, Aqueous two-phase partitioning of microsomes. Equal amounts of protein from upper (U3) and lower (L3) phases were separated by SDS-PAGE and subjected to immunoblot analysis with antibodies specific for Myc, H+-ATPase (PM), and BiP2 (ER). B, Microsomal membranes were fractionated on linear 20% to 50% (w/w) Suc gradients in the presence of Mg2+ (+) to stabilize ribosomes at the ER or in the absence of Mg2+ (−) to dissociate ribosomes from the ER. Samples (50–100 μL) of each fraction were analyzed by immunoblot using antibodies specific for Myc, H+-ATPase, BiP2, V-ATPase (Vac, vacuole membrane marker), Arf1 (tGN, trans-Golgi network marker), and COXII (Mito, mitochondria marker). Latent UDPase activity was used as an enzymatic marker for the Golgi apparatus, and chlorophyll was used to localize thylakoid membranes. In both cases, the highest measured enzyme activity (A650) and chlorophyll content (A652) were set at 100%. S, Free soluble BiP2, which has been released during processing and remains at the top of the gradient.
Figure 5.
Localization of AHK2-Myc in fractionated membranes from Arabidopsis seedlings expressing PAHK2:AHK2-Myc. A, Aqueous two-phase partitioning of microsomes from 6-d-old seedlings. Equal amounts of protein from upper (U3) and lower (L3) phases were separated by SDS-PAGE and subjected to immunoblot analysis with antibodies specific for Myc, H+-ATPase (PM), and BiP2 (ER). B, Microsomal membranes from shoots of 18-d-old plants were fractionated on linear 20% to 50% (w/w) Suc gradients in the presence of Mg2+ (+) to stabilize ribosomes at the ER or in the absence of Mg2+ (−) to dissociate ribosomes from the ER. Samples (50–100 μL) of each fraction were analyzed by immunoblot using antibodies specific for Myc, H+-ATPase, BiP2, V-ATPase (Vac, vacuole membrane marker), Arf1 (Golgi, trans-Golgi network marker), and COXII (Mito, mitochondria marker). Latent UDPase activity was used as an enzymatic marker for the Golgi apparatus, and chlorophyll was used to localize thylakoid membranes. In both cases, the highest measured enzyme activity (A650) and chlorophyll content (A652) were set at 100%. S, Free soluble BiP2, which has been released during processing and remains at the top of the gradient.
In order to identify the intracellular membrane fraction(s) to which AHK3-Myc and AHK2-Myc localize more precisely, the microsomal fraction was further separated by Suc gradient centrifugation either in the presence or absence of Mg2+ ions. The rationale behind this approach is that binding of ribosomes to the ER is dependent on the presence of Mg2+ ions (Lord, 1987). In the absence of Mg2+ ions (e.g. if chelators like EDTA are in the buffer), the release of ribosomes from the ER lowers the density of the ER membrane and thus leads to a shift within the Suc gradient. In contrast, other organelles are not significantly affected in their density by Mg2+ ions. The marker proteins belonging to different membrane types were detected by immunoblotting.
To study the intracellular distribution of AHK3-Myc, 6-d-old Arabidopsis seedlings and shoots of 18-d-old plants were used as starting material (Figs. 3 and 4). The position of the membrane-bound fraction of the ER marker BiP2 was clearly shifted upon depletion of Mg2+ ions from the extraction buffer, whereas the pattern obtained for the markers of the PM and the vacuolar membrane were largely independent of the presence of Mg2+ ions (Figs. 3B and 4B). Chlorophyll absorbance, which was measured as a marker for the chloroplast thylakoid membranes, also did not show the characteristic Mg2+ shift of BiP2. The position of AHK3-Myc in the gradient was similar to that of BiP2 in showing a clear shift upon Mg2+ depletion from the buffer. The position in the gradient of latent UDPase, an enzyme marker for the Golgi apparatus, was also dependent on Mg2+, but its position and peak distribution in the fractions were less well correlated with the positions of AHK3-Myc and BiP2, especially visible in 18-d-old shoots (Fig. 4B). Cytochrome c oxidase subunit II (COXII), a marker for mitochondria, and ADP-ribosylation factor1 (Arf1), a marker for the trans-Golgi network, were also tested in this material. The positions of these markers shifted only marginally to fractions of lower Suc concentrations in the Mg2+-free buffer (Fig. 4B). Taken together, the behavior of proteins isolated from 6-d-old seedlings and shoots of 18-d-old plants was qualitatively similar, indicating that the localization of AHK3-Myc to the ER is independent of the plant age.
The localization study of AHK2-Myc yielded similar results. After fractionation in a Suc gradient in the presence or absence of Mg2+ ions, a diagnostic shift of the AHK2-Myc position was detected that resembled the one of the membrane fraction of BiP2. The markers for other membranes showed a much weaker shift in the absence of Mg2+ ions (Fig. 5B) and/or their positions in the gradient were not correlated with the position of AHK2-Myc.
DISCUSSION
Cytokinin Receptors Are Located Mainly to the ER
Three different experimental approaches consistently support the localization of the bulk of cytokinin receptors in the internal membrane system of plant cells, notably in the ER. Importantly, the KD of microsomal binding sites for trans-zeatin and the ligand specificity of binding matched well those of Arabidopsis cytokinin receptors cloned in Escherichia coli (Romanov et al., 2005, 2006; Stolz et al., 2011). Moreover, cytokinin binding was significantly reduced in double receptor mutants but was very high in plant cells overexpressing a receptor. Visualization of receptor-GFP fusions and BiFC analysis using Split-YFP clearly supported the localization of the majority of receptors in the ER. Finally, immunoblot analysis of Myc-tagged receptors upon membrane fractionation in a Suc gradient demonstrated ER localization for AHK2 and AHK3.
The ER localization is consistent with the biochemical properties of cytokinin receptors (Romanov et al., 2006) and with earlier studies that identified high-affinity cytokinin-binding proteins in particulate/microsomal fractions of plant cells (Gardner et al., 1978; Kobayashi et al., 1981; Brault et al., 1999). The ER localization of the cytokinin receptors contrasts with the previous prediction that the cytokinin-binding CHASE domain is apoplastic (Inoue et al., 2001; Ueguchi et al., 2001) and with current models of cytokinin signaling assuming the localization of cytokinin receptors at the PM (Heyl and Schmülling, 2003; Hwang and Sakakibara, 2006; Müller and Sheen, 2007; To and Kieber, 2008). However, it should be noted that a localization of a minor but functionally relevant part of the receptor pool at the PM cannot be excluded (Fig. 1, D–G).
The mechanism for the localization of cytokinin receptors is not clear, as they do not possess a known ER retention signal, such as the cytosolic di-Lys motif [K(X)KXX] that is found at the C-terminal end of numerous type I membrane proteins of yeast and animal cells (Pagny et al., 1999). A di-Arg motif (i.e. RR, RXR, or RXXR) in the first five N-terminal amino acids of type II membrane proteins, which was first described in yeast but was recently also shown to be present in a plant type II membrane protein (Boulaflous et al., 2009), is present at the N terminus of CRE1/AHK4 (i.e. MRRD) but is lacking in the other two receptors. Whether this motif functions as an ER retention signal for CRE1/AHK4 and which other as yet unknown protein sequence(s) may fulfill similar functions as an ER retention signal for the cytokinin receptors are not known.
The membrane topology of cytokinin receptors is also highly relevant but currently unknown. The ligand-binding CHASE domain will most likely be on the extracytosolic side and the His kinase domain will most likely be oriented toward the cytoplasm and/or nuclear compartment, enabling them to interact with the AHP proteins (Suzuki et al., 2001b; Dortay et al., 2006), which are exclusively located in the cytoplasm and the nucleus (Hwang and Sheen, 2001; Punwani et al., 2010).
Are the ER-Localized Cytokinin Receptors Biologically Active?
The presence of cytokinin receptors in the ER raises the question of whether they are active in this compartment or whether it is used as a place for resting or recycling of cytokinin receptors that otherwise exert their functions at the PM. We favor the idea that the ER-localized cytokinin receptors have a function in cytokinin perception and signaling. This is underpinned by the fact that Myc-tagged receptors complementing the receptor loss-of-function phenotype were located to the ER (Figs. 3–5) and the strongly enhanced cytokinin binding of the microsomal fraction of cells harboring the AHK3-GFP fusion protein (Supplemental Fig. S1). It is also consistent with optimal cytokinin binding at neutral pH (Romanov et al., 2006), that is, in conditions close to those found in the cytoplasm and ER (Tian et al., 1995), and the proven activity of the cytokinin receptors in diverse membrane systems, including bacteria and yeast (Inoue et al., 2001; Suzuki et al., 2001a). The functionality of ER-located cytokinin receptors is also supported by different active cytokinin pools in plant cells. Not much is known about different subcellular pools of cytokinins, but cytokinin-synthesizing and cytokinin-degrading enzymes are located in different subcellular compartments (Werner et al., 2003; Sakakibara, 2006), indicating a relevance of such compartmentation for cytokinin action. Part of the cytokinin degradation in particular is compartmentalized to the ER, the extracellular space, and the cytoplasm (Motyka et al., 1996; Werner et al., 2003; Köllmer, 2009; T. Werner, personal communication), indicating that cytokinin homeostasis needs to be controlled at these different sites. Enhanced degradation of cytokinin at these different sites has partly different qualitative and/or quantitative consequences for plant growth and development, suggesting that different signaling pathways are affected (Werner et al., 2001, 2003; Köllmer, 2009). Thus, it could be that specification of the cytokinin signal is partly achieved through different cytokinin metabolites with distinct subcellular localizations. In this context, the question arises of how the cytokinin transport inside the cell is achieved, as intracellular cytokinin transporters have not yet been identified (Hirose et al., 2008). However, in the case of the ER, the problem of cytokinin supply is possibly alleviated by the fact that the ER membrane is permeable to small molecules (Le Gall et al., 2004).
A Modified Model for Cytokinin Signaling
The current model for cytokinin signaling needs to be amended to accommodate the novel predominant ER localization of the cytokinin receptors. Cytokinin receptors located on the ER membrane could transmit the cytokinin signal via AHP to the nucleus, similar to the way it is proposed for signal transmission from the PM (Schaller et al., 1995; Hwang and Sheen, 2001; Heyl and Schmülling, 2003; Kakimoto, 2003; Bishopp et al., 2006). One benefit of the localization of receptors on endomembranes is that it may alleviate disadvantages caused by the variable intracellular distances that signaling chains have to bridge (Geldner and Robatzek, 2008). The large central vacuole of plant cells leads to the close apposition of the nucleus to a small region of the PM, whereas other parts of the PM reside at a considerable distance. This could cause a strong bias in signaling impact on gene activation in the nucleus, depending on the position of the activated receptor. Signaling from intracellular compartments, such as the ER, could overcome this limitation (Geldner and Robatzek, 2008). Cytokinin receptor location in the ER would also offer opportunities for cross talk, as numerous other plant receptor proteins are at least partially located in the ER (Geldner and Robatzek, 2008; Irani and Russinova, 2009); notably, this includes the structurally related ethylene receptors (Chen et al., 2002). Noteworthy, intracellular distance could be overcome even more effectively if receptors were localized on the internal nuclear membrane. Cytokinin signaling from the perinuclear space would be in accordance with the cytokinin-independent presence of phosphotransmitter proteins in the cell nucleus and the interaction of receptors with transcription factors in yeast two-hybrid and coimmunoprecipitation assays (Dortay et al., 2006, 2008). Perinuclear signals were obtained with the receptor-GFP fusion proteins (Fig. 2), but these do not allow the distinction between the outer and inner nuclear membrane as a signal source; therefore, in the absence of more experimental proof, this remains a hypothetical possibility.
The possibility that a small part of the cytokinin receptors signal from the PM should also be considered. The signaling of the receptors from the outer membrane is consistent with the developmental consequences of the extracellular degradation of cytokinin (Werner et al., 2003) and the observation that CRE1/AHK4 interacts in a yeast-two hybrid assay with GNOM, ADL1, and adaptin, which are all proteins involved in vesicular trafficking, arguing for an intracellular transport of cytokinin receptors between the PM and endosome (Dortay et al., 2008). However, attempts to demonstrate an influence of cytokinin or the inhibitor of endosomal recycling brefeldin A on the localization of cytokinin receptor-GFP fusion proteins have not been successful so far (Wulfetange, 2010). Thus, a model involving the recycling of the receptors between the ER and the PM, as has been described for BRI1 and the receptor-like kinase CRINKLY4 (Gifford et al., 2005; Geldner et al., 2007), is currently not supported experimentally for the cytokinin receptors.
In summary, the results presented here give clear evidence for a predominant ER localization of the cytokinin receptors in Arabidopsis. In addition, ER localization of the cytokinin receptor ZmHK1 of Zea mays has also been demonstrated very recently (Lomin et al., 2011). This highlights the important role of this cellular compartment in cytokinin signaling and, more generally, the necessity of considering the subcellular topology of hormone metabolism and signaling as a relevant factor and additional layer of regulation in hormone biology.
MATERIALS AND METHODS
Recombinant DNA Techniques
The coding region of the cytokinin receptor AHK4/CRE1 (At2g01830.1) was amplified by PCR using a cDNA library from Arabidopsis (Arabidopsis thaliana) C24 as a template (Minet et al., 1992) and cloned into the Gateway entry vector pDONR201 by BP recombination reaction (Invitrogen). The coding sequence of AHK3 (At1g27320.1) was amplified via reverse transcription-PCR on total RNA extracted from the primary leaves of Arabidopsis ecotype Columbia using appropriate primers (Supplemental Table S1) and cloned into the entry vector pDONR222. AHK2 (At5g35750.1) was amplified with the stop codon from the genomic DNA of bacterial artificial chromosome clone MXH1 (Liu et al., 1995) using the appropriate primers (Supplemental Table S1) and cloned into pDONR221. To construct C-terminal fusions of AHK2 and AHK3 with GFP or 4× Myc tag, the stop codons were removed by site-directed mutagenesis using the Quick Change II site-directed mutagenesis kit (Stratagene) with the primers listed in Supplemental Table S1. The correct identities of all entry clones and mutated clones were verified by sequencing.
For transient GFP expression studies in tobacco (Nicotiana benthamiana), AHK3 and CRE1/AHK4 were shuttled into the destination vectors pB7FWG2 and pB7WGF2 (Karimi et al., 2002), which allow C- and N-terminal fusions with GFP, respectively, and thus were positioned under the control of the 35S promoter. The genomic fragment of AHK2 was also amplified without the stop codon (for primers, see Supplemental Table S1) and then cloned into the vectors pSPYNE-35S and pSPYCE-35S (Walter et al., 2004), forming C-terminal fusions with the N- and C-terminal fragments of YFP, respectively, and used for BiFC analysis.
Fusion constructs with a 4× Myc tag were created under the control of their respective promoters for AHK2 and AHK3. The MultiSite Gateway Three Fragment Vector Construction Kit was used for cloning and recombination reactions were performed according to the manufacturer’s manual (Invitrogen). Promoter sequences for AHK2 (2,124 bp) and AHK3 (2,063 bp) were amplified using the appropriate primers (Supplemental Table S1; Stolz et al., 2011) and recombined into the entry vector pDONR P4-P1R. The 4× Myc tag was amplified in a two-step PCR from the binary vector pGWB17 (Nakagawa et al., 2007) using the corresponding primers (Supplemental Table S1) and recombined into the entry vector pDONR P2R-P3. All three fragments (promoter, gene, and tag) were shuttled into the destination vector pB7m34GW (Karimi et al., 2005) to obtain PAHK2:AHK2-Myc and PAHK3:AHK3-Myc.
Plant Materials and Growth Conditions
Arabidopsis ecotype Columbia was used as the wild type. The cytokinin receptor double mutants have been previously described (Riefler et al., 2006). Arabidopsis plants were grown in the greenhouse on soil at 22°C under long-day conditions (16 h of light/8 h of dark). Plants were grown in vitro for the selection of primary transformants and segregation analyses. Seeds were surface sterilized and, after cold treatment for 2 to 3 d, exposed to white light (approximately 75 μE) in growth chambers. Seeds were grown at 23°C on horizontal plates containing half-strength MS medium (Murashige and Skoog, 1962) including 0.9% agar (Merck). The medium was supplemented with 3% Suc, 100 mg L−1 myoinositol, 20 mg L−1 thiamine, 1 mg L−1 nicotinic acid, 1 mg L−1 pyridoxine, 1 mg L−1 biotin, and 10 mm MES, pH adjusted to 5.7 with KOH. Tobacco plants were cultivated in a greenhouse on soil at 25°C under long-day conditions as described above.
Plant Transformation
Transgenic Arabidopsis plants were generated by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101. Transformed seedlings were selected on medium supplemented with 10 μg mL−1 phosphinotricin or on soil sprayed with Basta (0.1%; Bayer CropScience).
For the transient expression of GFP and BiFC fusion proteins, A. tumefaciens strain GV3101 carrying GFP or BiFC constructs was used together with the agrobacterial strain p19 (Voinnet et al., 2003) to infiltrate leaves of 4- to 6-week-old tobacco plants, according to the protocol described by Sparkes et al. (2006).
Membrane Fractionation
Arabidopsis plants used for membrane fractionation were grown in liquid half-strength MS medium (Murashige and Skoog, 1962) for 6 d or on soil for 18 d. Tissues were homogenized in buffer containing 50 mm Tris (pH 7.6 at 22°C), 20% glycerol, and 150 mm NaCl with either 5 mm MgCl2 and 2 mm EGTA (for subsequent Suc gradient separation in the presence of Mg2+) or with only 2 mm EDTA (for subsequent Suc gradient separation without Mg2+ and two-phase partitioning). Protease inhibitors (15 μm leupeptin, 77 nm aprotinin, 1 μm pepstatin A, and 0.5 mm phenylmethylsulfonyl fluoride) were added from stock solutions just before homogenization. Six-day-old seedlings were homogenized with a 3-fold amount of buffer using a pestle and mortar. Eighteen-day-old shoots were homogenized three times at low speed in a blender (Waring Laboratory & Science) with a 9-fold amount of buffer. Homogenates were filtered through four layers of cheesecloth and two layers of Miracloth (Merck) and then centrifuged for 10 min at 10,000g at 4°C. Supernatants were centrifuged again for 30 min at 100,000g at 4°C. The resulting microsome pellets were resupended in 2.5 mL of phase buffer (250 mm Suc, 5 mm potassium phosphate buffer, pH 7.8, and 2 mm dithiothreitol [DTT]) for aqueous polymer two-phase partitioning or in 10 mm Tris (pH 7.6 at 22°C), 10% Suc, and 1 mm DTT either with 2 mm EDTA or 5 mm MgCl2 for Suc gradient fractionation. The pellets were resuspended using a glass homogenizer. Microsome suspension was added to the polymer two-phase solution for aqueous two-phase partitioning.
For binding assays, two-phase partitioning was performed in a 5.8% (w/w) Dextran 500000/polyethylene glycol 3350 mixture, and for protein gel-blot analysis, it was performed in 5.9% (6-d-old seedlings) or 6.1% (shoots of 18-d-old plants) mixture. The aqueous polymer two-phase solution (5.8%, w/w) included 2.9 g of 20% (w/w) Dextran 500000, 1.45 g of 40% (w/w) polyethylene glycol 3350, 1.875 mL of 1 m Suc in 20 mm potassium phosphate buffer, pH 7.8, 250 μL of 0.7 m NaCl, and 2.5 mL of membrane suspension adjusted with water to 10 g. After mixing, the probes were centrifuged in a bucket rotor for 10 min at 1,000g at 4°C. The separation was performed three times using pure upper and lower phase solutions (without membranes) obtained in parallel by the same centrifugation step (Hodges and Mills, 1986). Then, the purified upper (PM-enriched) and lower (endomembrane-enriched) phases were 10-fold diluted by phase buffer and centrifuged for 30 min at 100,000g at 4°C. The resulting pellets were resuspended in 50 m MES-KOH, pH 7, 150 mm NaCl, 32 mm KCl, and 27 mm NH4Cl for binding assays or in 10 mm Tris (pH 7.6 at 22°C), 10% Suc, 1 mm DTT, and 2 mm EDTA for protein gel-blot analysis. The Suc gradient was obtained by layering 16 mL of 17% Suc solution onto 16 mL of 50% Suc solution in a 32-mL ultracentrifuge tube and subsequent horizontal incubation of the tube for 13 h at 4°C. Leupeptin (15 μm) was added to the gradient-forming solutions just before forming the gradient. Then, the membrane suspension was layered on the 20% to 50% (w/w) Suc gradient in 10 mm Tris (pH 7.6 at 22°C) and 1 mm DTT with either 2 mm EDTA or 5 mm MgCl2. Gradients were centrifuged in a bucket rotor for 16 h at 100,000g at 4°C. Two-milliliter fractions were collected from the bottom by peristaltic pump. The fractions were stored at −80°C before using.
Immunoblot Analysis
Anti-Myc tag monoclonal antibodies (mouse) were obtained from Millipore, and goat anti-mouse and anti-rabbit IgG peroxidase conjugates were obtained from Merck. The proteins were separated by 10% Tricine-SDS-PAGE as described (Schägger, 2006). Prior to Tricine-SDS-PAGE, the protein samples were mixed with SDS-PAGE loading buffer and incubated at 37°C for 1 h. The proteins were transferred afterward to Immobilon-P transfer membranes (Millipore). The antibodies were diluted 1:300 for the detection of Myc-tagged receptors. Secondary antibodies were diluted 1:10,000 for receptor detection and 1:30,000 for membrane marker detection. Membrane with transferred proteins was preincubated in phosphate-buffered saline (PBS) with 5% skim milk for 1 h, and the incubation with primary antibodies was performed in PBS with 5% skim milk for 2 h, then the membrane was washed three times in PBS with 0.05% Tween 20. Incubation with secondary antibodies was performed for 1 h followed by washing three times in PBS with 0.05% Tween 20. The peroxidase reaction was performed using SuperSignalWest Pico Chemiluminescent Substrate (Thermo Fisher Scientific), and the product was detected by CL-XPosure Clear Blue X-Ray Film (Thermo Fisher Scientific).
Membrane Markers
The membrane type was identified by the use of a set of polyclonal rabbit antibodies (Agrisera) against specific markers: ER luminal BiP2, PM marker H+-ATPase, tonoplast marker ε-subunit of tonoplast H+-ATPase, mitochondria marker COXII, and trans-Golgi network/Golgi apparatus marker Arf1. Antibodies were used in 1:2,000, 1:1,000, 1:2,000, 1:1,000, and 1:1,000 dilutions, respectively. Thylakoid membranes were identified by spectrophotometric analysis of the chlorophyll content (Arnon, 1949). The identification of Golgi membranes was based on the measurement of Triton X-100-stimulated latent UDPase activity (Nagahashi and Nagahashi, 1982).
Radioligand-Binding Assay
Highly labeled (592 GBq mmol−1) trans[2-3H]zeatin was obtained from the Isotope Laboratory at the Institute of Experimental Botany in Prague. Radiochemical purity was greater than 99%. For one probe, 3.8 nm [3H]trans-zeatin was used. The addition of 750 μL of membrane suspension to a tube containing 2.5 μL of labeled trans-zeatin with and without a 500-fold excess of unlabeled trans-zeatin was used for the determination of nonspecific and total binding, respectively (Romanov et al., 2005). To study the ligand specificity of binding, various unlabeled cytokinins at different concentrations were added together with radiolabeled trans-zeatin. Incubation was carried out on ice for 20 min. Then, the mixture was centrifuged for 20 min at 16,000g at 4°C. The supernatant was thoroughly removed by a vacuum pump. A total of 200 μL of 96% ethanol was added to the pellet, and the extraction was carried out for at least 16 h. Extracted radioactivity was measured by a scintillation counter in an ACS-II scintillation cocktail (Amersham Bioscience).
Confocal Microscopy
For the microscopic analyses of infiltrated tobacco plants, leaf discs were cut 4 to 5 d after infiltration. Confocal laser-scanning microscopy was performed using an inverted fluorescence microscope (Leica DMI 6000 CS) equipped with a Leica TCS SP5 laser scan unit (Leica Microsystems) and operated with the Leica Application Software. All confocal laser-scanning microscopy images were obtained using a HC PL APO 20X/0.70 IMM CORR water-immersion objective at constant imaging conditions (e.g. gain, offset, and exposure time). The GFP signal was acquired by excitation with the argon laser line at 488 nm and detection between 500 and 530 nm. YFP fluorescence was acquired by excitation at 514 nm and detection between 520 and 550 nm. The images were processed with ImageJ (http://rsbweb.nih.gov/ij/). The staining of the PM in infiltrated tobacco leaf epidermal cells was carried out by using the dye FM4-64 (Invitrogen); for this, the dye solution (50 μm) was infiltrated into transformed tobacco leaves. The infiltrated leaves were immediately scanned for fluorescence. The 488-nm laser line was used for the excitation of FM4-64 fluorescence, and emission was detected between 625 and 665 nm (Bolte et al., 2004).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Increase in specific binding of [3H]trans-zeatin by microsomes isolated from tobacco leaves transformed with the P35S:AHK3-GFP construct.
Supplemental Figure S2. Complementation of the small leaf rosette phenotype of ahk2 ahk3 mutants upon transformation with the PAHK3:AHK3-Myc or PAHK2:AHK2-Myc gene construct.
Supplemental Table S1. Primers used in this study.
References
- Argueso CT, Ferreira FJ, Kieber JJ. (2009) Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ 32: 1147–1160 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I. (2000) A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A, Mähönen AP, Helariutta Y. (2006) Signs of change: hormone receptors that regulate plant development. Development 133: 1857–1869 [DOI] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. (1998) Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Boevink P, SantaCruz S, Hawes C, Harris N, Oparka KJ. (1996) Virus-mediated delivery of the green fluorescent protein to the endoplasmic reticulum of plant cells. Plant J 10: 935–941 [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B. (2004) FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Boulaflous A, Saint-Jore-Dupas C, Herranz-Gordo MC, Pagny-Salehabadi S, Plasson C, Garidou F, Kiefer-Meyer MC, Ritzenthaler C, Faye L, Gomord V. (2009) Cytosolic N-terminal arginine-based signals together with a luminal signal target a type II membrane protein to the plant ER. BMC Plant Biol 9: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault M, Caiveau O, Pédron J, Maldiney R, Sotta B, Miginiac E. (1999) Detection of membrane-bound cytokinin-binding proteins in Arabidopsis thaliana cells. Eur J Biochem 260: 512–519 [DOI] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE. (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277: 19861–19866 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dortay H, Gruhn N, Pfeifer A, Schwerdtner M, Schmülling T, Heyl A. (2008) Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J Proteome Res 7: 3649–3660 [DOI] [PubMed] [Google Scholar]
- Dortay H, Mehnert N, Bürkle L, Schmülling T, Heyl A. (2006) Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J 273: 4631–4644 [DOI] [PubMed] [Google Scholar]
- Gardner G, Sussman MR, Kende H. (1978) In vitro cytokinin binding to a particulate cell fraction from protonemata of Funaria hygrometrica. Planta 143: 67–73 [DOI] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. (2007) Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Robatzek S. (2008) Plant receptors go endosomal: a moving view on signal transduction. Plant Physiol 147: 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Robertson FC, Soares DC, Ingram GC. (2005) ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell 17: 1154–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Ramireddy E, Brenner WG, Riefler M, Allemeersch J, Schmülling T. (2008) The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol 147: 1380–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Riefler M, Romanov GA, Schmülling T. (May 10, 2011) Properties, functions and evolution of cytokinin receptors. Eur J Cell Biol http://dx.doi.org/10.1016/j.ejcb.2011.02.009 [DOI] [PubMed] [Google Scholar]
- Heyl A, Schmülling T. (2003) Cytokinin signal perception and transduction. Curr Opin Plant Biol 6: 480–488 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. (2008) Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 59: 75–83 [DOI] [PubMed] [Google Scholar]
- Hodges TK, Mills D. (1986) Isolation of the plasma membrane. Methods Enzymol 118: 41–54 [Google Scholar]
- Hwang I, Sakakibara H. (2006) Cytokinin biosynthesis and perception. Planta 126: 528–538 [Google Scholar]
- Hwang I, Sheen J. (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Irani NG, Russinova E. (2009) Receptor endocytosis and signaling in plants. Curr Opin Plant Biol 12: 653–659 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T. (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (2003) Biosynthesis of cytokinins. J Plant Res 116: 233–239 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Zbell B, Reinert J. (1981) A high-affinity binding-site for cytokinin to a particulate fraction in carrot suspension cells. Protoplasma 106: 145–155 [Google Scholar]
- Köllmer I. (2009) Funktionelle Charakterisierung von CKX7 und cytokininregulierten Transkriptionsfaktorgenen in Arabidopsis thaliana. Doctoral thesis. Freie Universität Berlin, Berlin [Google Scholar]
- Larsson C, Widell S, Kjellbom P. (1987) Preparation of high-purity plasma membranes. Methods Enzymol 148: 558–568 [Google Scholar]
- Le Gall S, Neuhof A, Rapoport T. (2004) The endoplasmic reticulum membrane is permeable to small molecules. Mol Biol Cell 15: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Vazquez-Tello A, Whittier RF. (1995) Generation of a high-quality P1 library of Arabidopsis suitable for chromosome walking. Plant J 7: 351–358 [Google Scholar]
- Lomin SN, Yonekura-Sakakibara K, Romanov GA, Sakakibara H. (July 21, 2011) Ligand-binding properties and subcellular localization of maize cytokinin receptors. J Exp Bot http://dx.doi.org/10.1093/jxb/err220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord MJ. (1987) Isolation of endoplasmic reticulum: general principles, enzymatic markers, and endoplasmic reticulum-bound polysomes. Methods Enzymol 148: 576–584 [Google Scholar]
- Mathur J. (2007) The illuminated plant cell. Trends Plant Sci 12: 506–513 [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F. (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2: 417–422 [DOI] [PubMed] [Google Scholar]
- Moore I, Murphy A. (2009) Validating the location of fluorescent protein fusions in the endomembrane system. Plant Cell 21: 1632–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motyka V, Faiss M, Strand M, Kaminek M, Schmülling T. (1996) Changes in cytokinin content and cytokinin oxidase activity in response to derepression of ipt gene transcription in transgenic tobacco calli and plants. Plant Physiol 112: 1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J. (2007) Cytokinin signaling pathway. Sci STKE 2007: cm4. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nagahashi J, Nagahashi SL. (1982) Triton-stimulated nucleoside diphosphatase: characterization. Protoplasma 112: 174–180 [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagny S, Lerouge P, Faye L, Gomord V. (1999) Signals and mechanisms for protein retention in the endoplasmic reticulum. J Exp Bot 50: 157–164 [Google Scholar]
- Punwani JA, Hutchison CE, Schaller GE, Kieber JJ. (2010) The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J 62: 473–482 [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov GA, Lomin SN, Schmülling T. (2006) Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J Exp Bot 57: 4051–4058 [DOI] [PubMed] [Google Scholar]
- Romanov GA, Spíchal L, Lomin SN, Strnad M, Schmülling T. (2005) A live cell hormone-binding assay on transgenic bacteria expressing a eukaryotic receptor protein. Anal Biochem 347: 129–134 [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Schägger H. (2006) Tricine-SDS-PAGE. Nat Protoc 1: 16–22 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB. (1995) The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem 270: 12526–12530 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Spíchal L, Rakova NY, Riefler M, Mizuno T, Romanov GA, Strnad M, Schmülling T. (2004) Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol 45: 1299–1305 [DOI] [PubMed] [Google Scholar]
- Stolz A, Riefler M, Lomin SN, Achazi K, Romanov GA, Schmülling T. (2011) The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J 67: 157–168 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T. (2001a) The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Sakurai K, Ueguchi C, Mizuno T. (2001b) Two types of putative nuclear factors that physically interact with histidine-containing phosphotransfer (Hpt) domains, signaling mediators in His-to-Asp phosphorelay, in Arabidopsis thaliana. Plant Cell Physiol 42: 37–45 [DOI] [PubMed] [Google Scholar]
- Tian H, Klämbt D, Jones AM. (1995) Auxin-binding protein 1 does not bind auxin within the endoplasmic reticulum despite this being the predominant subcellular location for this hormone receptor. J Biol Chem 270: 26962–26969 [DOI] [PubMed] [Google Scholar]
- To JP, Kieber JJ. (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Sato S, Kato T, Tabata S. (2001) The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol 42: 751–755 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T. (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Schmülling T. (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Wulfetange K. (2010) Strukturelle und funktionelle Characterisierung des Cytokininrezeptors AHK4/CRE1 aus Arabidopsis thaliana. Doctoral thesis. Freie Universität Berlin, Berlin [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T. (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42: 1017–1023 [DOI] [PubMed] [Google Scholar]