Abstract
Vascular wilts caused by soil-borne fungal species of the Verticillium genus are devastating plant diseases. The most common species, Verticillium dahliae and Verticillium albo-atrum, have broad host ranges and are notoriously difficult to control. Therefore, genetic resistance is the preferred method for disease control. Only from tomato (Solanum lycopersicum) has a Verticillium resistance locus been cloned, comprising the Ve1 gene that encodes a receptor-like protein-type cell surface receptor. Due to lack of a suitable model for receptor-like protein (RLP)-mediated resistance signaling in Arabidopsis (Arabidopsis thaliana), so far relatively little is known about RLP signaling in pathogen resistance. Here, we show that Ve1 remains fully functional after interfamily transfer to Arabidopsis and that Ve1-transgenic Arabidopsis is resistant to race 1 but not to race 2 strains of V. dahliae and V. albo-atrum, nor to the Brassicaceae-specific pathogen Verticillium longisporum. Furthermore, we show that signaling components utilized by Ve1 in Arabidopsis to establish Verticillium resistance overlap with those required in tomato and include SERK3/BAK1, EDS1, and NDR1, which strongly suggests that critical components for resistance signaling are conserved. We subsequently investigated the requirement of SERK family members for Ve1 resistance in Arabidopsis, revealing that SERK1 is required in addition to SERK3/BAK1. Using virus-induced gene silencing, the requirement of SERK1 for Ve1-mediated resistance was confirmed in tomato. Moreover, we show the requirement of SERK1 for resistance against the foliar fungal pathogen Cladosporium fulvum mediated by the RLP Cf-4. Our results demonstrate that Arabidopsis can be used as model to unravel the genetics of Ve1-mediated resistance.
Verticillium wilts caused by soil-borne fungal species of the Verticillium genus, of which Verticillium dahliae and Verticillium albo-atrum are the most common, are devastating vascular plant diseases that occur on a wide host range of over 200 dicotyledonous plant species in temperate and subtropical regions (Fradin and Thomma, 2006; Klosterman et al., 2009). Verticillium wilt fungi are notoriously difficult to combat due to extremely persistent resting structures that reside in the soil and that are difficult to eradicate since the only effective control measure, soil fumigation, is expensive and has harmful environmental effects. Furthermore, the broad host ranges of Verticillium spp. make crop rotation ineffective, and fungicides to cure infected plants are not available (Wilhelm, 1955; Rowe et al., 1987; Fradin and Thomma, 2006).
Presently, genetic resistance is the preferred method to control Verticillium wilt diseases, and Verticillium resistance has been described in several plant species (Schaible et al., 1951; Putt, 1964; Huang, 2003; Simko et al., 2004b; Bolek et al., 2005; Zebrowska et al., 2006). However, only from tomato (Solanum lycopersicum) has a Verticillium resistance locus been cloned (Kawchuk et al., 2001; Fradin et al., 2009). This Ve locus mediates resistance against race 1 strains of V. dahliae and V. albo-atrum, and strains that are not contained by this locus are assigned to race 2 (Schaible et al., 1951; Fradin et al., 2009). The Ve locus comprises two genes, Ve1 and Ve2, and although it was initially reported that both Ve1 and Ve2 confer resistance when expressed in the close relative potato (Solanum tuberosum; Kawchuk et al., 2001), only Ve1 provides resistance in tomato (Fradin et al., 2009).
Both Ve1 and Ve2 encode cell surface receptors that belong to the extracellular leucine-rich repeat class of receptor-like proteins (LRR-RLPs), cell surface receptors with extracellular LRRs that lack a cytoplasmic signaling domain (Kawchuk et al., 2001; Wang et al., 2010a). This class of resistance (R) proteins was identified originally in tomato as Cf resistance proteins that provide resistance against the leaf mold pathogen Cladosporium fulvum (Jones et al., 1994; Thomma et al., 2005). Furthermore, this class of R proteins includes the apple HcrVf proteins that confer resistance to the scab fungus Venturia inaequalis (Vinatzer et al., 2001; Belfanti et al., 2004). In addition to race-specific R proteins, the RLP family harbors receptors that act in basal defense, including the tomato LeEIX receptors for the ethylene-inducible xylanase produced by Trichoderma biocontrol fungi (Ron and Avni, 2004) and Arabidopsis (Arabidopsis thaliana) AtRLP52 and AtRLP30 that play roles in basal defense against the powdery mildew fungus Erysiphe cichoracearum and the bacterium Pseudomonas syringae pv phaseolicola, respectively (Ramonell et al., 2005; Wang et al., 2008). Finally, AtRLP51 was found to regulate defense against the downy mildew pathogen Hyaloperonospora arabidopsidis and P. syringae pv tomato (Zhang et al., 2010). Apart from defense against pathogens, RLPs also play significant roles in plant development (Wang et al., 2008, 2010a, 2010b).
The interaction between C. fulvum and tomato has been the most exploited model to study the genetics of RLP-mediated resistance (Thomma et al., 2005; Wulff et al., 2009), and several components required for the Cf-mediated hypersensitive response or resistance against C. fulvum were identified, including the thioredoxin CITRX, the protein kinase ACIK1, the nucleotide binding (NB)-LRR protein NRC1, the U-box protein CMPG1, the mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3, the F-box protein ACRE189/ACIF1, and members of the phospholipase C family (Rivas et al., 2004; Rowland et al., 2005; González-Lamothe et al., 2006; Gabriëls et al., 2007; Stulemeijer et al., 2007; van den Burg et al., 2008; Vossen et al., 2010). Based on their involvement in Cf signaling, a number of candidate genes were tested for a role in Ve1 signaling, revealing that Ve1- and Cf-mediated resistance signaling only partially overlap (Fradin et al., 2009; Vossen et al., 2010). Intriguingly, in addition to NRC1, ACIF1, MEK2, and SERK3/BAK1, both EDS1 and NDR1 were found to be required for Ve1 signaling (Fradin et al., 2009).
Despite the knowledge obtained from tomato, relatively little is known about signaling mediated by RLP-type pathogen receptors (Wang et al., 2010a). This can partially be explained by the lack of a suitable model for RLP-mediated resistance signaling in Arabidopsis, the first plant species for which a genome sequence was released and large mutant collections are available covering nearly every gene in the genome. In the Arabidopsis genome, 57 putative RLP genes (AtRLPs) were identified, most of which encode orphan proteins, and attempts to assign biological functions to these genes have met with little success (Ellendorff et al., 2008; Wang et al., 2008). Since Arabidopsis is a host for Verticillium infection, we attempted to develop a model for RLP-mediated resistance signaling in Arabidopsis by transfer of the tomato gene encoding Ve1. We show that Ve1 remains fully functional after transfer to Arabidopsis and that Ve1-transgenic Arabidopsis is resistant to race 1, but not race 2, strains of Verticillium. Furthermore, we show that the signaling components utilized by Ve1 in Arabidopsis to establish Verticillium resistance overlap with those that are exploited in tomato (Fradin et al., 2009). We show that the blueprint for resistance signaling is conserved between tomato and Arabidopsis and, thus, that Arabidopsis can be used as a model to unravel the genetics of resistance signaling mediated by the RLP Ve1.
RESULTS
Verticillium Is a Vascular Pathogen of Arabidopsis
Arabidopsis is a widely used model for genetic characterization of disease signaling (Thomma et al., 2001; Nishimura and Dangl, 2010). Although several studies have used Arabidopsis as a host for V. dahliae (Veronese et al., 2003; Tjamos et al., 2005; Ellendorff et al., 2009), vascular colonization of Arabidopsis plants has not yet been demonstrated. Therefore, vascular colonization of Arabidopsis roots of the Columbia-0 (Col-0) ecotype by a GFP-transgenic V. dahliae strain was studied (Supplemental Materials and Methods S1). Clear GFP signals were observed within xylem vessels (Supplemental Fig. S1), demonstrating that V. dahliae is a vascular pathogen of Arabidopsis. This observation was confirmed by plating of stem sections of inoculated and noninoculated plants, showing fungal outgrowth only from stem sections of inoculated plants (Supplemental Fig. S1). This eliminates the possibility that Verticillium disease symptoms on Arabidopsis are inflicted by pathogen toxins or effectors that are taken up by the plant in the absence of pathogen colonization.
Comparison of Ve1 with AtRLPs
We queried the Arabidopsis genome for the presence of putative Ve1 orthologs. Previously, 57 AtRLPs were identified (Wang et al., 2008), of which full-length protein sequences were compared with that of Ve1. Furthermore, the sequences of Ve2, four putative Ve orthologs from Solanum torvum (StVe), Solanum lycopersicoides (SlVe1), Mentha longifolia (MlVe1), and Mentha spicata (MsVe1), and tomato Cf-4, Cf-9, LeEIX1, and -2 were added. Phylogenetic analysis demonstrated that Ve1 clusters in a separate clade with Ve homologs from Solanaceae as well as non-Solanaceae species (Supplemental Fig. S2). Remarkably, none of the 57 AtRLPs clustered with Ve1. Moreover, pairwise amino acid sequence comparison with Ve1 revealed low overall sequence identity between Ve1 and the AtRLPs, with a maximum of only 28% identity between Ve1 and At2g15080 (Supplemental Fig. S2).
Tomato Ve1 Is Functional against V. dahliae and V. albo-atrum, But Not against V. longisporum in Arabidopsis
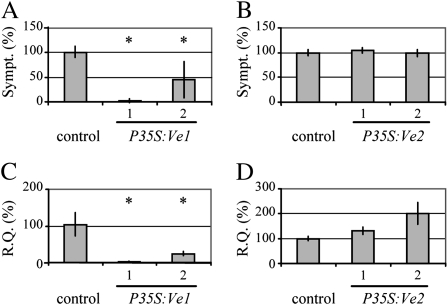
In an attempt to develop a model for RLP-mediated resistance signaling, wild-type Arabidopsis plants of the ecotypes Col-0 and Wassilewskija-0 (Ws-0) were transformed with the tomato Ve1 coding sequence (CDS) driven by the constitutive cauliflower mosaic virus (CaMV) 35S promoter (P35S:Ve1; Fradin et al., 2009; Supplemental Figs. S3 and S4). As a control, the 35S-driven tomato Ve2 CDS (P35S:Ve2) was used (Fradin et al., 2009). In no case were developmental alterations observed (Fig. 1); subsequently, two transgenic lines were assayed for Verticillium resistance. Interestingly, transgenic plants expressing Ve1 were clearly resistant to V. dahliae and V. albo-atrum race 1 strains, and only few, if any, symptoms were observed (Figs. 1A and 2A). In contrast, Ve2 transgenic plants were as diseased as nontransgenic plants upon challenge with these race 1 strains and displayed typical Verticillium symptoms, including stunting, wilting, anthocyanin accumulation, chlorosis, early senescence, and necrosis (Figs. 1A and 2B). As expected, Ve1 and Ve2 transgenic lines were fully susceptible when challenged with V. dahliae and V. albo-atrum race 2 strains (Fig. 1A; Supplemental Fig. S5, A and B). The phenotypes correlated with the degree of Verticillium colonization, as determined by real-time PCR (Fig. 2, C and D; Supplemental Fig. S5, C and D). In conclusion, the functionality of Ve1 and Ve2 in tomato (Fradin et al., 2009) is fully maintained upon expression in Arabidopsis, as only Ve1 mediates resistance against race 1, but not against race 2 strains of V. dahliae and V. albo-atrum.
Figure 1.
Transgenic expression of Ve1, but not of Ve2, mediates Verticillium resistance in Arabidopsis. Arabidopsis engineered to express tomato CaMV 35S-driven Ve1 or Ve2 (P35S:Ve1 and P35S:Ve2). A, Typical appearance of nontransgenic control and transgenic lines upon mock inoculation or inoculation with race 1 or race 2 strains of V. dahliae and V. albo-atrum (V. a-a) at 21 d after inoculation. B, Typical appearance of nontransgenic Ws-0 control and P35S:Ve1 at 21 d after inoculation with four V. longisporum strains (1–4). [See online article for color version of this figure.]
Figure 2.
Transgenic expression of Ve1, but not of Ve2, reduces Verticillium wilt symptoms and fungal biomass upon inoculation with V. dahliae race 1. Quantification of Verticillium wilt symptoms (Sympt.) in Arabidopsis Col-0 engineered to express CaMV 35S-driven tomato Ve1 (A) or Ve2 (B) at 21 d after inoculation. Bars represent quantification of symptom development as percentage of diseased rosette leaves with sd. Col-0 (control) is set to 100%. Fungal biomass determined by quantitative real-time PCR (R.Q.) in Arabidopsis Col-0 engineered to express CaMV 35S-driven Ve1 (C) or Ve2 (D). Bars represent Verticillium ITS transcript levels relative to Arabidopsis Rubisco transcript levels (for equilibration) with sd in a sample of four pooled plants. Col-0 (control) is set to 100%. A to D, Two transgenic lines per construct are shown (1 and 2). Asterisks indicate significant differences when compared with Col-0 (P < 0.05).
While V. dahliae and V. albo-atrum are the most prevalent pathogenic Verticillium species that occur on broad host ranges, V. longisporum particularly infects Brassicaceous hosts (Koike et al., 1994; Karapapa et al., 1997; Barbara and Clewes, 2003). Ve1-transgenic plants showed typical Verticillium wilt symptoms and were as diseased as wild-type and Ve2-transgenic plants when challenged with four different V. longisporum strains (Fig. 1B). This suggests that Ve1 does not control V. longisporum.
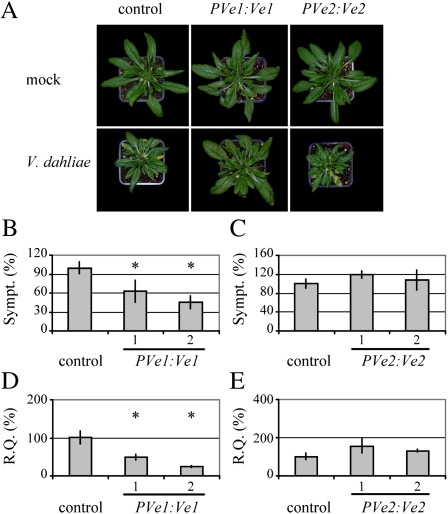
Functional Ve1 Driven by the Tomato Native Promoter
The functionality of the Ve1 gene in Arabidopsis was further investigated upon expression driven by the tomato native promoter (PVe1:Ve1; Fradin et al., 2009; Supplemental Fig. S3). To this end, Col-0 plants were transformed with PVe1:Ve1, while a similar construct for Ve2 (PVe2:Ve2) was used as a control (Supplemental Fig. S3; Fradin et al., 2009). For each construct, two transgenic lines were challenged with race 1 V. dahliae. Plants transgenic for PVe1:Ve1 showed significantly less Verticillium wilt symptoms and fungal biomass accumulation when compared with PVe2:Ve2-transgenic plants and nontransgenic control plants (Fig. 3), showing that the tomato Ve1 promoter is functional in Arabidopsis. However, the resistance in PVe1:Ve1-transgenic lines was not as robust as in P35S:Ve1-transgenic lines as the PVe1:Ve1-transgenic lines displayed more symptoms and accumulated more fungal biomass than the P35S:Ve1-transgenic lines (compare Figs. 2 and 3).
Figure 3.
Expression of Ve1, but not of Ve2, driven by their respective tomato native promoters reduces Verticillium wilt disease in Arabidopsis. Arabidopsis Col-0 engineered to express tomato Ve1 or Ve2 driven by their respective native promoters (PVe1:Ve1 and PVe2:Ve2, respectively). A, Typical appearance of nontransgenic Col-0 (control) and transgenic lines upon mock inoculation or inoculation with V. dahliae race 1 at 21 d after inoculation. Quantification of Verticillium wilt symptoms (Sympt.) in Arabidopsis Col-0 engineered to express tomato Ve1 (B) or Ve2 (C). Bars represent quantification of symptom development shown as percentage of diseased rosette leaves with sd. Col-0 (control) is set to 100%. Fungal biomass determined by quantitative real-time PCR (R.Q.) in Arabidopsis Col-0 engineered to express tomato Ve1 (D) or Ve2 (E). Bars represent Verticillium ITS transcript levels relative to Arabidopsis Rubisco transcript levels (for equilibration) with sd in a sample of four pooled plants. Col-0 (control) is set to 100%. B to E, Two transgenic lines are shown per construct (1 and 2). Asterisks indicate significant differences when compared with Col-0 (P < 0.05). [See online article for color version of this figure.]
Genetic Requirements for Ve1 Signaling in Arabidopsis
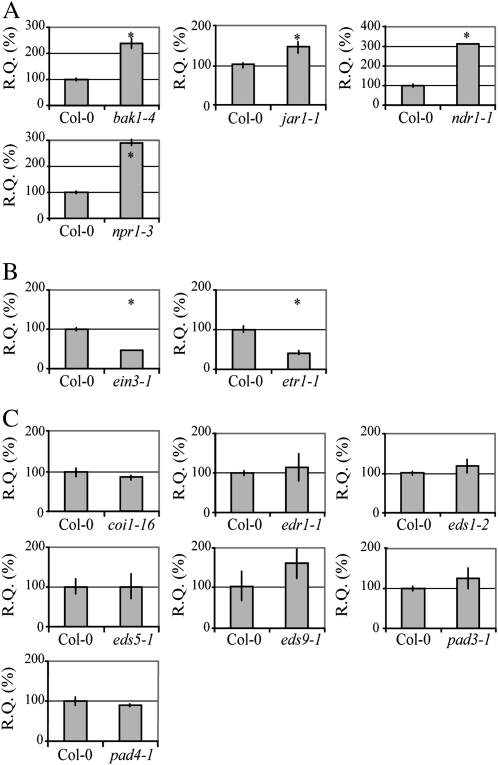
The use of Arabidopsis allows exploiting widely available mutant collections. To allow determination of the role of known resistance signaling components in Ve1 signaling, first the role of these components in basal defense against Verticillium was evaluated. The genotypes that were used included mutants deficient in salicylic acid (SA) signaling (eds1-2, eds5-1, npr1-3, and pad4-1), ethylene (ET) signaling (ein3-1 and etr1-1), jasmonic acid (JA) signaling (coi1-16 and jar1-1), phytoalexin biosynthesis (pad3-1 and pad4-1), and pathogen receptor signaling (bak1-4, eds1-2, and ndr1-1; Supplemental Table S1). All genotypes were challenged with race 1 V. dahliae and colonization was determined. When compared with Col-0, no difference in susceptibility was observed for the coi1-16, eds1-2, eds5-1, eds9-1, edr1-1, pad3-1, and pad4-1 mutants (Fig. 4; Supplemental Fig. S6). However, whereas the mutants bak1-4, jar1-1, ndr1-1, and npr1-3 showed enhanced V. dahliae susceptibility (Fig. 4; Supplemental Fig. S6), the ET mutants etr1-1 and ein3-1 showed enhanced resistance (Fig. 4; Supplemental Fig. S6).
Figure 4.
Quantification of V. dahliae biomass in Arabidopsis defense signaling mutants. Fungal biomass was determined by quantitative real-time PCR (R.Q.) in Col-0 and defense signaling mutants at 21 d after inoculation. Mutants that show enhanced (A) or reduced (B) susceptibility towards V. dahliae. C, Mutants for which fungal biomass is comparable to Col-0. A to C, Bars represent Verticillium ITS transcript levels relative to Arabidopsis Rubisco transcript levels (for equilibration) with sd in a sample of four pooled plants. Col-0 is set to 100%. Asterisks indicate significant differences when compared with Col-0 (P < 0.05).
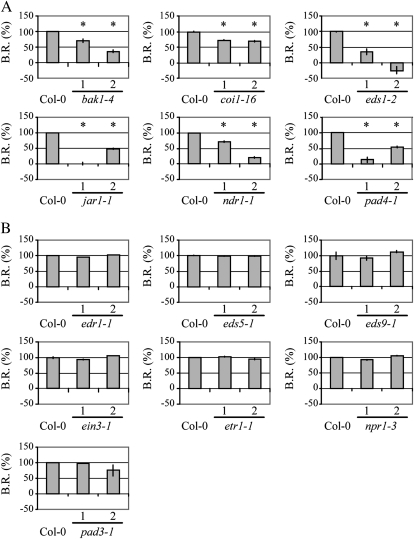
To evaluate the contribution of the various signaling components to Ve1-mediated signaling in Arabidopsis, all mutants were transformed with the P35S:Ve1 construct (Supplemental Fig. S7). For each mutant, two independent Ve1-transgenic lines were challenged with race 1 V. dahliae and evaluated for Ve1-mediated disease resistance (Supplemental Fig. S6). To confirm the observed phenotypes, the fungal biomass was determined by real-time PCR in each transgenic line and normalized to the biomass in the respective nontransgenic progenitors to determine the Ve1-mediated biomass reduction. This reduction was then compared to the biomass reduction determined in Ve1-transgenic Col-0 when compared with nontransgenic Col-0. This analysis showed that Ve1-mediated resistance was not compromised in edr1-1, eds5-1, eds9-1, ein3-1, etr1-1, npr1-3, and pad3-1 mutants as the Ve1-mediated fungal biomass reduction in these mutants was comparable to the reduction in Col-0 (Fig. 5; Supplemental Fig. S6). In contrast, Ve1-mediated resistance was compromised in the bak1-4, coi1-16, eds1-2, jar1-1, ndr1-1, and pad4-1 mutants as the Ve1-transgenic lines showed significantly less fungal biomass reduction when compared to the Ve1-mediated biomass reduction in Col-0 (Fig. 5; Supplemental Fig. S6).
Figure 5.
Ve1-mediated reduction of V. dahliae biomass in defense signaling mutants. Mutants for which Ve1-mediated resistance is compromised (A) or not compromised (B). Fungal biomass was determined by quantitative real-time PCR and represents Verticillium ITS transcript levels relative to Arabidopsis Rubisco transcript levels (for equilibration). Bars represent the percentage of Ve1-mediated fungal biomass reduction (B.R.) in Ve1-expressing lines when compared to the fungal biomass accumulated in the respective nontransformed progenitors, with sd in a sample of four pooled plants. Ve1-mediated fungal biomass reduction in Col-0 is set to 100%. Two independent transgenic lines expressing Ve1 are shown per construct (1 and 2). Asterisks indicate significant differences when compared with Col-0 (P < 0.05).
SERK1 Is Required for Ve1 Signaling in Arabidopsis and Tomato
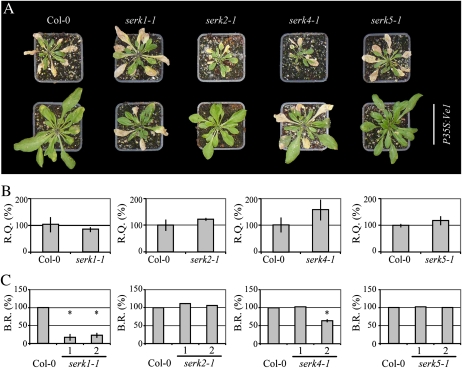
The somatic embryogenesis receptor-like protein kinase (SERK) was identified in carrot (Daucus carota) as a marker for the transition from somatic to embryogenic cells in carrot cell culture (Schmidt et al., 1997). In Arabidopsis, five SERK homologs have been identified with both significantly overlapping and distinct functions (Hecht et al., 2001; Albrecht et al., 2008). Of these five, SERK3 (also known as brassinosteroid insensitive 1 associated receptor kinase 1 [BAK1]) and SERK4 have previously been implicated in pathogen immunity and cell death signaling (Chinchilla et al., 2007; He et al., 2007; Heese et al., 2007; Kemmerling et al., 2007). To investigate whether other SERK family members in addition to SERK3/BAK1 are required for Ve1-mediated resistance, we transformed mutants of Serk1, Serk2, Serk4, and Serk5 with the P35S:Ve1 construct (Supplemental Fig. S8; Supplemental Table S1). For each mutant, two independent Ve1-transgenic lines were challenged with race 1 V. dahliae, and resistance was evaluated together with the nontransgenic mutants (Fig. 6). To confirm the observed phenotypes, the fungal biomass was determined by real-time PCR in each transgenic line and normalized to the biomass in the respective nontransgenic progenitors to determine the Ve1-mediated biomass reduction. This analysis showed that none of the Serk mutants was compromised in basal defense against Verticillium as the nontransgenic progenitors showed similar levels of susceptibility as Col-0 plants. Furthermore, Ve1-mediated resistance was not compromised in serk2-1 and serk5-1 mutants as the Ve1-mediated fungal biomass reduction in these mutants was comparable to the reduction in Col-0 (Fig. 6). In contrast, Ve1-mediated resistance was compromised in the serk1-1 mutant and, albeit to a lesser extent and not consistently, possibly also in the serk4-1 mutant (Fig. 6).
Figure 6.
Overview of Ve1-transgenic Serk mutants challenged with V. dahliae race 1. A, Typical appearance of nontransgenic (top row) and Ve1-transgenic (bottom row; P35S:Ve1) Col-0 and Serk mutant plants at 21 d after Verticillium inoculation. B, Quantification of V. dahliae biomass in nontransgenic Serk mutants when compared with Col-0. Bars represent Verticillium quantification (R.Q.) with sd in a sample of four pooled plants. Col-0 is set to 100%. C, Ve1-mediated reduction of V. dahliae biomass in Serk mutants when compared with Col-0. Bars represent the percentage of Ve1-mediated fungal biomass reduction (B.R.) in Ve1-expressing lines when compared to the fungal biomass accumulated in the respective nontransformed progenitors, with sd in a sample of four pooled plants. B and C, Fungal biomass was determined by quantitative real-time PCR and represents Verticillium ITS transcript levels relative to Arabidopsis Rubisco transcript levels (for equilibration). Ve1-mediated fungal biomass reduction in Col-0 is set to 100%. Two independent transgenic lines expressing Ve1 are shown per construct (1 and 2). Asterisks indicate significant differences when compared with Col-0 (P < 0.05). [See online article for color version of this figure.]
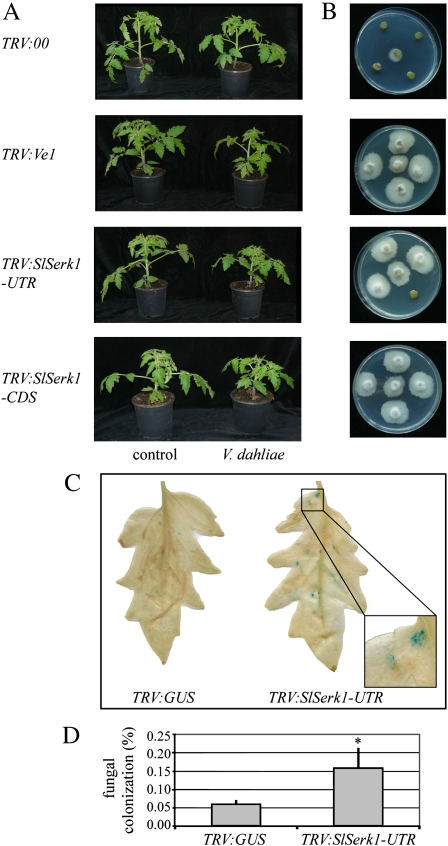
Based on the finding that SERK1 is required for Ve1 signaling in Arabidopsis, we assessed the role of Ve1 signaling in tomato. To this end, the tomato Serk1 (SlSerk1) homolog was identified by BLAST analysis using Arabidopsis SERK1 to query the tomato genome sequence (http://mips.helmholtz-muenchen.de/plant/tomato/index.jsp). One clear Serk1 homolog was identified (SGN-E623106), and its expression was targeted with virus-induced gene silencing (VIGS; Fradin et al., 2009). To this end, two recombinant tobacco rattle virus (TRV) vectors were designed: one based on the SlSerk1 CDS and one based on the 3′-untranslated region (3′-UTR). As controls, an empty TRV construct (TRV:00) and a construct targeting Ve1 expression were used (Fradin et al., 2009). Subsequently, the recombinant TRV vectors were inoculated onto tomato, and 2 weeks later, half of the plants were inoculated with a race 1 V. dahliae strain, while the other half were mock inoculated. Two weeks after inoculation, Verticillium resistance was assessed by evaluating the degree of stunting (height of the plant and length of the leaves) as an indicator of disease progression. Upon Verticillium inoculation of TRV:00-treated plants, little stunting was observed when compared with mock-inoculated plants, while Verticillium inoculation of TRV:Ve1-treated plants showed clear and consistent stunting (Fig. 7; Supplemental Table S2). Interestingly, targeting of SlSerk1 expression also resulted in compromised Verticillium resistance, irrespective of whether the CDS or the 3′-UTR was targeted, demonstrating that Serk1 is required for Ve1-mediated Verticillium resistance in tomato.
Figure 7.
VIGS of SlSerk1 impairs Ve1-mediated Verticillium resistance and Cf-4-mediated Cladosporium resistance in tomato. A, Motelle (Ve1/Ve1; resistant) plants were treated with an empty recombinant TRV vector (TRV:00), a TRV vector targeting Ve1 (TRV:Ve1), the 3′-UTR of SlSerk1 (TRV:SlSerk1-UTR), or the CDS of SlSerk1 (TRV:SlSerk1-CDS). Two weeks after treatment, the plants were mock inoculated (control) or inoculated with a race 1 strain of V. dahliae. Photographs were taken 14 d after V. dahliae inoculation, and compromised resistance is shown by a stunted appearance of the V. dahliae-inoculated plants when compared with mock-inoculated control plants. B, Two weeks after V. dahliae inoculation, stem sections were plated, allowing fungal outgrowth as a measure for fungal colonization. Photographs were taken at 14 d after plating. C, Cf4 tomato plants were treated with a TRV vector targeting GUS (TRV:GUS) as a control or a TRV vector targeting the 3′-UTR of SlSerk1 (TRV:SlSerk1-UTR) and challenged with transgenic C. fulvum expressing GUS. Representative leaflets after destaining are shown, revealing that full C. fulvum resistance is compromised when Serk1 is targeted. D, Quantitation of fungal growth in TRV:GUS and TRV:SlSerk1-UTR treated plants. Bars represent the degree of fungal colonization, expressed as the ratio between blue and total leaf area, in leaves from four independent experiments with se. The asterisk indicates a statistically significant difference (P < 0.05). [See online article for color version of this figure.]
We previously demonstrated that Ve1 signaling only partially overlaps with signaling mediated by Cf proteins that similarly belong to the RLP class of resistance proteins (Fradin et al., 2009; Vossen et al., 2010). To investigate the role of SlSERK1 in Cf signaling, we evaluated the resistance of Cf-4 tomato plants upon silencing of SlSerk1 using the construct that targets the UTR and inoculation with a C. fulvum strain expressing Avr4 as well as the constitutively expressed transgenic marker GUS (Vossen et al., 2010). Although no obvious macroscopic disease symptoms were observed, GUS staining clearly indicated colonization of intercellular spaces in the TRV:Serk1-inoculated plants and not in the control plants (Fig. 7). These histological data strongly suggest that SlSERK1 is required for full Cf-4-mediated resistance.
DISCUSSION
A Model for Ve1 Signaling in Arabidopsis
Resistance to race 1 Verticillium strains in tomato is conferred by the single dominant Ve locus that was introduced in cultivated varieties in the 1950s (Schaible et al., 1951) and that is still carried by most commercial tomato varieties. It was reported that both genes that reside in this locus, Ve1 and Ve2, confer resistance against the same race 1 strain when expressed in potato (Kawchuk et al., 2001). However, we subsequently failed to demonstrate a role for Ve2 in tomato resistance against various race 1 Verticillium strains (Fradin et al., 2009). In this study, Ve1-transgenic Arabidopsis plants of the Col-0 and Ws-0 ecotypes were found to be resistant to race 1 strains of both V. dahliae and V. albo-atrum since the plants displayed little to no wilt symptoms and accumulated significantly decreased amounts of fungal biomass when compared with their nontransgenic progenitors. The resistance could not be attributed to generally enhanced pathogen resistance, as Ve1-transgenic plants were as susceptible as the control plants towards race 2 strains and four different strains of V. longisporum. Furthermore, again we could not confirm a role for Ve2 in Verticillium resistance, as all Ve2 transgenic lines showed unaltered susceptibility towards V. dahliae and V. albo-atrum.
The interaction between C. fulvum and tomato is the most exploited model to unravel the genetics of RLP-mediated resistance (Thomma et al., 2005; Wulff et al., 2009). In addition, a number of studies have addressed the genetics of Ve1 signaling in tomato (Hu et al., 2005; Fradin et al., 2009; Vossen et al., 2010). These studies have demonstrated that Ve1-mediated resistance signaling only partially overlaps with signaling mediated by Cf proteins. Furthermore, a role for both Eds1 and Ndr1 in Ve1 signaling was shown (Hu et al., 2005; Fradin et al., 2009). This is remarkable since differential requirement of Eds1 and Ndr1 was shown for cytoplasmic NB-LRR disease resistance proteins in Arabidopsis, as EDS1 generally mediates signaling initiated by Toll and interleukin 1 receptor-NB-LRRs, whereas NDR1 mediates signaling initiated by coiled coil-NB-LRRs (Century et al., 1995; Aarts et al., 1998). Interestingly, we now show that Ve1 requires both Eds1 and Ndr1 also in Arabidopsis, thus suggesting that the signaling cascade exploited by Ve1 in Arabidopsis is homologous to the native signaling cascade in tomato. This is further supported by the observation that, similar to tomato (Fradin et al., 2009), Ve1 signaling in Arabidopsis requires Bak1 and does not require Npr1.
Basal defense towards V. dahliae was compromised in the mutants bak1-4, jar1-1, ndr1-1, and npr1-3, whereas the ET mutants etr1-1 and ein3-1 showed enhanced resistance. A role for BAK1 in basal defense was previously demonstrated in Nicotiana benthamiana and Arabidopsis (Heese et al., 2007; Kemmerling et al., 2007). The finding that Jar1 and Npr1 play a role in basal defense cannot be translated into a general requirement of SA and JA for Verticillium defense, as other SA and JA signaling components, such as Eds1, Pad4, and Coi1, were not required for Verticillium defense. Interestingly, Ve1-mediated resistance required the basal defense signaling components Bak1, Jar1, and Ndr1, as well as the components Coi1, Eds1, and Pad4, which are not required for basal defense against Verticillium. These findings suggest that JA signaling is required for Ve1-mediated resistance, while SA and ET signaling are not required. We speculated that studies to dissect Ve1 signaling in Arabidopsis, based on candidate genes and on random mutagenesis, would identify genes that would similarly play a role in Verticillium resistance in tomato. To test this hypothesis, we investigated the role of the five Arabidopsis AtSERK genes in Ve1-mediated resistance. Over the years, it has become evident that AtSERK family members differentially function in signaling pathways with roles that range from development to defense. While AtSERK1 and AtSERK2 play roles in anther development and male gametophyte maturation (Albrecht et al., 2005; Colcombet et al., 2005), AtSERK1 and AtSERK3/BAK1 act in receptor complexes for brassinosteroid perception (Li et al., 2002; Nam and Li, 2002; Karlova et al., 2006), and AtSERK3 and AtSERK4 have been implicated in programmed cell death responses in development and defense (He et al., 2007; Kemmerling et al., 2007). Here, we show that AtSERK1, AtSERK3, and, to a lesser extent, AtSERK4 are required for full Ve1-mediated resistance in Arabidopsis. We have previously shown that SlSERK3/BAK1 is required for Ve1-mediated resistance in tomato (Fradin et al., 2009), and with VIGS, we now confirm that also SlSERK1 is required for resistance in tomato. Together with the recent finding that SlSERK1 is required for aphid resistance mediated by the NB-LRR Mi-1 (Mantelin et al., 2011), our results suggest that SERK1 contributes to host defense mediated by extracellular and cytoplasmic immune receptors and extend the notion that SERK proteins are versatile regulators of various physiological processes in plants. Furthermore, our data demonstrate that Ve1-transgenic Arabidopsis can be used as a tool to identify critical signaling components for Ve1 signaling in tomato.
Interfamily Transfer of Verticillium Resistance
The transfer of race-specific R genes across species boundaries has been mostly successful between phylogenetically related donor and recipient species, while interfamily transfer has generally met little success (Stuiver and Custers, 2001; Hammond-Kosack and Parker, 2003; Gurr and Rushton, 2005; Gust et al., 2010; Wulff et al., 2011). It can be speculated that interfamily transfer of receptors (generally known as pattern recognition receptors [PRRs]) for conserved microbial structures (generally known as pathogen-associated molecular patterns [PAMPs]) could be more successful, as exemplified by the transfer of the Arabidopsis PRRs EFR for bacterial EF-Tu and FLS2 for bacterial flagellin from Arabidopsis to N. benthamiana and tomato (Gómez-Gómez and Boller, 2000; Chinchilla et al., 2006; Zipfel et al., 2006; Lacombe et al., 2010). As PRRs are considered to be more ancient than race-specific R proteins, the blueprint of their signaling cascade to establish resistance may be conserved across species and even families, while for R proteins that evolved after speciation, essential signaling components may be lacking. Although Ve1 is considered to encode a race-specific R protein (Schaible et al., 1951; Kawchuk et al., 2001), several observations support the hypothesis that Ve1 is an ancient pathogen receptor with traits of typical PRRs. First, Ve1-mediated race 1 resistance affects two distinct fungal species, V. dahliae and V. albo-atrum, suggesting that the yet unidentified elicitor is conserved between species. Second, putative Ve orthologs have been identified within (Chai et al., 2003; Fei et al., 2004; Simko et al., 2004a) and outside the Solanaceae family (Vining et al., 2007; Vining and Davis, 2009). Third, the receptor-like kinase BAK1/SERK3 that is crucial for various PAMP-triggered responses (Chinchilla et al., 2007; Heese et al., 2007; Kemmerling et al., 2007) is required for Ve1-mediated resistance of tomato (Fradin et al., 2009) and Arabidopsis. Fourth, typical for PRR-mediated resistance, which is generally considered as a weaker variant of R protein-mediated resistance (Tsuda and Katagiri, 2010), Ve1-mediated Verticillium resistance is rather weak and allows low-level proliferation of race 1 Verticillium strains even in resistant plants (Gold and Robb, 1995; Chen et al., 2004; Fradin et al., 2009). All these observations argue against a role for Ve1 as race-specific R protein and for a role as PRR. Furthermore, it may be argued that the yet unidentified Verticillium activator of Ve1 resistance is a PAMP, as this activator is likely to be conserved across species, reminiscent of Ax21 that is conserved across Xanthomonas spp. (Lee et al., 2009). Finally, the functionality of Ve1 after transfer across plant families suggests that it directly recognizes a pathogen component rather than that it guards a host target. This may explain why efforts to identify the Ve1 elicitor, which focused on typical characteristics of effector molecules, have failed thus far.
The current paradigm states that the first line of active plant defense in plant immunity is formed by PRRs that recognize PAMPs and activate PAMP-triggered immunity (PTI). Successful pathogens developed effectors that suppress PTI responses, resulting in effector-triggered susceptibility. Subsequently, some plants developed R proteins to recognize these effectors and activate effector-triggered immunity. Over recent years, several examples illustrate that classifying a particular pathogen molecule as PAMP or effector, or host molecule as PRR or R protein, has become a nebulous exercise (Thomma et al., 2011). The results of this study further illustrate the impossibility to mark Ve1 as PRR or R protein and argue for the existence of a continuum between PTI and effector-triggered immunity.
MATERIALS AND METHODS
All experiments have been performed a minimum of three times yielding similar results.
Plant Material and Manipulations
Plants were grown in soil in the greenhouse or in the climate chamber at 21°C/19°C during 16-h/8-h day/night periods, respectively, with 70% relative humidity and 100 W/m2 supplemental light when the intensity dropped below 150 W/m2. Arabidopsis (Arabidopsis thaliana) transformations were performed as described (Clough and Bent, 1998). Homozygous single insert transgenic lines were selected by analyzing the segregation of antibiotic resistance. For Verticillium inoculations, 2- to 3-week-old Arabidopsis plants were uprooted, and the roots were rinsed in water. Subsequently, the roots were dipped for 3 min in a suspension of 106 conidia per milliliter of potato dextrose broth (Difco) and harvested from 1- to 2-week-old Verticillium cultures on potato dextrose agar (Oxoid). Control plants were treated similarly, but their roots were dipped in potato dextrose broth without conidiospores. After replanting in fresh soil, disease development was monitored up to 21 d after inoculation. The following strains were used: Verticillium dahliae strains JR2 (race 1) and M050414 (race 2), Verticillium albo-atrum strains CBS385.91 (race 1) and VA1 (race 2), and Verticillium longisporum strains O1, 43, Boc74, and CBS649.85.
In Planta V. dahliae Biomass Quantification
Quantification of V. dahliae biomass was performed as described previously (Ellendorff et al., 2009). Essentially, 21 d after inoculation, four V. dahliae-inoculated plants per genotype were harvested and pooled. The samples were ground to powder, and DNA was extracted from 100 mg of powder. V. dahliae biomass was determined by real-time PCR using the qPCR Core kit for SYBR Green I (Eurogentec). To assess V. dahliae biomass, the internal transcribed spacer (ITS) region of the ribosomal DNA was targeted using the fungus-specific ITS1-F primer in combination with the V. dahliae-specific reverse primer ST-Ve1-R (Supplemental Table S3). For sample calibration, the Arabidopsis large subunit of the Rubisco gene was targeted using the primer pair AtRuBisCo-F3 and -R3 (Supplemental Table S3). Real-time PCR conditions consisted of an initial denaturation step of 10 min at 95°C, followed by denaturation for 15 s at 95°C, annealing for 30 s at 62°C, and extension for 30 s at 72°C for 40 cycles.
Bioinformatic Analysis
The tomato (Solanum lycopersicum) Ve1 full-length protein sequence (ACR33105) was compared to the tomato Ve2 sequence (ACR33107; Fradin et al., 2009), the putative Ve homologs from Solanum torvum (StVe, AAQ8205), Solanum lycopersicoides (SlVe1, AAP20229), Mentha longifolia (MlVe1, ACB99682), Mentha spicata (MsVe1, ACB99693), Cf-4 (CAA05268), Cf-9 (AAA65235), LeEIX1 (AAR28377), and LeEIX2 (AAR28378) and the 57 AtRLPs (Jones et al., 1994; Thomas, 1997; Chai et al, 2003; Fei et al., 2004; Ron and Avni, 2004; Wang et al., 2008; Vining and Davis, 2009). Multiple sequence alignment and phylogenetic analysis were conducted using MEGA4 version 4 (Tamura et al., 2007). The multiple sequence alignment was performed using the ClustalW function, using the Gonnet protein weight matrix, a gap opening penalty of 10, and a gap extension penalty of 0.2. The obtained alignment was used as input for the phylogenetic analysis. This analysis was performed using the neighbor-joining method (Saitou and Nei, 1987), p-distance was used as parameter, positions containing alignment gaps were eliminated with the pairwise deletion option, and validity of the analysis was tested by 1000 bootstrap replicates (Felsenstein, 1985). To assess the percentage of protein identity and similarity between tomato Ve1 and the other RLP sequences, the full-length sequences were uploaded in BioEdit. Percentages were calculated based on a pairwise alignment using the Gonnet similarity matrix.
Virus-Induced Gene Silencing followed by Pathogen Inoculation
To amplify the 3′-UTR of SlSerk1, 3′-RACE-PCR using the primers SERK1-UTR-F and SERK1-UTR-R (Supplemental Table S3) was used on tomato cDNA. The amplicon was cloned into the pGEMT plasmid (Promega) and transformed to Escherichia coli. Plasmid DNA was isolated from single colonies, and the correct SlSerk1 insert sequence was verified through sequencing. Subsequently, primers UTR-F and UTR-R (Supplemental Table S3) were designed to amplify the 3′-UTR of SlSerk1 containing EcoRI and KpnI restriction sites, respectively. Using the EcoRI and KpnI restriction sites, the amplicon was ligated into the TRV2 plasmid (Liu et al., 2002) and subsequently transformed to electro-competent Agrobacterium tumefaciens strain GV3101. The TRV construct targeting the SlSERK1 LRR region was generated in a similar fashion using primers SERK1-LRR-F and SERK1-LRR-R (Supplemental Table S3) on tomato cDNA and LRR-F and LRR-R (Supplemental Table S3) to generate the TRV clone. The VIGS procedure followed by inoculation with V. dahliae (Fradin et al., 2009) and with Cladosporium fulvum (Vossen et al., 2010) was performed as described previously.
To determine the degree of leaf colonization by C. fulvum, a transgenic C. fulvum strain that constitutively expresses GUS was used. Briefly, leaflets were harvested from inoculated tomato plants, stained with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and destained in 70% ethanol to remove natural pigments and facilitate detection of the GUS stain. Images were made from individual leaflets using a flatbed photoscanner, and quantitation of the total leaf area and the total blue area was carried out using the image processing plugin Phenotype Quant of the program ImageJ. The ratio between the total blue area and total leaf area was calculated. Data from four independent experiments were used for statistical analysis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Vascular colonization of Arabidopsis by GFP-expressing V. dahliae.
Supplemental Figure S2. None of the AtRLPs cluster with tomato Ve1.
Supplemental Figure S3. Constructs used for transgenic expression of Ve1 and Ve2.
Supplemental Figure S4. RT-PCR of Ve1 and Ve2 expression in transgenic Arabidopsis lines.
Supplemental Figure S5. Transgenic expression of neither Ve1 nor Ve2 reduces Verticillium wilt symptoms and fungal biomass upon inoculation with V. albo-atrum race 2.
Supplemental Figure S6. Overview of Ve1-transgenic Arabidopsis defense signaling mutants challenged with V. dahliae race 1 at 21 days after inoculation.
Supplemental Figure S7. Phenotypic appearance of mock-inoculated Ve1-transgenic Arabidopsis defense signaling mutants.
Supplemental Figure S8. Phenotypic appearance of mock-inoculated Ve1-transgenic Arabidopsis Serk mutants.
Supplemental Table S1. Arabidopsis mutants used in this study.
Supplemental Table S2. VIGS analysis of SlSerk1 in resistant Motelle plants.
Supplemental Table S3. Primers used in this study.
Acknowledgments
We thank Dr. Birgit Kemmerling and Dr. Sacco de Vries for providing Arabidopsis Serk mutants and Jeyseelan Baskarathevan, Tine Depaepe, Irene Hanenburg, Damien Ponassié, and Zhao Zhang for technical assistance. Bert Essenstam and Henk Smid are acknowledged for excellent plant care.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries SC. (2005) The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17: 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Kemmerling B, Kwaaitaal M, de Vries SC. (2008) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol 148: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbara DJ, Clewes E. (2003) Plant pathogenic Verticillium species: How many of them are there? Mol Plant Pathol 4: 297–305 [DOI] [PubMed] [Google Scholar]
- Belfanti E, Silfverberg-Dilworth E, Tartarini S, Patocchi A, Barbieri M, Zhu J, Vinatzer BA, Gianfranceschi L, Gessler C, Sansavini S. (2004) The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc Natl Acad Sci USA 101: 886–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolek Y, El-Zik KM, Pepper AE, Bell AA, Magill CW, Thaxton PM, Reddy OUK. (2005) Mapping of Verticillium wilt resistance genes in cotton. Plant Sci 168: 1581–1590 [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ. (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA 92: 6597–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Zhao L, Liao Z, Sun X, Zuo K, Zhang L, Wang S, Tang K. (2003) Molecular cloning of a potential Verticillium dahliae resistance gene SlVe1 with multi-site polyadenylation from Solanum lycopersicoides. DNA Seq 14: 375–384 [DOI] [PubMed] [Google Scholar]
- Chen P, Lee B, Robb J. (2004) Tolerance to a non-host isolate of Verticillium dahliae in tomato. Physiol Mol Plant Pathol 64: 283–291 [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. (2005) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17: 3350–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Ellendorff U, Fradin EF, De Jonge R, Thomma BPHJ. (2009) RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J Exp Bot 60: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellendorff U, Zhang Z, Thomma BPHJ. (2008) Gene silencing to investigate the roles of receptor-like proteins in Arabidopsis. Plant Signal Behav 3: 893–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Chai Y, Wang J, Lin J, Sun X, Sun C, Zuo K, Tang K. (2004) cDNA cloning and characterization of the Ve homologue gene StVe from Solanum torvum Swartz. DNA Seq 15: 88–95 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Fradin EF, Thomma BPHJ. (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol 7: 71–86 [DOI] [PubMed] [Google Scholar]
- Fradin EF, Zhang Z, Ayala JCJ, Castroverde CDM, Nazar RN, Robb J, Liu CM, Thomma BPHJ. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol 150: 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriëls SHEJ, Vossen JH, Ekengren SK, Ooijen GV, Abd-El-Haliem AM, Berg GCMVD, Rainey DY, Martin GB, Takken FLW, Wit PJGMD, et al. (2007) An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J 50: 14–28 [DOI] [PubMed] [Google Scholar]
- Gold J, Robb J. (1995) The role of the coating response in Craigella tomatoes infected with Verticillium dahliae, races 1 and 2. Physiol Mol Plant Pathol 47: 141–157 [Google Scholar]
- Gómez-Gómez L, Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- González-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JDG. (2006) The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurr SJ, Rushton PJ. (2005) Engineering plants with increased disease resistance: What are we going to express? Trends Biotechnol 23: 275–282 [DOI] [PubMed] [Google Scholar]
- Gust AA, Brunner F, Nürnberger T. (2010) Biotechnological concepts for improving plant innate immunity. Curr Opin Biotechnol 21: 204–210 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Parker JE. (2003) Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14: 177–193 [DOI] [PubMed] [Google Scholar]
- He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J. (2007) BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol 17: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt EDL, Boutilier K, Grossniklaus U, de Vries SC. (2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127: 803–816 [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu GS, deHart AKA, Li YS, Ustach C, Handley V, Navarre R, Hwang CF, Aegerter BJ, Williamson VM, Baker B. (2005) EDS1 in tomato is required for resistance mediated by TIR-class R genes and the receptor-like R gene Ve. Plant J 42: 376–391 [DOI] [PubMed] [Google Scholar]
- Huang HC. (2003) Verticillium wilt of alfalfa: epidemiology and control strategies. Can J Plant Pathol 25: 328–338 [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG. (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793 [DOI] [PubMed] [Google Scholar]
- Karapapa VK, Bainbridge BW, Heale JB. (1997) Morphological and molecular characterization of Verticillium longisporum comb. nov., pathogenic to oilseed rape. Mycol Res 101: 1281–1294 [Google Scholar]
- Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries SC. (2006) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, Fischer R, Prüfer D. (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA 98: 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Müssig C, et al. (2007) The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol 17: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV. (2009) Diversity, pathogenicity, and management of Verticillium species. Annu Rev Phytopathol 47: 39–62 [DOI] [PubMed] [Google Scholar]
- Koike ST, Subbarao KV, Davis RM, Gordon TR, Hubbard JC. (1994) Verticillium wilt of cauliflower in California. Plant Dis 78: 1116–1121 [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, Van Esse HP, Smoker M, Rallapalli G, Thomma BPHJ, Staskawicz B, et al. (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 28: 365–369 [DOI] [PubMed] [Google Scholar]
- Lee SW, Han S-H, Sririyanum M, Park C-J, Seo Y-S, Ronald PC. (2009) A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326: 850–853 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Mantelin S, Peng HC, Li B, Atamian HS, Takken FL, Kaloshian I. (2011) The receptor-like kinase SlSERK1 is required for Mi-1-mediated resistance to potato aphids in tomato. Plant J http://dx.doi.org/10.1111/j.1365-313X.2011.04609.x [DOI] [PubMed] [Google Scholar]
- Nam KH, Li JM. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Dangl JL. (2010) Arabidopsis and the plant immune system. Plant J 61: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putt ED. (1964) Breeding behavior of resistance to leaf mottle or Verticillium in sunflower. Crop Sci 4: 177–179 [Google Scholar]
- Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S. (2005) Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 138: 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas S, Rougon-Cardoso A, Smoker M, Schauser L, Yoshioka H, Jones JD. (2004) CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J 23: 2156–2165 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ron M, Avni A. (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe RC, Davis JR, Powelson ML, Rouse DI. (1987) Potato early dying: causal agents and management strategies. Plant Dis 71: 482–489 [Google Scholar]
- Rowland O, Ludwig AA, Merrick CJ, Baillieul F, Tracy FE, Durrant WE, Fritz-Laylin L, Nekrasov V, Sjölander K, Yoshioka H, et al. (2005) Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell 17: 295–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schaible L, Cannon OS, Waddoups V. (1951) Inheritance of resistance to Verticillium wilt in a tomato cross. Phytopathology 41: 986–990 [Google Scholar]
- Schmidt EDL, Guzzo F, Toonen MAJ, de Vries SC. (1997) A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124: 2049–2062 [DOI] [PubMed] [Google Scholar]
- Simko I, Costanzo S, Haynes KG, Christ BJ, Jones RW. (2004a) Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor Appl Genet 108: 217–224 [DOI] [PubMed] [Google Scholar]
- Simko I, Haynes KG, Ewing EE, Costanzo S, Christ BJ, Jones RW. (2004b) Mapping genes for resistance to Verticillium albo-atrum in tetraploid and diploid potato populations using haplotype association tests and genetic linkage analysis. Mol Genet Genomics 271: 522–531 [DOI] [PubMed] [Google Scholar]
- Stuiver MH, Custers JHHV. (2001) Engineering disease resistance in plants. Nature 411: 865–868 [DOI] [PubMed] [Google Scholar]
- Stulemeijer IJE, Stratmann JW, Joosten MHAJ. (2007) Tomato mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol 144: 1481–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thomas CM. (1997) Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Nürnberger T, Joosten MHAJ. (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Penninckx IAMA, Broekaert WF, Cammue BPA. (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13: 63–68 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, van Esse HP, Crous PW, de Wit PJGM. (2005) Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol Plant Pathol 6: 379–393 [DOI] [PubMed] [Google Scholar]
- Tjamos SE, Flemetakis E, Paplomatas EJ, Katinakis P. (2005) Induction of resistance to Verticillium dahliae in Arabidopsis thaliana by the biocontrol agent K-165 and pathogenesis-related proteins gene expression. Mol Plant Microbe Interact 18: 555–561 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Katagiri F. (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13: 459–465 [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Tsitsigiannis DI, Rowland O, Lo J, Rallapalli G, MacLean D, Takken FLW, Jones JDG. (2008) The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 20: 697–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P, Narasimhan ML, Stevenson RA, Zhu JK, Weller SC, Subbarao KV, Bressan RA. (2003) Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana. Plant J 35: 574–587 [DOI] [PubMed] [Google Scholar]
- Vinatzer BA, Patocchi A, Gianfranceschi L, Tartarini S, Zhang HB, Gessler C, Sansavini S. (2001) Apple contains receptor-like genes homologous to the Cladosporium fulvum resistance gene family of tomato with a cluster of genes cosegregating with Vf apple scab resistance. Mol Plant Microbe Interact 14: 508–515 [DOI] [PubMed] [Google Scholar]
- Vining K, Davis T. (2009) Isolation of a Ve homolog, mVe1, and its relationship to Verticillium wilt resistance in Mentha longifolia (L.) Huds. Mol Genet Genomics 282: 173–184 [DOI] [PubMed] [Google Scholar]
- Vining KJ, Zhang Q, Smith CA, Davis TM. (2007) Identification of resistance gene analogs and Verticillium wilt resistance-like sequences in Mentha longifolia. J Am Soc Hortic Sci 132: 541–550 [Google Scholar]
- Vossen JH, Abd-El-Haliem A, Fradin EF, Van Den Berg GCM, Ekengren SK, Meijer HJG, Seifi A, Bai Y, Ten Have A, Munnik T, et al. (2010) Identification of tomato phosphatidylinositol-specific phospholipase-C (PI-PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance. Plant J 62: 224–239 [DOI] [PubMed] [Google Scholar]
- Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, Mitchell K, Bastas K, Liu CM, Woods-Tör A, Zipfel C, et al. (2008) A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol 147: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Fiers M, Ellendorff U, Wang Z, de Wit PJGM, Angenent GC, Thomma BPHJ. (2010a) The diverse roles of extracellular leucine-rich repeat-containing receptor-like proteins in plants. Crit Rev Plant Sci 29: 285–299 [Google Scholar]
- Wang G, Long Y, Thomma BPHJ, de Wit PJGM, Angenent GC, Fiers M. (2010b) Identification and functional analyses of equivalents of CLV2, and domains that contribute to CLV2 specificity. Plant Physiol 152: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S. (1955) Longevity of the Verticillium wilt fungus in the laboratory and field. Phytopathology 45: 180–181 [Google Scholar]
- Wulff BB, Chakrabarti A, Jones DA. (2009) Recognitional specificity and evolution in the tomato-Cladosporium fulvum pathosystem. Mol Plant Microbe Interact 22: 1191–1202 [DOI] [PubMed] [Google Scholar]
- Wulff BB, Horvath DM, Ward ER. (2011) Improving immunity in crops: new tactics in an old game. Curr Opin Plant Biol http://dx.doi.org/10.1016/j.pbi.2011.04.002 [DOI] [PubMed] [Google Scholar]
- Zebrowska J, Hortyäski J, Cholewa T, Honcz K. (2006) Resistance to Verticillium dahliae (Kleb.) in the strawberry breeding lines. Commun Agric Appl Biol Sci 71: 1031–1036 [PubMed] [Google Scholar]
- Zhang Y, Yang Y, Fang B, Gannon P, Ding P, Li X, Zhang Y. (2010) Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRK70. Plant Cell 22: 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]