Abstract
Many plant host factors are known to interact with viral proteins during pathogenesis, but how a plant virus induces a specific disease symptom still needs further research. A lily strain of Cucumber mosaic virus (CMV-HL) can induce discrete necrotic spots on infected Arabidopsis (Arabidopsis thaliana) plants; other CMV strains can induce similar spots, but they are not as distinct as those induced by CMV-HL. The CMV 2b protein (2b), a known RNA-silencing suppressor, is involved in viral movement and symptom induction. Using in situ proximity ligation assay immunostaining and the protoplast assays, we report here that CMV 2b interacts directly with Catalase3 (CAT3) in infected tissues, a key enzyme in the breakdown of toxic hydrogen peroxide. Interestingly, CAT3, normally localized in the cytoplasm (glyoxysome), was recruited to the nucleus by an interaction between 2b and CAT3. Although overexpression of CAT3 in transgenic plants decreased the accumulation of CMV and delayed viral symptom development to some extent, 2b seems to neutralize the cellular catalase contributing to the host defense response, thus favoring viral infection. Our results thus provide evidence that, in addition to altering the type of symptom by disturbing microRNA pathways, 2b can directly bind to a host factor that is important in scavenging cellular hydrogen peroxide and thus interfere specifically with that host factor, leading to the induction of a specific necrosis.
Cucumber mosaic virus (CMV) belongs to the genus Cucumovirus and contains tripartite, single-stranded sense RNAs designated RNA1 to RNA3 in decreasing order of molecular mass. RNA1 and RNA2 encode replicase proteins 1a and 2a, respectively, and RNA3 encodes the movement protein 3a. A subgenomic RNA, RNA4, encodes the coat protein. The 2b protein (2b) is encoded in a subgenomic RNA, RNA4A, which has a 3′-proximal open reading frame (ORF) in RNA2 (Ding et al., 1994).
The CMV 2b protein is involved in local and systemic movement of CMV and inhibits host defense through RNA silencing (Ding et al., 1995; Ji and Ding, 2001; Soards et al., 2002; Shi et al., 2003) and also acts as a symptom determinant, which is apparently associated with its RNA-silencing suppressor (RSS) activity (Brigneti et al., 1998; Lucy et al., 2000). The 2b protein contains two nuclear localization signals (NLSs) in its N terminus (Lucy et al., 2000; Goto et al., 2007) and has been confirmed to be actually localized in the nucleus in infected tissues; the NLSs of the 2b protein are required for viral symptom induction and RSS activity (Lucy et al., 2000; Lewsey et al., 2009). Recently, 2b was found to bind to Argonaute1 (AGO1), necessary for the plant antiviral machinery, and to AGO4, necessary for the DNA methylation process that controls heterochromatin formation and transcription (Zhang et al., 2006; González et al., 2010). In addition, 2b was also reported to directly bind to small interfering RNAs (Goto et al., 2007; Kanazawa et al., 2011).
Hydrogen peroxide (H2O2), one of the reactive oxygen species, plays an important role in many physiological phenomena such as senescence, photorespiration and photosynthesis, growth and development, and resistance responses against pathogens (Noctor and Foyer, 1998; Foreman and Tang, 2003; Peng et al., 2005). In addition, H2O2 is generated as a primary messenger to induce hypersensitive cell death in the plant host. Catalase, which breaks down H2O2 and functions as a cellular sink for H2O2, is an enzyme found in nearly all living organisms. In Arabidopsis (Arabidopsis thaliana), the catalase gene family is composed of three members, CAT1, CAT2, and CAT3 (Frugoli et al., 1996). CAT2 and CAT3 are localized in the peroxisome and are the major H2O2 scavengers that control the reactive oxygen species homeostasis in plants (Du et al., 2008a). The mutants cat2 and cat3 typically display patches of chlorosis and necrotic lesions (Contento and Bassham, 2010).
Although the lily strain of CMV (CMV-HL) can induce a necrosis in many Arabidopsis ecotypes such as Columbia (Col-0) and Landsberg erecta, we chose Col-0 because many mutant lines are available for future study. In Arabidopsis Col-0, CMV induces a distinct necrosis, which is associated with the H2O2 that is generated after infection. In this paper, we investigated a direct link between the CMV-induced necrosis and the 2b-CAT3 interaction and found that cell death was indeed induced by 2b’s binding to CAT3. Here, on the basis of a simple and specific protein-protein interaction between 2b and CAT3, we can begin to explain the molecular mechanism underlying CMV-induced necrosis in Arabidopsis.
RESULTS
Leaf Necrosis in CMV-HL-Infected Arabidopsis Plants
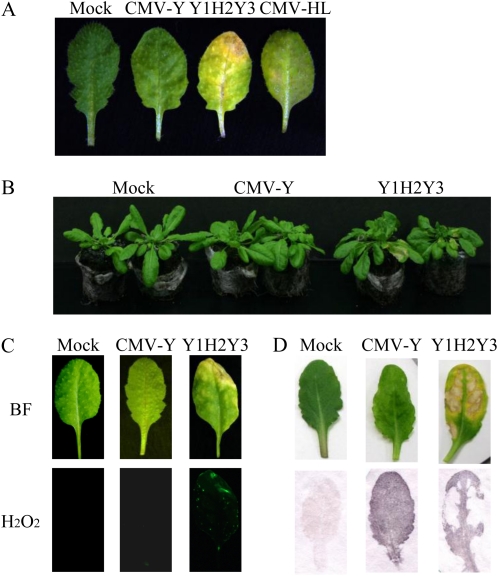
CMV can systemically infect Arabidopsis and causes a variety of symptoms depending on the particular combination of viral strain and Arabidopsis ecotype. After the Col-0 plants were inoculated with CMV Y strain (CMV-Y), they developed systemic mosaic symptoms 10 d after inoculation, and at 3 weeks after inoculation, necrotic lesions were sometimes observed on the upper leaves. When Col-0 plants were inoculated with CMV-HL, the virus was initially localized within the inoculated leaves and spread very slowly to the upper leaves compared with CMV-Y, but it induced more severe necrosis on the upper leaves than CMV-Y did (Fig. 1A). To identify the viral factor(s) for the necrosis induction, we created pseudorecombinants between CMV-Y and CMV-HL. The three genomic RNAs of CMV-Y and CMV-HL were designated Y1 to Y3 and H1 to H3, respectively. Among the different pseudorecombinants between CMV-Y and CMV-HL, Y1H2Y3 induced clear necrosis on the inoculated plants, suggesting that RNA2 is important for the symptom. Instead of the original CMV-HL, we chose Y1H2Y3 for further analysis of the observed necrosis, because unlike CMV-HL, it could systemically move in Col-0 as fast as CMV-Y and induce very distinct necrosis on the upper leaves (Fig. 1; data not shown).
Figure 1.
Necrosis induction and H2O2 generation by Y1H2Y3, a pseudorecombinant between CMV-Y and CMV-HL, in plants of Arabidopsis Col-0. A, Necrotic symptoms on the plants infected with CMV-HL and Y1H2Y3 at 18 dpi. B, Symptoms on CMV-infected plants at 10 dpi. C, Detection of H2O2 in the Col-0 plants infected with Y1H2Y3 using the H2O2-sensitive fluorescent probe H2DCFDA. Leaves were harvested at 10 dpi and photographed using either bright-field light microscopy (BF) or fluorescence microscopy (H2O2). Note that necrosis was associated with the sites that fluoresced. D, Symptomatic Col-0 leaves (top panels) were subjected to tissue printing (bottom panels) to localize CMV.
Y1H2Y3 induced distinct necrosis on the upper leaves at 10 d postinoculation (dpi), while CMV-Y did not at this stage (Fig. 1B). The necrotic spots induced by Y1H2Y3 infection were found to be associated with an increase in H2O2 production, as measured by oxidation of H2DCFDA; 2′,7′-dichloro fluorescein diacetate (Fig. 1C). Because H2O2 acts as an inducer of defense genes such as pathogenesis-related protein (PR) genes, we analyzed the expression of some PR genes. As a result, levels of PR1 and PR5 were enhanced, implying that the necrosis might be a defense response (Supplemental Fig. S1). In tissue-printing experiments, both CMV-Y and Y1H2Y3 were distributed throughout the upper leaves, but the virus was not detected in the centers of the coalescing necrotic spots (Fig. 1D). Because we found that RNA2 of CMV-HL (H2) was important for the necrosis, we then asked whether the RNA2-encoded proteins (2a and/or 2b) are involved in the necrosis. We focused on 2b because it has been suggested that 2b is implicated in CMV symptoms including necrosis (Du et al., 2008b).
HL2b Interacts with CAT3 in the Yeast Two-Hybrid Assay
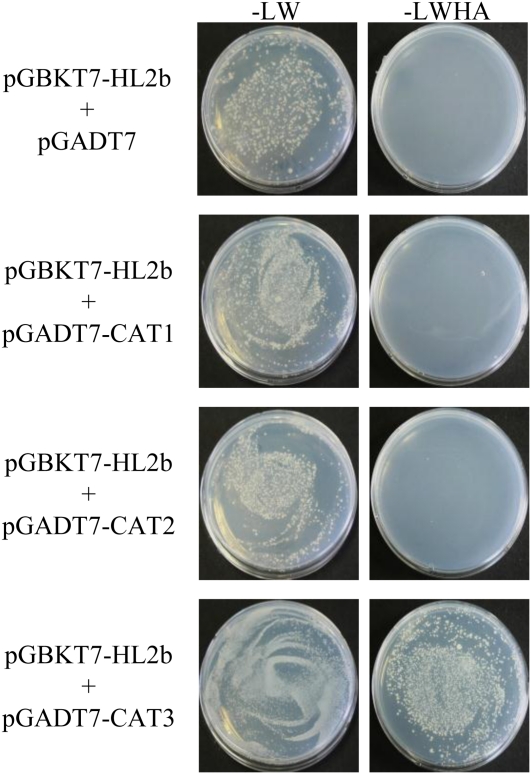
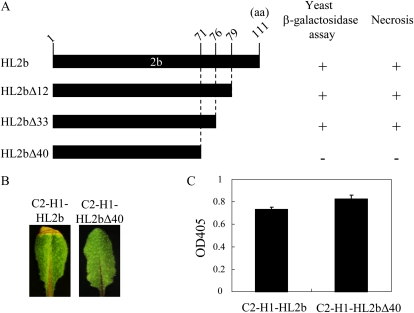
To analyze how CMV 2b is involved in the necrosis induction, we started with the isolation of host factors that interact with CMV 2b. We screened an Arabidopsis cDNA library for the yeast two-hybrid assay in the GAL4 activation domain vector pGADT7 using the GAL4 binding domain bait vector that expresses the HL2b in-frame fusion with the binding domain (pGBKT7-HL2b). Although CMV 2bs had been reported to act as a transcriptional activator in yeast (Ham et al., 1999; Sueda et al., 2010), fortunately, HL2b did not have this function. We screened about 5.0 × 106 independent yeast transformants and eventually obtained eight candidate clones. One of the clones that had strong β-galactosidase activity in the two-hybrid assay was selected for sequencing and found to be the CAT3 gene (AT1G20620). Because the CAT3 gene isolated from the pGADT7 library was not complete, to verify the interaction between HL2b and CAT3, we then cloned the full-length ORF of the CAT3 gene and reexamined it by the yeast two-hybrid assay; we here included two other Arabidopsis catalase genes (CAT1 and CAT2). The catalase genes (CAT1, CAT2, and CAT3) were cloned into the pGADT7 vector to create pGADT7-CAT1, pGADT7-CAT2, and pGADT7-CAT3, respectively. Each construct was cotransferred with pGBKT7-HL2b into Saccharomyces cerevisiae strain AH109. The results revealed that HL2b indeed interacted with CAT3 but not with the other two catalases (CAT1 and CAT2; Fig. 2). To determine which domain of 2b is important for the interaction with CAT3, we created several HL2b deletion mutants that lack the C-terminal amino acids. When the C-terminal amino acids were deleted by 40 amino acid residues, creating HL2bΔ40, the mutant 2b lost the ability to bind to CAT3 in the two-hybrid assay, and the virus (C2-H1)-containing HL2bΔ40 (C2-H1-HL2bΔ40) also lost the ability to induce necrosis, indicating a link between the binding of 2b to CAT3 and necrotic induction (Fig. 3). C2-H1 is a CMV-Y-based virus vector in which a multiple cloning site replaces the 2b gene (Matsuo et al., 2007). Therefore, the C-terminal region of HL2b is important for the interaction between 2b and CAT3.
Figure 2.
Interaction between HL2b and catalase in the yeast two-hybrid assay. Each plasmid (pGADT7, pGADT7-CAT1, pGADT7-CAT2, or pGADT7-CAT3) was cotransferred with pGBKT7-HL2b into yeast strain AH109. The transformed AH109 cells were plated on the growth medium without Leu and Trp (−LW; left panel) or without Leu, Trp, His, and adenine (−LWHA; right panel). Only when the expressed proteins bind to each other can the cells grow on the synthetic medium. [See online article for color version of this figure.]
Figure 3.
CMV-HL2b interacts with Arabidopsis CAT3. A, Schematic representation of the C-terminal deletion mutants of HL2b for the yeast two-hybrid assay. Deleted forms of the HL2b gene were fused to the binding domain in the pGBKT7 vector (Clontech), and the CAT3 ORF was inserted in pGADT7 activation domain vector (Clontech). Transcriptional activity of β-galactosidase was assayed. Plus (+) indicates transcriptional activation, while minus (−) indicates a negative result. aa, Amino acids. B, Symptoms in the Col-0 plants infected with C2-H1-HL2b or C2-H1-HL2bΔ40. The recombinant HL2bs (HL2b, HL2bΔ12, HL2bΔ33, and HL2bΔ40) were inserted in the CMV vector, C2-H1, for virus inoculation experiments. No necrotic symptom was observed in the Col-0 plants infected with C2-H1-HL2bΔ40. C, CMV accumulation levels. ELISA was conducted to analyze the CMV accumulation in Col-0 plants. Error bars represent se of the means. OD405, Optical density at 405 nm. [See online article for color version of this figure.]
Catalase Activity and Necrosis in CMV-Infected Plants and Transgenic Plants Expressing 2b
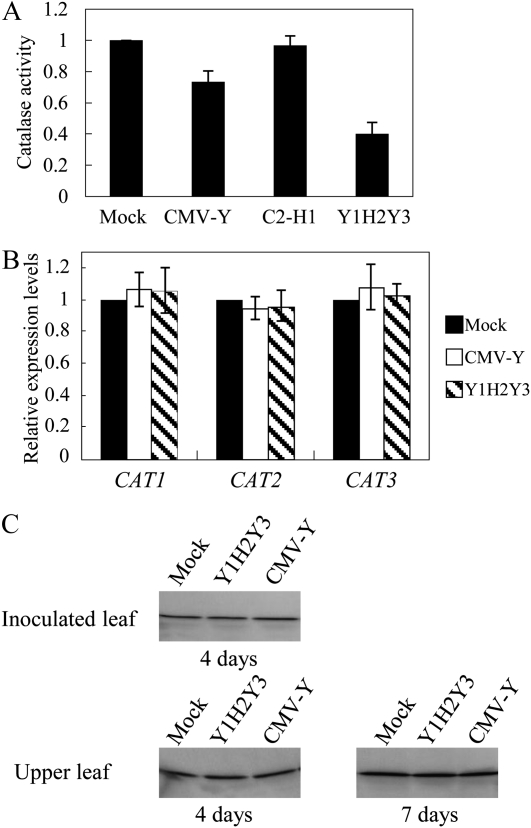
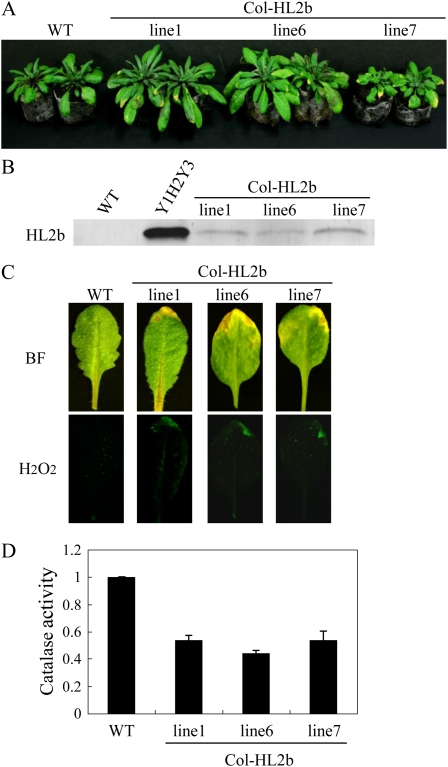
Because we had an idea that the interaction between 2b and CAT3 could induce the necrosis, we then measured catalase activity in virus-infected tissues. CAT2 and CAT3 are highly expressed in leaves and thus account for most of the catalase activity. Both cat2 and cat3 knockout plants displayed patches of necrotic lesions, suggesting that reduction in either CAT2 or CAT3 causes necrosis in leaves (Contento and Bassham, 2010). In fact, catalase activity was significantly decreased by 10 dpi in the Y1H2Y3-infected leaves when compared with the mock- or CMV-Y-inoculated control plants (Fig. 4A). To investigate whether the mRNA levels of the Arabidopsis catalase genes (CAT1–CAT3) were decreased in CMV infection, we measured those by real-time reverse transcription (RT)-PCR and found that there was no difference in any catalase genes at the mRNA level (Fig. 4B). Western blots using CAT3-specific antibodies revealed that CAT3 was also not changed at the protein level (Fig. 4C). To examine whether 2b is the determinant for the necrosis induction by Y1H2Y3 in Col-0 leaves, we then generated transgenic Arabidopsis plants that express the HL2b gene (Col-HL2b; Fig. 5). In those transgenic lines, we confirmed the accumulation of 2b by western-blot analyses using anti-2b antibodies (Fig. 5B). Three transgenic lines developed necrosis on leaves at the seedling stage in association with H2O2 formation, just as observed for CMV-infected leaves (Fig. 5C). These results suggest that the expression of 2b alone causes necrosis by generating H2O2 in Col-0 leaves, which is analogous to that observed in CMV-infected Arabidopsis. We also measured catalase activities in the Col-HL2b plants and found that catalase activities in all the lines were indeed reduced below 50% of the catalase activity of nontransgenic plants (Fig. 5D). Therefore, we considered that 2b expression caused a decrease in catalase activity in the transgenic cells, which likely caused H2O2 generation, leading to necrosis.
Figure 4.
Catalase activity, transcripts, and protein in the CMV-infected Col-0 plants. A, Catalase activity in CMV-Y-, mock-, and Y1H2Y3-inoculated plants was determined by measuring the decrease in A240 at 10 dpi. Catalase activity was expressed as mmol min−1 mg−1 protein. Three replicates were done. Error bars represent se of the means. C2-H1 lacks the entire 2b. B, CAT (CAT1, CAT2, and CAT3) expression levels in the Col-0 plants inoculated with Y1H2Y3 were determined by quantitative RT-PCR 2 weeks after mock and CMV inoculation. Error bars represent se of three replicates. C, Western-blot analysis was performed using the specific anti-CAT3 antibody. Total protein was extracted from the CMV-inoculated leaves at 4 dpi and from the upper leaves at 4 and 7 dpi. Note that there was little difference in the expression level of CAT3 between mock- and CMV-inoculated plants, indicating that CMV does not affect CAT3 accumulation.
Figure 5.
Necrosis induction and H2O2 generation in transgenic Col-0 plants expressing HL2b. A, Necrotic symptoms in the leaves of transgenic plants expressing HL2b (Col-HL2b). WT, Wild-type Col-0. B, Western-blot analysis of the 2b protein in the transgenic plants. Total proteins (30 μg), which were extracted from Col-0 (WT), CMV-infected leaves (Y1H2Y3), and Col-HL2b (lines 1, 6, and 7), were separated by SDS-PAGE and blotted onto a polyvinylidene difluoride membrane. The blots were then subjected to immunodetection using anti-2b antibodies. C, Detection of H2O2 in the Col-HL2b lines using the H2O2-sensitive fluorescent probe H2DCFDA. Note that necrosis on the edge of the leaves that were harvested from the transgenic plants was associated with fluorescent spots. Leaves were harvested and photographed using either bright-field light microscopy (BF) or fluorescence microscopy (H2O2). D, Catalase activities in the Col-HL2b lines. Catalase activity was determined as described for Figure 4.
HL2b Interacts with CAT3 in Col-0 Cells and Alters the Cellular Location of CAT3
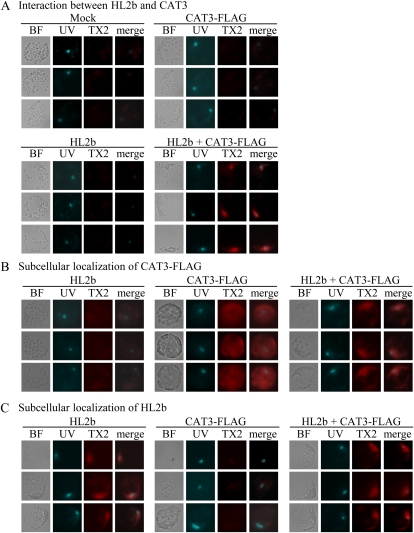
To confirm further the results of the two-hybrid assay and to determine whether HL2b associates with CAT3 in Col-0 cells, we performed the Duolink in situ proximity ligation assay (PLA), a method to detect proteins using two primary antibodies for visualization and quantification of specific protein-protein interactions in situ (Söderberg et al., 2008). According to the assay principle, only when two target proteins are close to each other will a fluorescent signal be detected. Col-0 protoplasts were transfected with the plasmid constructs expressing HL2b, CAT3 with a FLAG tag (CAT3-FLAG), and GFP (control). In the cells transfected with HL2b + CAT3-FLAG, strong fluorescent signals were localized in the nuclei (Fig. 6A), whereas no signal was detected in the cells transfected with either of the plasmids alone, indicating that the 2b-CAT interaction was in the nuclei. Compared with the HL2b-CAT3 interaction, we observed relatively weaker signals between Y2b and CAT3, indicating that Y2b binds more weakly than HL2b to CAT3 (Supplemental Fig. S2). This is consistent with the observation that CMV-Y did not induce necrosis on Arabidopsis as strongly as CMV-HL did (Fig. 1). Subcellular localization of 2b and CAT3 was analyzed by the single recognition method in PLA using one primary antibody. We investigated whether the interaction between 2b and CAT3 alters the original localization of 2b or CAT3. In the protoplasts transfected with CAT3-FLAG alone, CAT3 signals visualized by anti-FLAG antibodies diffused through the cytoplasm (Fig. 6B). When protoplasts were transfected with HL2b + CAT3-FLAG, the CAT3 signals were not detected in the cytoplasm but found in the nucleus, where 2b originally is localized (Fig. 6, B and C), suggesting that CAT3 was translocated into the nucleus with 2b. We also obtained similar results using Y2b instead of HL2b, suggesting that the 2b proteins generally interact with CAT3 in vivo, although they differ in their intensity of interaction (Supplemental Fig. S2).
Figure 6.
In situ association between HL2b and CAT3 in Arabidopsis cells. Protoplasts were transfected with HL2b, CAT3-FLAG, or HL2b + CAT3-FLAG. BF indicates images observed with bright-field light microscopy. Protoplast nuclei were visualized using Hoechst 33342 staining and UV light (UV). For detection of in situ molecular interaction, the Duolink in situ PLA kit was used. A, Interaction between HL2b and CAT3. Primary antibodies against FLAG and CMV 2b were used to detect CAT3 and 2b, respectively. With fluorescence microscopy, PLA red signals from TX Red (TX2) will theoretically be detected only when there is an in vivo interaction between 2b and CAT3. The images of UV and TX2 were merged (merge column) to identify the exact localization of the signals. B, Subcellular localization of CAT3-FLAG in protoplasts. CAT3 was detected using anti-FLAG antibodies as red signals of TX2. Note that CAT3 was detected in nuclei when HL2b was expressed with the CAT3 gene, while the protein was dispersed in cells without HL2b. C, Subcellular localization of HL2b in protoplasts. HL2b was detected using anti-2b antibodies, which yielded PLA red signals from TX2. As expected, HL2b was localized in nuclei.
CAT3 Overexpression Enhanced Tolerance to CMV Infection
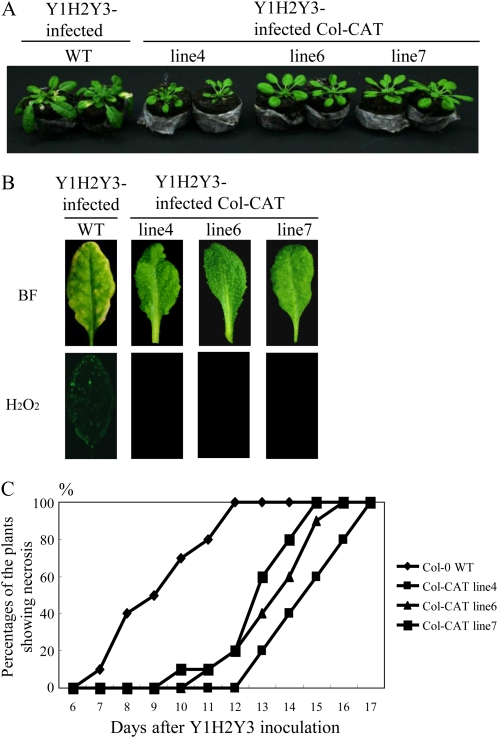
Because we found that a viral protein (2b) interacted with a host protein (CAT3) in vivo, we then investigated whether overexpression of CAT3 affects not only CMV 2b but also CMV infection (e.g. symptom appearance) in the transgenic plants. To obtain Arabidopsis plants that overexpress the CAT3 gene, we transformed Col-0 plants using the Ti-plasmid vector in which the CAT3 gene was inserted under the control of the 35S promoter. We finally selected three transgenic lines (4, 6, and 7), which grew normally, for inoculation experiments (Supplemental Fig. S3). By western-blot analyses, we confirmed enhanced levels of CAT3 in those transgenic plants. In agreement with the CAT3 levels, the catalase activities also increased in all three lines (Supplemental Fig. S3). When those transgenic lines were inoculated with Y1H2Y3, at 10 dpi, none of the transgenic plants had developed necrosis, while nontransgenic plants had distinct necrosis associated with H2O2 generation (Fig. 7, A and B). In addition, necrosis was delayed significantly according to a statistical analysis compared with the nontransgenic plants, although all plants eventually developed necrosis, suggesting that elevated levels of CAT3 only delayed the onset of necrosis (Fig. 7C).
Figure 7.
Characterization of Col-CAT transgenic (lines 4, 6, and 7) plants inoculated with Y1H2Y3. A, Delay of necrosis in Col-CAT transgenic plants compared with the wild-type Col-0 (WT) at 10 dpi. B, Detection of H2O2 in control and Col-CAT plants inoculated with Y1H2Y3 using H2DCFDA. Leaves were harvested at 10 dpi and photographed either by bright-field light microscopy (BF) or by fluorescence microscopy (H2O2). C, Development of necrotic symptoms in control and Col-CAT transgenic (lines 4, 6, and 7) plants. The percentage of symptomatic plants was calculated using 10 plants for each transgenic line.
CAT3 Expression Compromises Cell Death Induced by 2b in Protoplasts
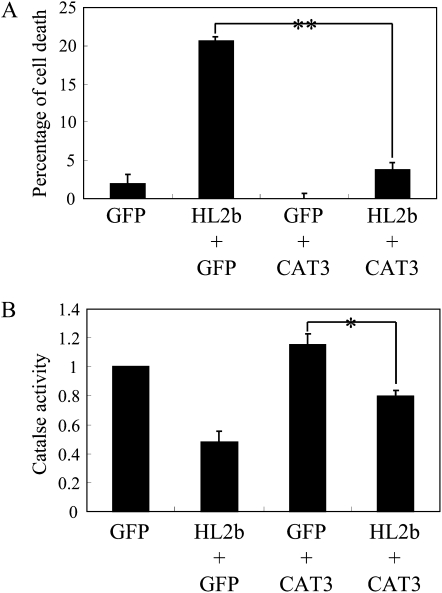
To determine at the cellular level whether HL2b expression causes cell death, we used the Arabidopsis protoplast system, which we had previously developed to analyze the hypersensitive response-like cell death induced by Turnip mosaic virus (Kim et al., 2010). When protoplasts were transfected with the Ti-plasmid expressing HL2b, the percentage of cell death was significantly increased. However, addition of the Ti-plasmid expressing CAT3 negated the cell death induced by HL2b (Fig. 8A). Coexpression of HL2b and CAT3 led to a decrease in catalase activity (Fig. 8B). These results thus suggest that HL2b can induce cell death by binding to CAT3 and impairing the catalase activity and that the HL2b-CAT3 interaction partially compromised the H2O2 scavenging by CAT3.
Figure 8.
Effect of the interaction between CAT3 and HL2b on cell death of Arabidopsis Col-0 protoplasts. A, Percentage of cell death for transfected Col-0 protoplasts at 20 h after transfection with GFP (control, 10 μg), HL2b (5 μg) + GFP (5 μg), GFP (5 μg) + CAT3 (5 μg), or HL2b (5 μg) + CAT3 (5 μg). Cells that were stained by Evans blue were scored as dead. Data are percentages of cell death after the control percentage was subtracted from the background. Error bars represent se of the means of three replicates. Differences in the means were evaluated with a t test; ** significant at the 1% level. B, Catalase activities in Col-0 protoplasts transfected with GFP (control), HL2b + GFP, GFP + CAT3, or HL2b + CAT3 at 20 h after transfection. Activity was measured spectrophotometrically. Error bars represent se of the means of three replicates. Differences in means among groups were evaluated by t test; * significant at the 5% level.
Effect of CAT3 on the Silencing Suppressor Activity of 2b and CMV Accumulation Levels in Protoplasts
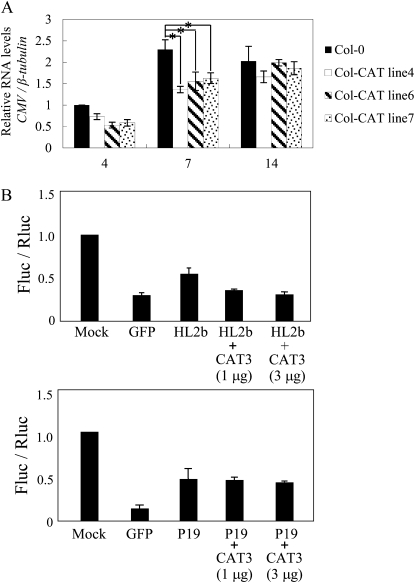
Figure 9A shows that enhanced levels of CAT3 enabled the transgenic lines to suppress CMV multiplication at least until 7 dpi. However, at 14 dpi, the levels of CMV reached those of the nontransgenic control plants for all three lines. These results indicate that overexpression of CAT3 could confer partial tolerance at an initial stage of viral infection. We hypothesized that CAT3 may inactivate the RSS activity of 2b by binding with 2b, thus inhibiting CMV multiplication. To investigate the effect of CAT3 on the RSS activity of 2b, we used a protoplast assay that we have previously developed to measure viral RSS activities on the cellular level (Shimura et al., 2008). As shown in Figure 9B, CAT3 expression actually decreased the RSS activity of 2b by 30% to 40%, but it did not affect another virus RSS, P19. On the contrary, CMV infection did not down-regulate CAT3; when nontransgenic plants were inoculated with CMV, viral infection did not affect CAT3 either at the mRNA or the protein level (Fig. 4).
Figure 9.
Enhanced resistance to CMV by CAT3 overexpression in protoplasts and Col-CAT transgenic plants. A, Effect of CAT3 expression on the accumulation of Y1H2Y3 in Col-CAT plants. Quantitative RT-PCR was performed to measure the accumulation of CMV in Col-CAT transgenic plants inoculated with Y1H2Y3. The values were normalized using the mRNA levels of the β-tubulin gene. The value for the nontransgenic Col-0 plant at 4 dpi was set at 1.0. The values represent means with se obtained from three replicates. Differences in means among groups were evaluated by t test; * significant at the 5% level. B, Effect of CAT3 overexpression on the RSS activity of HL2b. N. benthamiana protoplasts were cotransfected with pBI-Fluc (0.3 μg), pE-Rluc (0.03 μg), dsFluc (0.03 μg), and viral suppressor (HL2b or P19; 3 μg of plasmid) together with CAT3 (0, 1, and 3 μg of plasmid). GFP (3 μg) was used as a control. At 20 h after transfection, Fluc activities in the treated protoplasts were measured. Fluc activity was then divided by Rluc activity, and the value for the mock treatment was set at 1.0. The values represent means with sd from three replicates. Note that the addition of CAT3 decreased the RSS activity of HL2b but did not affect that of P19.
The Binding of 2b to CAT3 Is Important for Cell Death But the Translocation of CAT3 to the Nucleus Is Not
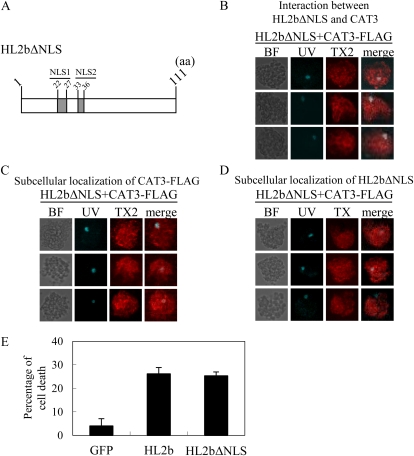
Finally, to answer the question of which is important for the 2b-induced cell death, the binding of 2b to CAT3 or the subsequent translocation of CAT3 to the nucleus, we created a mutant HL2b (HL2bΔNLS) that lacks its NLSs (NLS1 and NLS2; Fig. 10A). We then conducted the protoplast assays to observe cell death and the localization of CAT3 and HL2bΔNLS. In situ PLA revealed that neither HL2bΔNLS nor CAT3 was localized in the nucleus, but the two proteins bound to each other (Fig. 10, B–D). As shown in Figure 10E, HL2bΔNLS could still induce cell death as efficiently as the authentic HL2b. These results thus indicate that the binding of 2b to CAT3 is per se important to inhibit catalase activity.
Figure 10.
Cellular localization of CAT3 and cell death induction in protoplasts transfected with HL2bΔNLS. A, Schematic representation of HL2bΔNLS. The mutants had deletions of the two NLSs (NLS1 and NLS2). aa, Amino acids. B, In situ association between HL2bΔNLS and CAT3 in Arabidopsis cells. Protoplasts isolated from Col-0 plants were transfected with HL2bΔNLS+CAT3 and harvested after 20 h. The TX2 signals are theoretically detected only when 2b and CAT3 interact in vivo. C, Subcellular localization of CAT3-FLAG in protoplasts. CAT3 was detected using anti-FLAG antibodies as red signals of TX2. D, Subcellular localization of HL2bΔNLS in protoplasts. HL2bΔNLS was detected using anti-CMV 2b antibodies as red signals of TX2. E, Cell death of Col-0 protoplasts transfected with HL2bΔNLS. The protoplasts were transfected with GFP (control), HL2b, or HL2bΔNLS. At 20 h after transfection, cell death was scored by staining the protoplasts with Evans blue. Data are presented as a percentage of cell death. Three replicates were done. Error bars represent se. BF, Bright-field light microscopy.
DISCUSSION
Direct Interactions between a Viral Factor and Host Factor in Symptom Induction
Many attempts to find host factors necessary for viral replication and movement, which actually interact with viral proteins, have been made to elucidate the molecular mechanisms for viral pathogenicity. For example, as reviewed by Ishibashi et al. (2010), more than 10 host factors for multiplication of tobamoviruses have been identified already. Those factors certainly play a role in viral pathogenicity as well as viral multiplication, apparently serving as a host range or symptom determinant. There are also many reports that describe symptom modifications as an indirect consequence of the disturbance of the cellular physiology of the plant, such as host factors that interact with viral RSS against the host RNA-silencing machinery, which is essential not only for defense against viruses but also plant development. For example, the CMV 2b protein can interfere with RNA silencing by binding to the host AGO1 protein, leading to disturbance of microRNA-directed physiology in CMV-infected plants (Zhang et al., 2006). However, there are only a few reports that link protein-protein interactions between viral and host factors to distinct symptom phenotypes as a direct result of a viral protein interfering with the intrinsic function of a specific host factor. In one such study, Zhu et al. (2005) demonstrated a direct interaction between the P2 protein of Rice dwarf virus (RDV) and rice (Oryza sativa) ent-kaurene oxidases, which play a role in the biosynthesis of GAs; this interaction led to a decrease in the hormones and stunting of the infected plants. More interestingly, they also raised the possibility that the interaction may actually decrease phytoalexin biosynthesis and make the plants more competent for viral replication, suggesting that the interaction apparently works in favor of the virus. We here provide more evidence strongly supporting a viral protein as a primary target for determining the type of viral symptom expression. It is quite reasonable to speculate that the necrosis formation in CMV-infected leaves is due to the decreased levels of CAT activity as a result of the 2b-CAT3 interaction. Other catalases in many natural hosts for CMV, including Nicotiana benthamiana, may also bind to 2b, because a database search revealed that their amino acid sequences were well conserved. In fact, in our preliminary experiments, the potato virus X vector overexpressing the intact Y2b but not a truncated Y2b lacking the C-terminal 40 amino acids could induce necrosis on N. benthamiana.
The 2b-CAT3 Interaction Serves as a Necrosis Determinant in Favor of CMV Infection
The interaction between RDV P2 and ent-kaurene oxidases, as we have described it, appears to work in favor of the virus RDV. Now, an interesting question is for which side does the 2b-CAT3 interaction work? Does it elevate or reduce host defense after all? We discuss this “two sides of the same coin” question taking the following observations into account: (1) 2b bound to CAT3 and changed the localization of CAT3 from the cytosol to the nucleus in Col-0 protoplasts; (2) the expression of CAT3 inhibited the silencing suppressor ability of the coexpressed 2b in N. benthamiana protoplasts and the necrosis induced by 2b in Col-0 protoplasts; and (3) the overexpression of CAT3 reduced the accumulation of CMV and delayed the necrosis in transgenic Col-0 plants. When these results are considered together, CAT3 seems to have the potential to inhibit CMV infection but normally not to sufficiently function as a natural defense protein. In fact, it is not even induced by CMV infection (Fig. 4); thus, Arabidopsis does not seem to have developed it as a defense system right against CMV. However, we will not presume to deny CAT3’s potential, because there is certainly the case where a host factor binds to a viral RSS to inhibit the RSS activity. For example, in Tobacco mosaic virus (TMV), the host factor TOM1 binds to the RSS (130k protein) of TMV and recruits it on the membrane, resulting in reduced RSS activity (Yamanaka et al., 2000).
The interaction between a host and a viral factor also must be discussed from the obverse side of defense. Catalase activity can be compromised by the binding of 2b to CAT3. We here demonstrated that the translocation of CAT3 to the nucleus was not essential. However, the CAT3 malfunction is likely to be irreversible after CAT3 is forced into the nucleus; thus, it may still contribute to the necrosis formation in the long run. Although this necrosis was shown to be hypersensitive response like, it is induced directly by a simple protein-protein interaction but not in a decent signal transduction for defense. Part of host defense responses may have been activated because some PR genes were elevated, but CMV was not restricted with the necrosis at all, suggesting that any coordinated defense system did not operate against CMV in association with this 2b-mediated necrosis.
It is conceivable, therefore, that a viral protein may bind to a host factor and/or further alter the localization of the host protein to specifically induce a viral symptom. Considering that most CMV strains do not cause necrosis in Arabidopsis, the necrosis-inducing CMVs such as CMV-HL may have developed a strategy to interfere with the catalase-mediated defense to more efficiently multiply by establishing the binding between 2b and catalase in viral evolution. In conclusion, our study here suggests that the HL2b-CAT3 interaction serves as a necrosis determinant, rather than a potential defense mechanism, perhaps in favor of the virus.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 and the transgenic plants expressing HL2b or CAT3 were used in these experiments. Seeds were directly sown in peat pellets and grown in an MLR-350 growth chamber (Sanyo) at 21°C with a 12-h photoperiod (150 μmol m−2 s−1).
Plant Transformation
The cDNAs of HL2b and CAT3 were inserted into the multiple cloning site of the plant expression vector pBE2113 (Mitsuhara et al., 1996). Agrobacterium tumefaciens strain EHA105 was first transformed with the vector construct by the freeze-thaw method (Holsters et al., 1978). Col-0 plants were then transformed with pBE2113-HL2b or -CAT3 by the vacuum infiltration method (Bechtold et al., 1993). Transformants were selected on a solid medium containing 2% Suc supplemented with half-strength Murashige and Skoog nutrients and kanamycin (50 μg mL−1).
Viral Inoculation and H2O2 Detection
The plasmids containing the full-length cDNAs of RNAs of CMV-Y or CMV-HL were transcribed in vitro. The leaves of 4-week-old plants of Nicotiana benthamiana were dusted with carborundum and rub inoculated with the in vitro-transcribed RNAs. For Arabidopsis, 3-week-old plants were inoculated with the sap of an infected N. benthamiana leaf. Successful infection was confirmed by conventional ELISA (Kim et al., 2010). Viral infection was confirmed using tissue prints and anti-CMV antibodies essentially as described by Kaneko et al. (2004). H2O2 generation in leaf tissues was detected using H2DCFDA (Kim et al., 2008).
Quantitative Real-Time RT-PCR for CMV RNAs
Total RNA was isolated using Trizol reagent (Invitrogen) as described previously (Kim et al., 2008). For the real-time RT-PCR, 1 μg of total RNA was used as a template for cDNA synthesis. The first-strand cDNA was synthesized by the Takara RNA PCR Kit with a random primer. Real-time RT-PCR was carried out using 1 μL of the RT reaction mixture and SYBR Green mixture (Takara) in the DNA Engine Opticon 2 System (MJ Research). A specific primer pair for the 3′-end sequence of CMV RNA3 (352 bp) was used to measure CMV RNAs (Yamaguchi et al., 2005). For the amplification of the Arabidopsis CAT genes, the following primer pairs were used: AtCAT1-5 (5′-ATCTGCTTGAGAAACTCGCTAACT-3′) and AtCAT1-3 (5′-ATGACAGTTGAGAAACGAACGATA-3′) for CAT1; AtCAT2-5 (5′-GGATGGTTCAGGTGTCAATACATA-3′) and AtCAT2-3 (5′-GATAAAGAGCTTCCATTCAGGGTA-3′) for CAT2; and AtCAT3-5 (5′-GGAATTCGAACAAAGTGCGT-3′) and AtCAT3-3 (5′-AAGGATCGATCAGCCTGAGA-3′) for CAT3. For all the Arabidopsis genes analyzed by real-time PCR, the β-tubulin sequence was amplified as an internal control using primer pair Attub-5-200 (5′-GAGGGAGCCATTGACAACATCTT-3′) and Attub-3-200 (5′-GCGAACAGTTCACAGCTATGTTCA-3′).
Western-Blot Analysis
Total proteins from Col-0, Col-HL2b, and Col-CAT were extracted as described by Masuta et al. (1995). The samples were first boiled for 5 min, separated on a 15% SDS page gel, and blotted to a polyvinylidene difluoride membrane (Millipore). The blots were probed with antibodies against CMV 2b or CAT3. Proteins were visualized using anti-mouse secondary antibodies conjugated to alkaline phosphatase, followed by treatment with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate as described previously (Sato et al., 2003).
Detection of Catalase Activities
Total protein of Arabidopsis seedlings was extracted from 0.1 g of tissue by grinding to a fine powder in liquid nitrogen and homogenized in 0.4 mL of the extraction buffer (50 mm phosphate buffer, pH 7.0, and 1% Triton X-100). After centrifugation at 12,000g for 10 min at 4°C, the proteins in the supernatant were quantified by a protein assay kit (Bio-Rad). In the catalase activities assay, 50 μg of total protein was added to 0.9 mL of the buffered substrate (50 mm phosphate buffer, pH 7.0, and 20 mm H2O2). The degradation of H2O2 was determined by a decline in A240.
Immunostaining by in Situ PLA
Protoplasts were isolated from 4-week-old seedlings of Col-0 and transfected with the plasmids (GFP, HL2b, Y2b, CAT3-FLAG, HL2b + CAT3-FLAG, or Y2b + CAT3-FLAG) using polyethylene glycol-calcium-mediated transformation (Kim et al., 2010). The cDNA clones of HL2b, Y2b, and CAT3-FLAG were inserted into the cloning site in pBE2113 (Mitsuhara et al., 1996), and the control GFP gene was also cloned in pE2113. These protoplasts were fixed with 2% paraformaldehyde on l-poly-Lys-coated slides (Tariq et al., 2003; Yoo et al., 2007) 24 h after transfection. After rehydration in phosphate-buffered saline (PBS), the fixed protoplasts on the slides were blocked in 2% bovine serum albumin in PBS (30 min, room temperature) and incubated for 1 h at room temperature in 0.5% bovine serum albumin in PBS containing primary antibodies. A protein-protein interaction was detected using antibodies against CMV 2b or FLAG with Duolink in situ PLA probes (OLINK Biosience). The images were observed with a fluorescence microscope (Leica DMI 6000B). Red signals (TX Red) were visualized with excitation at 543 nm (emission, 585–615 nm), and blue signals (UV) were visualized with excitation at 365 nm (emission, 420–470 nm).
Observation of Cell Death in Protoplasts
Cell death of protoplasts was assessed with Evans blue (0.04%, w/v) staining 20 h after transfection as described previously (Kim et al., 2010). Our average transfection efficiency was approximately 70%. We counted the number of dead cells in several fields at 400×. Protoplasts were exposed to the dye for 5 min and then observed with light microscopy (Olympus BX51).
Protoplast Assay for RSS Activity
Protoplasts were prepared from leaves of N. benthamiana and used to analyze RSS activity essentially as described by Shimura et al. (2008). For this assay, we used the Dual-Luciferase Reporter Assay System (Promega) using firefly luciferase (Fluc) and Renilla luciferase (Rluc). As a silencing inducer, double-stranded (ds) RNA of the Fluc gene (dsFluc) was used. RNA silencing in protoplasts was induced by cotransfection of the reporter genes and dsFluc with or without the plasmids containing a viral suppressor gene, CMV-HL2b or P19 of Tomato bushy stunt virus. The protoplasts were then harvested 24 h after transfection and analyzed for the luciferase activity.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression levels of defense-related genes in the Col-0 plants of Arabidopsis inoculated with Y1H2Y3.
Supplemental Figure S2. In situ association between Y2b and CAT3 in Arabidopsis cells.
Supplemental Figure S3. Characterization of the Col-CAT transgenic plants.
Acknowledgments
We thank Kae Sueda, Eri Yoshimoto, and Shuichi Tanaka for their technical assistance.
References
- Bechtold N, Ellis J, Pelletier G. (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194–1199 [Google Scholar]
- Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J 17: 6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Contento AL, Bassham DC. (2010) Increase in catalase-3 activity as a response to use of alternative catabolic substrates during sucrose starvation. Plant Physiol Biochem 48: 232–238 [DOI] [PubMed] [Google Scholar]
- Ding SW, Anderson BJ, Haase HR, Symons RH. (1994) New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198: 593–601 [DOI] [PubMed] [Google Scholar]
- Ding SW, Li WX, Symons RH. (1995) A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J 14: 5762–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du YY, Wang PC, Chen J, Song CP. (2008a) Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J Integr Plant Biol 50: 1318–1326 [DOI] [PubMed] [Google Scholar]
- Du Z, Chen F, Zhao Z, Liao Q, Palukaitis P, Chen J. (2008b) The 2b protein and the C-terminus of the 2a protein of cucumber mosaic virus subgroup I strains both play a role in viral RNA accumulation and induction of symptoms. Virology 380: 363–370 [DOI] [PubMed] [Google Scholar]
- Foreman KE, Tang J. (2003) Molecular mechanisms of replicative senescence in endothelial cells. Exp Gerontol 38: 1251–1257 [DOI] [PubMed] [Google Scholar]
- Frugoli JA, Zhong HH, Nuccio ML, McCourt P, McPeek MA, Thomas TL, McClung CR. (1996) Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol 112: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González I, Martínez L, Rakitina DV, Lewsey MG, Atencio FA, Llave C, Kalinina NO, Carr JP, Palukaitis P, Canto T. (2010) Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Mol Plant Microbe Interact 23: 294–303 [DOI] [PubMed] [Google Scholar]
- Goto K, Kobori T, Kosaka Y, Natsuaki T, Masuta C. (2007) Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol 48: 1050–1060 [DOI] [PubMed] [Google Scholar]
- Ham BK, Lee TH, You JS, Nam YW, Kim JK, Paek KH. (1999) Isolation of a putative tobacco host factor interacting with cucumber mosaic virus-encoded 2b protein by yeast two-hybrid screening. Mol Cells 9: 548–555 [PubMed] [Google Scholar]
- Holsters M, de Waele D, Depicker A, Messens E, van Montagu M, Schell J. (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163: 181–187 [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Nishikiori M, Ishikawa M. (2010) Interactions between tobamovirus replication proteins and cellular factors: their impacts on virus multiplication. Mol Plant Microbe Interact 23: 1413–1419 [DOI] [PubMed] [Google Scholar]
- Ji LH, Ding SW. (2001) The suppressor of transgene RNA silencing encoded by Cucumber mosaic virus interferes with salicylic acid-mediated virus resistance. Mol Plant Microbe Interact 14: 715–724 [DOI] [PubMed] [Google Scholar]
- Kanazawa A, Inaba J, Shimura H, Otagaki S, Tsukahara S, Matsuzawa A, Kim BM, Goto K, Masuta C. (2011) Virus-mediated efficient induction of epigenetic modifications of endogenous genes with phenotypic changes in plants. Plant J 65: 156–168 [DOI] [PubMed] [Google Scholar]
- Kaneko YH, Inukai T, Suehiro N, Natsuaki T, Masuta C. (2004) Fine genetic mapping of the TuNI locus causing systemic veinal necrosis by turnip mosaic virus infection in Arabidopsis thaliana. Theor Appl Genet 110: 33–40 [DOI] [PubMed] [Google Scholar]
- Kim BM, Masuta C, Matsuura H, Takahashi H, Inukai T. (2008) Veinal necrosis induced by turnip mosaic virus infection in Arabidopsis is a form of defense response accompanying HR-like cell death. Mol Plant Microbe Interact 21: 260–268 [DOI] [PubMed] [Google Scholar]
- Kim BM, Suehiro N, Natsuaki T, Inukai T, Masuta C. (2010) The P3 protein of turnip mosaic virus can alone induce hypersensitive response-like cell death in Arabidopsis thaliana carrying TuNI. Mol Plant Microbe Interact 23: 144–152 [DOI] [PubMed] [Google Scholar]
- Lewsey M, Surette M, Robertson FC, Ziebell H, Choi SH, Ryu KH, Canto T, Palukaitis P, Payne T, Walsh JA, et al. (2009) The role of the Cucumber mosaic virus 2b protein in viral movement and symptom induction. Mol Plant Microbe Interact 22: 642–654 [DOI] [PubMed] [Google Scholar]
- Lucy AP, Guo HS, Li WX, Ding SW. (2000) Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J 19: 1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuta C, Tanaka H, Uehara K, Kuwata S, Koiwai A, Noma M. (1995) Broad resistance to plant viruses in transgenic plants conferred by antisense inhibition of a host gene essential in S-adenosylmethionine-dependent transmethylation reactions. Proc Natl Acad Sci USA 92: 6117–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Hong JS, Tabayashi N, Ito A, Masuta C, Matsumura T. (2007) Development of Cucumber mosaic virus as a vector modifiable for different host species to produce therapeutic proteins. Planta 225: 277–286 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37: 49–59 [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Peng Y, Kwok KH, Yang PH, Ng SS, Liu J, Wong OG, He ML, Kung HF, Lin MC. (2005) Ascorbic acid inhibits ROS production, NF-kappa B activation and prevents ethanol-induced growth retardation and microencephaly. Neuropharmacology 48: 426–434 [DOI] [PubMed] [Google Scholar]
- Sato M, Masuta C, Uyeda I. (2003) Natural resistance to Clover yellow vein virus in beans controlled by a single recessive locus. Mol Plant Microbe Interact 16: 994–1002 [DOI] [PubMed] [Google Scholar]
- Shi BJ, Miller J, Symons RH, Palukaitis P. (2003) The 2b protein of cucumoviruses has a role in promoting the cell-to-cell movement of pseudorecombinant viruses. Mol Plant Microbe Interact 16: 261–267 [DOI] [PubMed] [Google Scholar]
- Shimura H, Fukagawa T, Meguro A, Yamada H, Oh-Hira M, Sano S, Masuta C. (2008) A strategy for screening an inhibitor of viral silencing suppressors, which attenuates symptom development of plant viruses. FEBS Lett 582: 4047–4052 [DOI] [PubMed] [Google Scholar]
- Soards AJ, Murphy AM, Palukaitis P, Carr JP. (2002) Virulence and differential local and systemic spread of cucumber mosaic virus in tobacco are affected by the CMV 2b protein. Mol Plant Microbe Interact 15: 647–653 [DOI] [PubMed] [Google Scholar]
- Söderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, Landegren U. (2008) Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45: 227–232 [DOI] [PubMed] [Google Scholar]
- Sueda K, Shimura H, Meguro A, Uchida T, Inaba J, Masuta C. (2010) The C-terminal residues of the 2b protein of Cucumber mosaic virus are important for efficient expression in Escherichia coli and DNA-binding. FEBS Lett 584: 945–950 [DOI] [PubMed] [Google Scholar]
- Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J. (2003) Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Seshimo Y, Yoshimoto E, Ahn HI, Ryu KH, Choi JK, Masuta C. (2005) Genetic mapping of the compatibility between a lily isolate of Cucumber mosaic virus and a satellite RNA. J Gen Virol 86: 2359–2369 [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Ohta T, Takahashi M, Meshi T, Schmidt R, Dean C, Naito S, Ishikawa M. (2000) TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci USA 97: 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SF, Gao F, Cao XS, Chen M, Ye GY, Wei CH, Li Y. (2005) The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol 139: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]