Abstract
Gene networks involved in inorganic phosphate (Pi) acquisition and homeostasis in woody perennial species able to form mycorrhizal symbioses are poorly known. Here, we describe the features of the 12 genes coding for Pi transporters of the Pht1 family in poplar (Populus trichocarpa). Individual Pht1 transporters play distinct roles in acquiring and translocating Pi in different tissues of mycorrhizal and nonmycorrhizal poplar during different growth conditions and developmental stages. Pi starvation triggered the up-regulation of most members of the Pht1 family, especially PtPT9 and PtPT11. PtPT9 and PtPT12 showed a striking up-regulation in ectomycorrhizas and endomycorrhizas, whereas PtPT1 and PtPT11 were strongly down-regulated. PtPT10 transcripts were highly abundant in arbuscular mycorrhiza (AM) roots only. PtPT8 and PtPT10 are phylogenetically associated to the AM-inducible Pht1 subfamily I. The analysis of promoter sequences revealed conserved motifs similar to other AM-inducible orthologs in PtPT10 only. To gain more insight into gene regulatory mechanisms governing the AM symbiosis in woody plant species, the activation of the poplar PtPT10 promoter was investigated and detected in AM of potato (Solanum tuberosum) roots. These results indicated that the regulation of AM-inducible Pi transporter genes is conserved between perennial woody and herbaceous plant species. Moreover, poplar has developed an alternative Pi uptake pathway distinct from AM plants, allowing ectomycorrhizal poplar to recruit PtPT9 and PtPT12 to cope with limiting Pi concentrations in forest soils.
Phosphorus (P) is required for most plant processes, being essential for development and metabolism. The primary source of P for plants is inorganic phosphate (Pi). Since Pi is often present in limited amounts in soils, it represents the second most frequently limiting factor for plant growth in a variety of natural ecosystems (Marschner, 1995; Poirier and Bucher, 2002; Misson et al., 2004). Pi concentration in the soil solution hardly reaches 1 mm in forest soils and may drop to micromolar or even submicromolar levels at the root-soil interface, where a depletion zone is generated by rapid Pi uptake of plants and rhizospheric microorganisms (Poirier and Bucher, 2002; Plassard and Dell, 2010).
Higher plants use a series of adaptive morphological and biochemical strategies to increase the acquisition of poorly available Pi. These adaptive strategies are often based on root cell-specific activation of genes, the regulation of which can be conserved between distantly related species. In vascular plants, the root epidermis including root hair cells participates in direct nutrient uptake from the soil solution at the root-soil interface (Marschner, 1995). The formation of the arbuscular mycorrhizal (AM) symbiosis allows the establishment of a second interface involved in plant nutrient acquisition (i.e. at the intraradical root-fungus interface in the inner cortex). Thus, the AM symbiosis evolved to improve the nutrient acquisition efficiency of especially Pi, by forming far-reaching extraradical mycelia, which operate as functional extensions of the plant root system (Paszkowski et al., 2002; Kara–ov and Bucher, 2005).
The ectomycorrhizal (EM) fungi are a type of root symbionts that have been shown to improve nutrient availability mainly in tree species (Martin and Nehls, 2009). A typical ectomycorrhiza is characterized by a dense sheath of fungal hypha formed mainly around young lateral roots. EM hyphae extend intercellularly into the outer root cortex, forming the Hartig net (Martin and Nehls, 2009). Extraradical EM fungal hyphae secrete enzymes able to hydrolyze soil organic P compounds that are normally unavailable to non-EM roots (Plassard and Dell, 2010).
Specific transport systems are essential for the uptake of Pi and for its partitioning within plants (Poirier and Bucher, 2002). Kinetic studies have shown the coexistence of two transport systems with low or high affinity for Pi (Dunlop et al., 1997; Misson et al., 2004; Miller et al., 2009). Transport of Pi through plant membranes is mediated by a number of transporter proteins, which belong to at least three distinct protein families, and serves Pi partitioning and recycling of Pi from, for example, senescing leaves throughout a plant’s life cycle. The low-affinity transporters are active in the remobilization of acquired P (Smith et al., 2001), whereas the high-affinity Pi transporters play an important role in the acquisition of Pi. In annual plants, Himelblau and Amasino (2001) have found that about 80% of P is removed from senescing Arabidopsis (Arabidopsis thaliana) leaves. In barley (Hordeum vulgare), expression of the Pi transporter HvPT6, likely involved in Pi remobilization (Preuss et al., 2010), was increased as leaves aged (Rae et al., 2003).
The Pht1 family, comprising nine members in the Arabidopsis genome, includes proteins involved in the uptake of Pi from the soil solution and the redistribution of Pi within the plant. Members of this family are H+/Pi symporters (Bucher, 2007; Javot et al., 2007). Most of the genes of the Pht1 family that are expressed in roots are up-regulated in P-deprived plants (Mudge et al., 2002). The Pht2 family proteins share high similarity with mammalian Na+/Pi cotransporters but function as H+/Pi cotransporters in plastids of plants (Versaw and Harrison, 2002; Bucher, 2007). Expression patterns, Pi transport activities, and subcellular localization suggest roles for Pht4 proteins in the transport of Pi in plastids and the Golgi apparatus (Guo et al., 2008). The expression pattern of 11 PHO1 homologs in the Arabidopsis genome indicated a likely role of the PHO1 proteins in the transfer of Pi to the vascular cylinder of various tissues but also in Pi acquisition into cells, such as pollen or root epidermal cells and cells of the cortex (Wang et al., 2004).
The first described AM-inducible Pi transporter gene, StPT3 from potato (Solanum tuberosum), encodes a high-affinity Pi transporter (Rausch et al., 2001). The 1.7-kb-long StPT3 promoter directed GUS expression specifically in cortex cells harboring arbuscules and coiled hyphae in transgenic roots of several herbaceous plant species, including potato, Petunia hybrida, Daucus carota, Medicago truncatula, and Lotus japonicus (Rausch et al., 2001; Kara–ov et al., 2004), indicating evolutionary conservation of gene regulatory mechanisms in symbiotic Pi transport of distantly related plant species.
The major aim of this study was to establish a comprehensive genome-wide inventory of Pi transporters of the Pht1 family (TC 2.A.1.9) in poplar (Populus trichocarpa ‘Nisqually’) and to study their expression. Pht1 transporters are homologs of the yeast PHO84 Pi transporter, and they belong to the major facilitator superfamily of proteins (Pao et al., 1998). The release of the genome sequence of this perennial species (Tuskan et al., 2006) and the presence of an extensive EST database enabled genome-wide screening and the identification of 12 Pht1 transporter genes and corresponding transcripts. The genome organization and evolutionary history of the poplar Pht1 genes are described along with their expression patterns in various tissues during Pi starvation, senescence, and EM and AM symbioses. In addition, the regulation of the AM-specific poplar PtPT10 gene is revealed in transgenic potato plants harboring the PtPT10 promoter/GUS chimeric gene. Reciprocally, the conservation of solanaceous root hair-specific LeExt1 and AM-specific StPT3 promoter regulation is investigated in transgenic poplar roots.
RESULTS
Annotation and Distribution of Poplar Pht1 Genes
Twelve gene models coding for putative Pht1 transporters were identified in the predicted gene catalog resulting from the automated annotation of the P. trichocarpa genome assembly (version 1.1; http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). The curated set of Pht1 genes (for P. trichocarpa Pi transporter family 1) were named according to the Commission on Plant Gene Nomenclature as POPtr;Pht1;1 through POPtr;Pht1;12; for simplification, they will be called PtPT1 through PtPT12 in this article. ESTs have been identified for these 12 Pht1 transporters, confirming that they were expressed. In the P. trichocarpa genome assembly (version 1.1), these 12 putative Pht1 genes were located on scaffolds 125, 197, and 273 and mapped to linkage groups (LG) I, II, V, and X. In the P. trichocarpa genome assembly (version 2.0; http://www.phytozome.net), the Pht1 transporters were integrated into LG XIX, XV, and X (Supplemental Table S1; Supplemental Fig. S1). In genome assembly version 2.0, three more Pht1 transporters were predicted (Supplemental Table S2). Examination of these gene models revealed one truncated gene model (POPTR_0005s24510.1), another (POPTR_0005s15860.1) sharing 100% sequence similarity with PtPT6, and gene model POPTR_0005s24510.1 having 97% sequence similarity with PtPT1. These similarities concerned not only the coding sequences but also the upstream and downstream noncoding parts of the genes, thus making these gene models questionable. Therefore, they were not included in this study.
PtPT4 and PtPT7, sharing 96% sequence similarity, were closely located on LG V (Supplemental Table S3). These two genes belonged to tandem repeat sequences, that is, multiple copies of a similar nucleotide sequence within a less than 10-kb region of the genome (here a 5.94-kb region). PtPT4, PtPT6, PtPT7, and PtPT11 and PtPT1, PtPT2, and PtPT3, respectively, located on LG V and LG X are not tandem repeat sequences. PtPT4/PtPT7 and PtPT9/PtPT11 have 96% and 85% homology, respectively, suggesting that they are duplicated genes (Supplemental Table S3).
Primary and Secondary Structures of Pht1 Proteins
All putative poplar Pht1 proteins contained 12 predicted transmembrane domains (TM) separated into two blocks of six TM spanners by a long hydrophilic loop and hydrophilic N and C termini (Supplemental Table S1; Pao et al., 1998). The Pht1 signature GGDYPLSATIxSE (Kara–ov and Bucher, 2005), conserved among all Pht1 proteins, was identified. In three of them, the signature was slightly modified: in PtPT6, a Ser (S) was exchanged with an Ala (A); in PtPT8, a Thr (T) was exchanged with a Val (V); and in PtPT10, a Pro (P) was exchanged with a Ser (S). TM10 of PtPT10 shared a high similarity with the WW domain signature, also known as rsp5 or WWP, between amino acids 400 and 425, as reported for LePT4 and StPT4 (Nagy et al., 2005). Eight out of 12 Pht1 genes are intronless in poplar (Supplemental Table S1). The percentage identity between poplar and Arabidopsis Pht1 proteins reached values from 83.5% (PtPT4 against AtPT2) to 50.2% (PtPT9 against AtPT2; Supplemental Table S1).
Phylogenetic and Functional Analysis of Pht1 Proteins
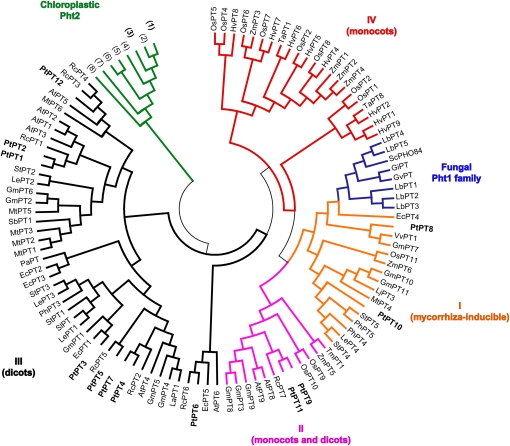
A neighbor-joining tree was constructed using a multiple sequence alignment of poplar Pht1 proteins and sequences from fungal and other plant Pi transporters. As in other dicot plant species, Pht1 proteins clustered into three subfamilies (Fig. 1). Poplar harbors two nonorthologous Pi transporters, PtPT8 and PtPT10, which belong to subfamily I. PtPT10 appeared to be closely related to AM-inducible StPT4 and LePT4 from two solanaceous species (potato and tomato [Solanum lycopersicum]; Nagy et al., 2005). Two Pht1 proteins belonged to subfamily II (PtPT9 and PtPT11), and eight Pht1 proteins belonged to subfamily III (PtPT1–PtPT7 and PtPT12). Growth of the Δpho84 yeast mutant expressing poplar Pht1 (PtPT1, PtPT2, PtPT4, PtPT6, PtPT9, or PtPT10) was assessed (Supplemental Fig. S2). As in other plants like tomato (Nagy et al., 2005) and maize (Zea mays; Nagy et al., 2006), we were not able to complement the yeast mutants for all poplar Pht1 proteins. Whereas the wild-type yeast strain could normally grow on both sufficient and low-Pi media, a Δpho84 strain exhibited a growth defect at low-Pi conditions. Yeast cells expressing PtPT1, PtPT2, PtPT4, PtPT9, and PtPT10 were not able to grow on low-Pi medium. On the contrary, yeast cells transformed with the pFL61 vector harboring PtPT6 cDNA showed partially restored growth. PHO84P was shown to function also as a high-affinity manganese transporter (Jensen et al., 2003). However, all the Pht1-expressing cells did not present any growth defect on toxic manganese medium (data not shown). Therefore, yeast complementation revealed that PtPT6 has high-affinity Pi transporter activity.
Figure 1.
Neighbor-joining tree for Pht1 transporters based on realigned sequences (745 amino acids ). Bootstrap values are from 500 replications. Sequence names consist of species code (first letter of genus and first letter of species name) and gene name. Accession numbers of the predicted proteins are given in Supplemental Materials and Methods S1. Roman numerals indicate four different Pht1 subfamilies thought to differ in evolutionary age (according to Nagy et al., 2005). Concerning chloroplastic Pht2 transporters, numbers correspond as follows: 1, POPtr;Pht2;2 (poplar); 2, LYCes;Pht2;1 (tomato); 3, POPtr;Pht2;1 (poplar); 4, ARAth;Pht2;1 (Arabidopsis); 5, TRIae;Pht2;1 (Triticum aestivum); 6, ORYsa;Pht2;1 (rice); 7, PHYpa;Pht2;1 (Physcomitrella patens); and 8, MEDtr;Pht2;1 (M. truncatula).
Gene Expression of Pht1 Transporters in Poplar Tissues
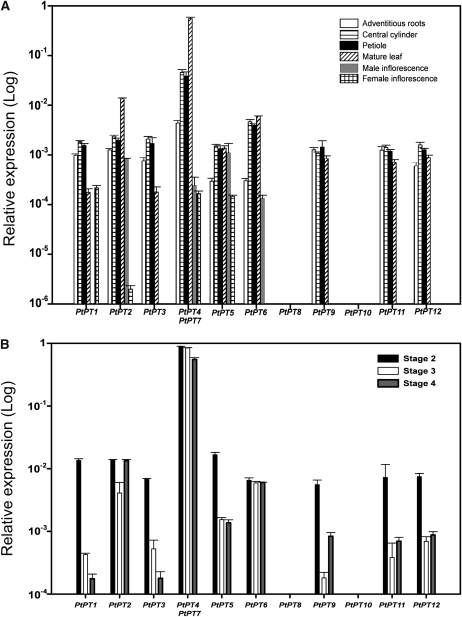
In poplar adventitious roots, PtPT1, PtPT2, PtPT3, PtPT4/PtPT7, PtPT9, PtPT11, and PtPT12 showed the highest expression levels, whereas PtPT5 and PtPT6 were expressed at 5-fold lower levels in the same tissue (Fig. 2A). In central cylinder, petiole, and mature leaves, the relative expression of PtPT1, PtPT2, PtPT3, PtPT5, PtPT6, PtPT9, and PtPT12 was similar, but PtPT4/PtPT7 were expressed at higher levels (from 5- to 100-fold). In inflorescences, PtPT3, PtPT9, PtPT11, and PtPT12 were not detectable. PtPT2, PtPT4/PtPT7, and PtPT5 were expressed both in female and male inflorescences. PtPT6 and PtPT1 were expressed only in male and female inflorescences, respectively (Fig. 2A). In three bud developmental stages, PtPT4/PtPT7 were the most highly expressed genes (Fig. 2B). PtPT2 and PtPT6 were lowly, but similarly, expressed in these three stages. PtPT1, PtPT3, PtPT5, PtPT9, PtPT11, and PtPT12 were expressed at higher levels in stage 2 than in stages 3 and 4. Finally, PtPT8 and PtPT10 expression was not detected in these tissues.
Figure 2.
Quantification by qRT-PCR of the transcript levels of the 12 poplar Pht1 genes in different developmental stages. Ubiquitin was used as the reference transcript. The values are means of five replicates, and each replicate represented a pool from at least three plants. Error bars represent sd. A, Tissues from plantlets grown in the greenhouse. B, Three stages during bud development according to Couturier et al. (2007).
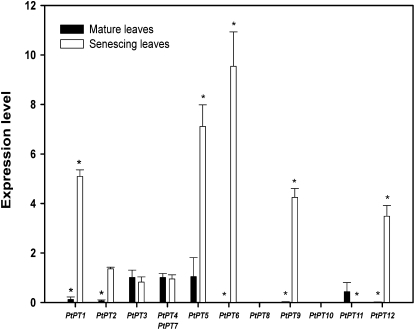
Young and mature poplar leaves showed similar transcript levels in PtPT3, PtPT4/PtPT7, PtPT5, and PtPT11 (Fig. 3). On the other hand, five Pht1 gene transcripts (PtPT1, PtPT5, PtPT6, PtPT9, and PtPT12) were significantly up-regulated (3-fold or greater) during autumnal leaf senescence. The highest increases (7.1- and 9.5-fold) corresponded to PtPT5 and PtPT6, respectively.
Figure 3.
Quantification by qRT-PCR of the transcript levels of the 12 poplar Pht1 genes upon autumnal leaf senescence. Transcript levels in young leaves were used as the control values. Ubiquitin was used as the reference transcript. The values are means of five replicates, and each replicate represented a pool from at least three plants. Error bars represent sd. Genes were considered to be regulated for expression when greater than or less than three. Student’s t test was performed for statistical analysis. Asterisks indicate P < 0.05.
Pht1 Expression Depends on Pi Availability and on Poplar Genotype
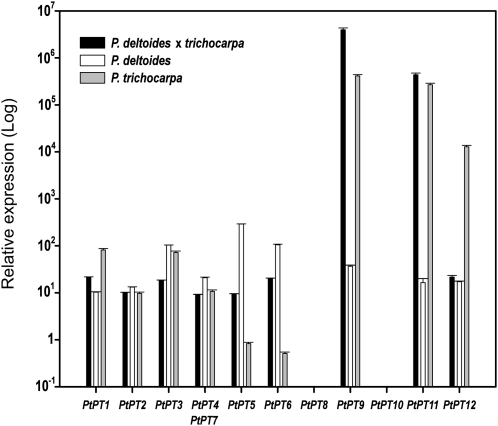
P. trichocarpa, Populus deltoides, and genotypes of the hybrid clone P. deltoides × P. trichocarpa were tested for their different responsiveness to Pi depletion (Fig. 4). As rooted cuttings were used, the root systems relied on both Pi uptake and Pi remobilized from perennial stem tissues. It was speculated, however, that the growth medium depletion would have a dramatic effect on Pi content of rapidly growing roots. P. deltoides was highly affected by Pi starvation, showing a low growth rate and a high number of necrotic spots on leaves, whereas the hybrid exhibited a high growth rate during Pi starvation and no indication of Pi deficiency in leaves (data not shown). Poplar exhibited an intermediate responsiveness to Pi deprivation. Plants growing in full-Pi nutrient solution were compared with plants growing in Pi-free nutrient solution. Removal of Pi enhanced the expression of most Pht1 genes in adventitious roots of the three different poplar genotypes (Fig. 4). In P. trichocarpa, PtPT9 and PtPT11 expression was up-regulated more than 10,000-fold, PtPT12 expression was about 1,000-fold higher, and PtPT1 and PtPT3 were up-regulated about 100-fold. In contrast, PtPT5 and PtPT6 were down-regulated about 2-fold at low-Pi conditions. In P. deltoides, the homologs of PtPT3, PtPT5, and PtPT6 were found to be up-regulated by more than 100-fold, whereas other homologs were moderately up-regulated by 10- to 40-fold. In the hybrid, the homologs of PtPT9 and PtPT11 exhibited transcript levels that were more than 100,000-fold higher than in control plants. Other Pht1 gene homologs were moderately up-regulated by 10- to 20-fold. Finally, expression of PtPT8 and PtPT10 was not detectable in control and Pi-depleted roots in any of the genotypes. Thus, there was an extensive variation in Pht1 gene expression levels in response to Pi deprivation between the two parental species and their hybrid.
Figure 4.
Quantification by qRT-PCR of the transcript levels of the 12 poplar Pht1 genes in response to Pi starvation. Analyses were performed in P. trichocarpa, P. deltoides, and the hybrid P. trichocarpa × P. deltoides. Transcript levels in roots with Pi were used as the control values. Ubiquitin was used as the reference transcript. The values are means of five replicates, and each replicate represented a pool from at least three plants. Error bars represent sd.
Coregulation and Interspecific Conversation in Pht1 Gene Expression in EM and AM Roots
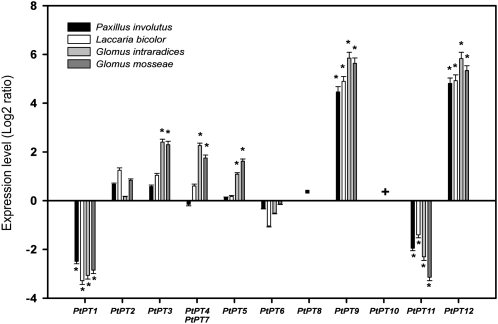
The expression level of each Pht1 gene was quantified in Populus tremula × Populus alba roots colonized either by EM fungi (Laccaria bicolor and Paxillus involutus) or AM fungi (Glomus intraradices and Glomus mosseae). Expression of PtPT1 and PtPT11 was repressed in both EM and AM roots, whereas expression of PtPT9 and PtPT12 was strongly enhanced in both symbiotic associations (Fig. 5). PtPT8 and PtPT10 transcripts were not detectable in both nonmycorrhizal and EM root tips. PtPT3, PtPT4/PtPT7, and PtPT5 expression was enhanced in AM roots only. In AM roots, transcripts coding for PtPT8 were not detected, whereas PtPT10 was only found to be expressed in these symbiotic tissues (Fig. 5).
Figure 5.
Quantification by qRT-PCR of the transcript levels of the 12 Pht1 genes in P. tremula × P. alba roots colonized either by EM (P. involutus and L. bicolor) or AM (G. intraradices and G. mosseae) fungi. Transcript levels in nonmycorrhizal roots were used as the control values. The values are means of four replicates, and error bars represent sd. The ubiquitin gene mRNA was used as the reference transcript. Genes were considered to be regulated for expression when greater than or less than three. Student’s t test was performed for statistical analysis. Asterisks indicate P < 0.05. −, PtPT8 transcripts were not detectable under these conditions; +, PtPT10 expression was induced specifically by AM fungi in mycorrhizal poplar roots.
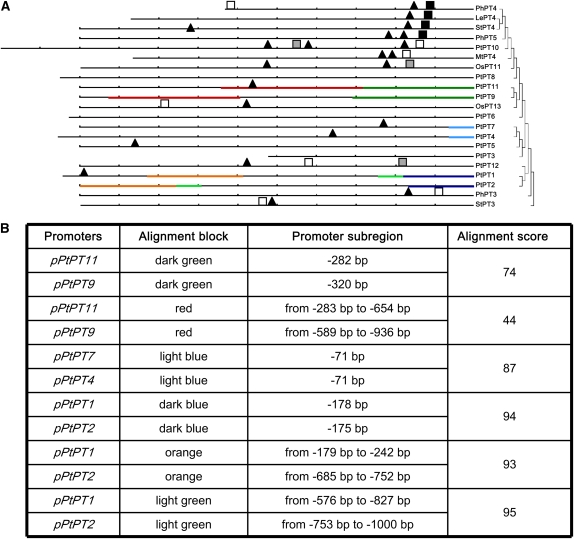
Pht1 gene regions of up to 2.4 kb upstream of their respective initiator ATG were analyzed and phylogenetically compared with orthologous Pht1 genes from other plant species (Fig. 6A). The P1BS cis-acting element (PHR1-binding site [GNATATNC]; Rubio et al., 2001; Schünmann et al., 2004) was found in the promoter region of PtPT1, PtPT4, PtPT7, PtPT10, PtPT11, and PtPT12. The P1BS element and the CTTC motif (CTTCTTGTTCTA; Kara–ov et al., 2004) were found in close proximity to each other within the 200 bp upstream of the ATG start codon of PtPT10 and PtPT12 and a set of other AM-specific Pi transporter genes (Fig. 6A). A second copy of the CTTC motif, exhibiting some sequence variation, was present in combination with the P1BS element in both the PtPT10 and PtPT12 promoters in a subregion between 400 and 600 bp upstream of the start ATG, similar to the StPT3 promoter (Kara–ov et al., 2004). Both the CTTC and P1BS motifs were present in the PtPT10 promoter sequences of both poplar Nisqually 1 and poplar clone INRA 101-74 in the same subregion. Moreover, the PtPT10 promoter sequence has three motifs out of 13 (TGTATTGATTTT, GTATTGATTTTT, and ATTTTAAAAAAA) similar with those from MtPT4 and StPT4 as well as a motif (CAATTATTT at −1,049) very similar to the motif reported in StPT3 (CAATTATTG at −1,227; Rausch et al., 2001).
Figure 6.
Phylogenetic footprinting of the Pht1 promoters from poplar and of mycorrhiza-inducible Pht1 transporters from other plant species (A), and similarity of the promoter subregions underlined in the phylogenetic footprinting (B). A, The distance between two tick marks represents 100 bp of promoter sequence upstream of the start ATG. The phylogram represents the phylogeny of the corresponding genes. The red lines represent the truncated CTTC motif TCTTGTTC, and the green lines represent the P1BS motif. Black shapes represent a 100% match to the extended motif. Gray shapes represent fewer changes in the motif. Blocks of similar promoter sequences (44% identity or greater) are underlined with the same color. B, The length of the alignment blocks in similar promoter subregions is indicated together with the percentage identity.
The other promoters of poplar Pht1 genes, and especially PtPT8 from Pht1 subfamily I (AM inducible), did not contain these motifs or variations of them, suggesting that they might not be regulated upon mycorrhization.
Phylogenetic footprinting also identified longer blocks of sequence homologies in closely related promoters (Fig. 6A). No similarity was detected in the promoter sequences of mycorrhiza-inducible Pht1 subfamily I genes PtPT8 and PtPT10 and in most of the poplar Pht1 gene promoters. In addition, the PtPT9 and PtPT11 promoters shared highly similar sequences in two different blocks (74% and 44%, respectively; Fig. 6B). About 300 bp separated the two blocks in the PtPT9 promoter, while there were no gaps in the PtPT11 promoter. A P1BS element was identified in the second block of the PtPT11 promoter but not in the PtPT9 promoter. High similarity was also found between the PtPT7 and PtPT4 promoters and the PtPT1 and PtPT2 promoters (Fig. 6B). Thus, computational promoter analysis suggested evolutionary conservation of AM-specific gene regulation in mycorrhizal tree and herbaceous plant species.
In view of reciprocal examination of AM-specific promoter activation in potato and poplar, the LeExt1 promoter mediating root hair-specific expression in solanaceous species (Bucher et al., 2002) was used to drive GUS expression in transgenic poplar. GUS staining was detected exclusively in root hair-bearing rhizodermal cells of P. tremula × Populus tremuloides roots grown in in vitro and aeroponic systems. GUS staining was absent in cells at the root tip and the root cap (Supplemental Fig. S3), which was consistent with the GUS expression pattern described in herbaceous species (Bucher et al., 2002). Overall, these results suggested that root cell type-specific cis-regulation of a solanaceous promoter is conserved in poplar.
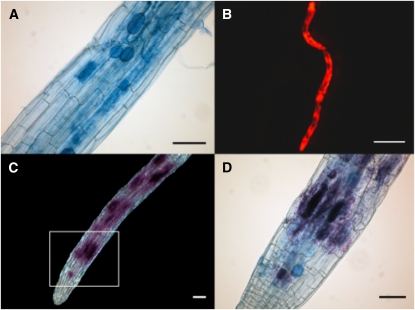
To study the evolutionary conservation of AM-specific Pi transporter gene regulation (Kara–ov et al., 2004), the activities of the 1.2-kb-long PtPT10 promoter and of the 1.7-kb-long AM-inducible StPT3 promoter were studied in transgenic hairy roots of potato and P. tremula × P. tremuloides, respectively. This cannot be assessed in P. trichocarpa, as this species is not yet amenable to routine genetic transformation. Mycorrhizal symbiosis was established between two potato hairy root genotypes (one wild type and one transgenic containing the 1.2-kb pPtPT10-GUS construct) and G. intraradices after 4 to 6 weeks (Fig. 7). There was no difference in colonization pattern between the two potato hairy root genotypes. GUS activity was colocalized with intracellular arbuscules and coiled hyphae in transgenic potato (Fig. 7, B–D), while GUS activity was absent in wild-type mycorrhizal potato (Fig. 7A) and in nonmycorrhizal wild-type and transgenic hairy roots from potato (data not shown). As in hairy roots of potato, there was no difference in colonization pattern between the two poplar hairy root genotypes (one wild type and one transgenic containing the 1.7-kb pSTPT3-GUS construct). However, mycorrhizal (Fig. 8, A–D) and nonmycorrhizal (data not shown) poplar hairy roots did not reveal GUS activity, indicating an absence of StPT3 promoter activity. In contrast, GUS activity colocalized with arbuscules and branched hyphae in transgenic potato root or hairy root controls both in vitro and in pot cultures (Fig. 8, E and F). Together, these results suggested that the mechanism involved in the regulation of the PtPT10 promoter is conserved between poplar and potato.
Figure 7.
Histochemical staining for GUS activity in mycorrhizal potato roots expressing the GUS reporter gene under the control of the PtPT10 promoter. A, Negative control showing mycorrhizal structures in nontransgenic (wild-type) roots after trypan blue staining (specific for fungal structures). No GUS staining of the hairy roots composite plants is visible, meaning that the fungus or any contaminating bacteria exhibit GUS staining in the absence of GUS expression. B, Mycorrhizal transgenic roots with Magenta-GUS staining (DsRED fluorescence) indicating transgenesis. C, Magenta-GUS staining colocalizes with trypan blue staining. D, Enlarged view of the white square in C. Bars = 0.1 mm except for 0.5 mm in B.
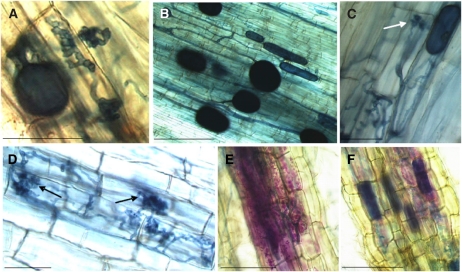
Figure 8.
Histochemical staining for GUS activity in mycorrhizal transgenic (A and B) and wild-type (C and D) P. tremula × P. tremuloides hairy roots and in mycorrhizal transgenic potato hairy roots (E and F) expressing the GUS reporter gene under the control of the StPT3 promoter. Mycorrhizal structures are visible in roots after ink blue staining (specific for fungal structures): finger-like coiled hyphae (A), vesicles (intracellular) and spores (extracellular; B), and arbusculate coils (arrows; C and D) in transgenic poplar plants and branched hyphae (E) and arbuscules (F) in transgenic potato plants. A and D show that Magenta-GUS staining does not colocalize with ink blue staining in transgenic and wild-type poplar hairy roots. E and F show a positive control, where Magenta-GUS staining colocalizes with ink blue staining in transgenic potato hairy roots. Bars = 200 μm.
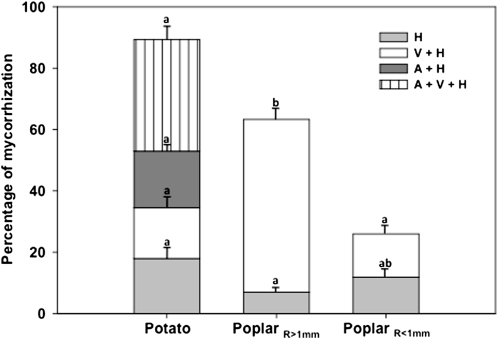
Next, we examined whether the AM colonization pattern in P. tremula × P. tremuloides revealed differences compared with that of potato. An intermediate Arum-Paris-type pattern of mycorrhizal colonization was observed both in poplar and potato. However, instead of dichotomous branching of hyphae that form the arbuscules, as in potato, poplar developed arbusculate coils (Fig. 8, A, C, and D) and coiled hyphae that occasionally ramified in finger-like structures (Fig. 8A). In vitro, the total percentage of colonization of hairy roots was about 32% in potato and 22% in poplar at both harvesting times (2 and 8 months; Supplemental Fig. S4). In pot culture, it was about 90% in potato and 44% in poplar. The detailed analysis of the mycorrhizal structures revealed a difference in the percentage of colonization (Fig. 9) between older-thick (PoplarR>1mm) and younger-thin (PoplarR<1mm) poplar roots. Overall colonization was almost 70% in thick roots and about 30% in thin roots. In potato, thick roots did not develop. The total percentage of root sectors containing arbuscules was significantly higher in potato (19%) than in poplar (1.7%; Fig. 9). The percentage of root sectors containing vesicles/spores and hyphae in thick poplar roots (56%; Fig. 9) was significantly higher than in potato (16%) and in thin poplar roots (14%; Fig. 9). In contrast, the percentage of root sectors containing vesicles/spores and hyphae in thin poplar roots was not statistically different from that of potato (Fig. 9). Thus, in pot culture, the percentage of root sectors containing vesicles/spores and hyphae is significantly higher in thick poplar roots compared with that of potato and thin poplar roots, while the percentage of root sectors containing vesicles/spores and hyphae in potato and in thin poplar roots is similar. In vitro, thick roots of poplar never developed, while in potato, all colonized roots stained positive irrespective of their thickness and age. Taken together, these results suggested that the mycorrhizal pattern of G. intraradices is dependent on the host plant species, and in the case of poplar grown in pot culture, also by the age/diameter of the roots. Thus, the absence of StPT3 promoter activation in poplar could be explained by the absence of arbuscules and a reduced number of coiled hyphae, which might have become inactive at the time of harvest.
Figure 9.
Colonization patterns of mycorrhizal hairy roots of potato and poplar harvested after 3 months of cocultivation with G. intraradices. Percentages of root segments containing hyphae (H), vesicles/spores and hyphae (V+H), arbuscules and hyphae (A+H), and arbuscules, vesicles/spores, and hyphae (A+V+H) are indicated and compared. Note that both in PoplarR>1mm and PoplarR<1mm, we observed 1.4% containing vesicles/spores and hyphae and 0.3% containing arbuscules, vesicles/spores, and hyphae. However, as data are low, it is not possible to show them clearly here. Different letters represent significantly different means between plant material (ANOVA lsd, P < 0.05).
Localization of PtPT10, an AM-Inducible Phosphate Transporter
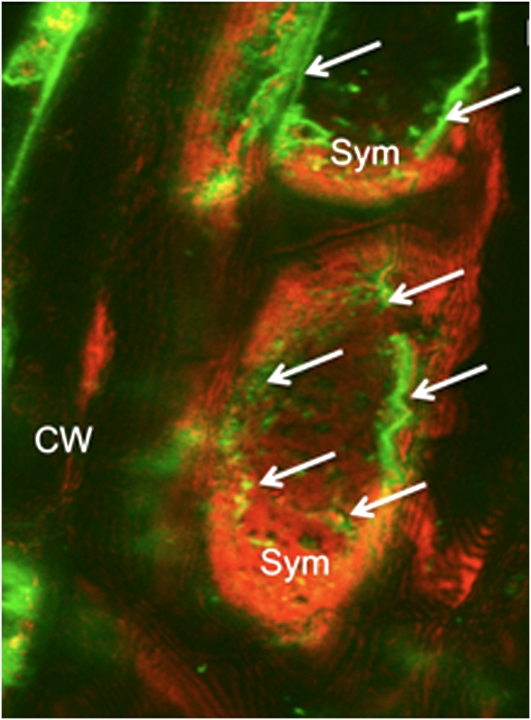
Two peptides were selected as antigens for the production of anti-PtPT10 antibodies. The selected peptides were not found in the deduced protein sequences of other poplar gene models. PtPT10 localization in G. intraradices-P. trichocarpa mycorrhizal root tips by indirect immunofluorescence is illustrated in Figure 10. Control images in which the mycorrhizal root sections were obtained by replacing primary anti-PtPT10 antibodies with preimmune immunoglobulin showed no signal. For cases in which AM roots were treated with anti-PtPT10 antibody followed by fluorescence-labeled secondary antibody, fluorescence was localized along the mycelial branches. The individual branches of the G. intraradices coiled hyphae were clearly visible (Fig. 10) into poplar cortical cells, and the PtPT10-related fluorescence signal colocalized with the symbiotic structures filling the cells. No signal was detected in noncolonized cortical cells.
Figure 10.
Immunolocalization of PtPT10 in symbiotic structures in root cortical cells of poplar colonized by the AM fungus G. intraradices. CW, Cell walls of cortical cells; Sym, G. intraradices symbiotic structures.
DISCUSSION
Little is known about the molecular basis of Pi transport in tree species and which components play a role in symbiosis-mediated Pi uptake. In this study, a comprehensive genome-wide inventory of the Pht1 family of Pi transporters in poplar cv Nisqually has been established, including expression regulation in different plant tissues and AM and EM symbioses.
Evolutionary History of Pht1 Subfamilies in Poplar
The phylogenetic analysis revealed the clustering of the 12 poplar transporters (Fig. 1) with members of the previously described Pht1 subfamilies (Kara–ov and Bucher, 2005; Nagy et al., 2005). Subfamily I clustered the AM-inducible Pi transporters from both monocot and dicot species, suggesting their evolution before the separation of the two plant groups (dicots versus monocots). Proteins encoded by the poplar EM- and AM-up-regulated Pht1 genes (PtPT9 and PtPT11) fall into the highly divergent subfamily II. Genes from dicot and monocot plants were found to be present in subfamilies III and IV, indicating their evolutionary divergence after the separation of these two primary groups of flowering plants from their common ancestor. In addition, the fact that all poplar members of subfamily III are intronless except PtPT3 suggested that they were generated by duplication from a common ancestral sequence. Whatever the subfamily, tandem duplications are clearly an important mechanism generating new gene copies in poplar (Tuskan et al., 2006). PtPT1/PtPT2, PtPT4/PtPT7, and PtPT9/PtPT11 are likely to be recent duplications, subsequently followed by divergence in gene expression levels and regulation of paralogous pairs. Moreover, this duplication is strongly supported by an analysis of promoter regions revealing that PtPT1/PtPT2, PtPT4/PtPT7, and PtPT9/PtPT11 have longer blocks of sequence similarities in promoter subregions.
Transcriptional Control of Poplar Pht1 Gene Expression
Homologous Pht1 genes exhibited substantial differences in their regulation in the various organs examined. PtPT4/PtPT7 showed high transcript levels in leaf and root tissues compared with other Pht1 genes (Fig. 2A). The transcripts coding for PtPT1, PtPT5, PtPT6, PtPT9, and PtPT12 showed strong up-regulation in the oldest leaves undergoing autumnal senescence (Fig. 3), suggesting that they play a role in Pi remobilization from source to sink tissues, which may be specific for the perennial lifestyle of poplar. During bud development, PtPT1 and PtPT2 were up-regulated in a similar way as their homolog AtPT1 from Arabidopsis Pht1 subfamily III (Figs. 1 and 2B; Mudge et al., 2002).
Upon Pi starvation, PtPT9 and PtPT11 showed dramatic up-regulation in poplar (Fig. 4), as did their orthologs in the hybrid P. trichocarpa × P. deltoides. PtPT9 and PtPT11 are highly homologous to AtPT8 and AtPT9, two genes from the Pht1 subfamily II (Fig. 1), which were also shown to be strongly up-regulated during Pi starvation (Mudge et al., 2002). Therefore, these Pht1 transporters likely play a role in Pi scavenging within the Pht1 family during conditions of low soil Pi availability. P1BS-like elements were identified in six (PtPT1, PtPT4, PtPT7, PtPT11, PtPT10, and PtPT12) of the 12 Pht1 promoter sequences, as in a range of Pi-regulated genes in dicots and monocots (Fig. 6A). In Arabidopsis, the MYB transcription factor PHR1 (At4g28610) binds to the P1BS element and is a key regulator in the Pi starvation response (Rubio et al., 2001). An ortholog of this MYB transcription factor (Joint Genome Institute protein identifer 711538), sharing 57% similarity with the PHR1 protein, was located on LG II of the poplar draft genome, suggesting PHR1-dependent regulation of a set of Pht1 genes in poplar.
Two nonorthologous mycorrhiza-inducible Pht1 genes were identified in solanaceous plant species (i.e. LePT3/LePT4 in tomato and StPT3/StPT4 in potato, as well as their orthologs from other solanaceous species; Nagy et al., 2005; Chen et al., 2007) belonging to Pht1 subfamily III and I, respectively, and a single gene or two genes in leguminous (MtPT4 and LjPT3 in M. truncatula and L. japonicus, respectively) and poaceous (OsPT11 and OsPT13 in rice [Oryza sativa] and ZmPT6 in maize) species were also identified (Harrison et al., 2002; Güimil et al., 2005; Maeda et al., 2006; Nagy et al., 2006). In poplar, PtPT8 and PtPT10, phylogenetically related to the AM-inducible Pht1 genes from subfamily I, are functionally different; only PtPT10 was specifically induced in poplar roots colonized by AM fungi. Analysis of the promoter region revealed the presence of the CTTC motif in PtPT10 but not in PtPT8 (Fig. 6A). This motif was shown to be highly conserved in AM-inducible Pht1 genes in dicots (Kara–ov et al., 2004; Chen et al., 2011) but was never described in trees and especially in Malphigiales, to which poplar belongs.
Interestingly, AM and EM development led to the up-regulation of two other Pht1 transporters, PtPT9 and PtPT12 of subfamily II and III, respectively, suggesting that they may have evolved with both symbiotic lifestyles and that different signaling pathways control Pi uptake in these mycorrhizal symbioses. Although the drastic accumulation of PtPT9, PtPT10, and PtPT12 transcripts upon mycorrhizal colonization could be symbiosis specific, it may also be the result of a secondary effect of enhanced Pi supply (i.e. Pi provided by the fungus). In poplar rooted cuttings grown on Pi-depleted medium, PtPT10 was not responsive to Pi availability whereas PtPT9 and PtPT12 were strongly up-regulated. Thus, PtPT10 is regulated during AM but not EM symbiosis, indicating that the regulation of AM-inducible Pht1 genes is conserved in woody and herbaceous species. Moreover, our data suggested that an alternative regulatory pathway distinct from the AM regulatory pathway developed during evolution, which allowed EM plants to recruit PtPT9 and PtPT12 for symbiotic Pi uptake in EM root tips. Alternatively, during both AM and EM development, PtPT9 and PtPT12 expression may be triggered by low-Pi conditions at the symbiotic interfaces. This latter scenario could explain the high level expression of the AM fungal Pi transporter gene GmosPTA from G. mosseae in arbuscules of mycorrhizal tomato roots (Balestrini et al., 2007) or the expression of EM fungal Pi transporter genes expressed in the hyphae emanating from and/or forming the extraradical mycelium (Tatry et al., 2009). These results showed that at least three poplar Pi transporters are involved in acquiring Pi from AM and EM fungi, emphasizing the complexity of Pi acquisition in perennial trees with access to several symbiotic and nonsymbiotic Pi uptake pathways. Our results also suggested that the mycorrhizal Pi uptake pathway in poplar is controlled to a large part by the host plant through the differential activation of symbiosis-specific Pi transporter genes. This mechanism is in agreement with the observation that the pattern of secretory enzyme activities of EMs is largely determined by the poplar genotype (Courty et al., 2011).
To summarize, the poplar Pht1 members within each subfamily are highly homologous to each other in sequence and predicted structure, but their differential expression patterns in response to developmental stages, Pi starvation, and mycorrhizal interactions indicated functional diversification.
Transcriptional Regulation of the AM-Inducible PtPT10 Transporter
The LeExt1 promoter from tomato was previously found to be specifically active in root hairs of several herbaceous species, including three Solanaceae species (potato, tobacco [Nicotiana tabacum], and tomato), M. truncatula, Manihot esculenta, and Arabidopsis (Bucher et al., 2002; Mirabella et al., 2004). Root hair-specific LeExt1.1 promoter activity in transgenic poplar (Supplemental Fig. S3) indicated conservation of promoter regulation in roots of solanaceous and poplar species, which justified the efforts to investigate the reciprocal analysis of AM-specific Pht1 gene promoter regulation in potato and poplar (see below). Moreover, this opened up new possibilities for the modification of the rhizosphere of poplar trees through genetic engineering of root properties like the secretion of exoenzymes in root hairs (Zimmermann et al., 2003).
Immunocytolocalization demonstrated that PtPT10 protein accumulated around the symbiotic mycelial structures developed by G. intraradices in colonized cortical cells of its host. GUS expression directed by the PtPT10 promoter was investigated in transgenic potato and found to be induced upon AM colonization. Thus, the signal perception and transduction pathway mediating the mycorrhiza-specific regulation of Pi transport in dicot species can be expanded from the Solanales, Apiales, and Fabales (Kara–ov et al., 2004) to the Malphigiales (this study). However we found that the StPT3 promoter was not active in poplar AMs (Fig. 8). The lack of StPT3 promoter activation in AM could be explained by a lack of functional AM structures detected in poplar roots at the time of harvesting. Although colonization of root cells by coiled hyphae induced StPT3 promoter-directed GUS activity in potato (Kara–ov et al., 2004), coiled structures formed in poplar roots in different cultivation systems were not associated with visible GUS staining. It has been previously shown that the StPT3 promoter is not permanently active in every cell harboring coils or arbuscules, suggesting that the induction of StPT3 is temporally controlled (Kara–ov et al., 2004). Sequence, number, and spacing of the two promoter motifs, CTTC and P1BS, which are likely to be involved in the regulation of AM-inducible Pi transporter genes including PtPT10, varied between the plant species investigated here (Fig. 6A). Interestingly, the function of and cooperativity between the two motifs have recently been investigated in tobacco (Chen et al., 2011). The function of the CTTC, P1BS, and other motifs of still unknown function in the regulation of AM- or EM-inducible Pht1 genes in poplar remains to be elucidated.
AM Symbiosis in Perennial and Annual Plants
The AM symbiosis in poplar is poorly studied, and our results give new insights into the process governing the developmental pattern of AM colonization in this tree model species. In particular, we found that, contrary to potato, Glomus species do not form arbuscules in poplar but rather arbusculate coils that occasionally ramified in finger-like structures (Fig. 8A) under the chosen experimental conditions. The fraction of roots containing vesicles/spores was much higher in poplar roots than in potato. In vitro, a longer cocultivation time (8 months) with G. intraradices affected the pattern of colonization of poplar hairy roots but not that of potato hairy roots, suggesting that the development of the symbiosis proceeds more slowly in poplar than in potato (Smith and Read, 2008). Thick poplar roots were characterized by a higher percentage of colonization compared with thin roots. Thick roots could have delivered carbohydrates for a longer time to sustain fungal growth compared with young thin roots (Brundrett and Kendrick, 1990). Taken together, these results showed distinct differences between the developmental patterns of mycorrhizal colonization in poplar and potato. It remains to be shown to what degree these differences affect global gene expression in the two species.
CONCLUSION
This study opened up the characterization of Pi transporters from a perennial plant and provided a survey of the complete assortment of Pht1 proteins in this species. The results of this study suggested that members of the Pht1 family are expressed in a diverse range of tissues, thus fulfilling different functions, which may, in part, be essential for the growth and development of perennial species, such as leaf senescence or symbioses with AM and EM fungal species. Further exploitation of this system should substantially contribute to a better understanding of the signaling pathways involved in Pi acquisition and symbiosis development.
MATERIALS AND METHODS
Plant Growth Conditions for Tissues and Pi Starvation Analysis
Experiments were performed on cuttings of Populus trichocarpa (clone INRA 101-74), except for Pi starvation, where cuttings of Populus deltoides (clone INRA 54B14) and the hybrid from the two parents, P. deltoides × P. trichocarpa (clone INRA B060-283), were used. The hybrids were selected as being representative of a range of genotypes with differential response to low levels of P when grown on Pi-depleted medium. Poplar cuttings (about 20 cm length, 1 cm diameter) were planted into pots containing sterile, acid-washed sand and were watered with deionized water and fertilized twice weekly with a Pi-free or full-Pi nutrient solution (Supplemental Materials and Methods S1). All experiments were independently repeated five times, using a new batch of plants for each experiment. Each sample represented a plant material pool from at least three plants. Roots were inspected with a compound microscope to ensure the absence of ectomycorrhiza and endomycorrhiza.
Mycorrhizal Symbiosis and Pht1 Gene Expression
Populus tremula × Populus alba (INRA clone 717-1B4) cuttings (propagation by cuttings of 1-cm pieces under sterile conditions) were placed in plastic containers containing 150 mL of Murashige and Skoog (MS) agar and then transferred into a growth chamber with a 16-h-light/8-h-dark cycle at 22°C. After 1 month, well-rooted plantlets were inoculated either with EM fungi (Laccaria bicolor S238N and Paxillus involutus) or with AM fungi (Glomus intraradices BEG-75 and Glomus mosseae ISCB14; Supplemental Materials and Methods S1). Within the 16 weeks of the experiment, plants (n = 4) were growing in a greenhouse. From the second month, a low-nitrogen, low-P nutrient solution was applied weekly (Frey-Klett et al., 1997).
Infectivity and functioning of the symbiosis were also checked. Transverse sections of EM root tips were stained with ink, and the presence of a well-developed Hartig net was observed. Sections of L. bicolor from the same plants were also provided in Figure 1 of Martin et al. (2008). A subsample of AM root tips was stained with ink (to avoid staining of poplar vascular tissues), and the colonization and the presence of arbuscules were checked with a microscope.
Growth Conditions of Wild-Type and Transgenic Poplar
Cuttings of wild-type plants and transformants of P. tremula × Populus tremuloides cv T89 containing the pLeExt1.1::GUS construct (Tuominen et al., 1995) were grown in sterile glass jars on half-strength solid MS medium with 10 g L−1 Suc and 0.8% agar-agar (Merck; Hampp et al., 1996). After 1 month, 20 transgenic and wild-type poplar plants were transferred in the greenhouse to aeroponic systems. Thirty-two-week-old transgenic plants and six wild-type poplar plants were used in in vitro systems (Supplemental Materials and Methods S1). In both systems, plants were harvested after 3 to 4 weeks for subsequent GUS staining.
Generation and Growth of Wild-Type and Transgenic Potato
Wild-type and transformant potato plants (Solanum tuberosum var Désirée) containing the 1.7-kb pStPT3::GUS construct (Rausch et al., 2001) were propagated in vitro in glass jars (E+H Services AG Däniken) in 2× MS medium in a growth chamber. The plants were then used for the generation of hairy roots in vitro or transferred to a miniature greenhouse for mycorrhizal experiments (Supplemental Materials and Methods S1).
Growth of Hairy Roots of Potato and Poplar
The generation of poplar and potato hairy roots is described in Supplemental Materials and Methods S1. Wild-type and transgenic hairy roots from potato and P. tremula × P. tremuloides cv T89 containing the 1.7-kb pSTPT3-GUS construct or the 1.2-kb pPtPT10-GUS construct were grown on petri plates either alone or together with Daucus carota hairy roots colonized with G. intraradices BEG-75.
AM Inoculation of Wild-Type and Transgenic Poplar and Potato
In the in vitro systems, P. tremula × P. tremuloides cv T89 and potato hairy roots were inoculated with a hairy root culture of D. carota colonized with G. intraradices BEG-75 using bicompartmented petri dishes. Hairy roots of poplar and potato (a minimum of two plates) were harvested after 2 and 8 months of cocultivation. In pot systems, two plants per each of the transgenic poplar lines (16 in total) and two wild-type plants were grown alone or in cocultivation with mycorrhizal Plantago lanceolata as nurse plants in pots in a greenhouse for 3 months. Three transgenic and three wild-type potato plants were transferred from tissue culture to pots and infected with root pieces of P. lanceolata infected with G. intraradices BEG-75 as a fungal inoculum. The same nutrient solution was applied to all three plant species (Jentschke et al., 1999). For the mycorrhization experiments, the nutrient solution was totally depleted of NaH2PO4. Nonmycorrhizal control plants from both species were also grown as controls.
Histochemical GUS Staining and Staining of AM Fungi in Plant Roots
Samples of hairy roots or roots from P. tremula × P. tremuloides cv T89 and potato were collected and rinsed in water. GUS assays were performed as described (Kara–ov et al., 2004). The percentage of colonization was quantified according to the grid-line-intersect method as described (Brundrett et al., 1984). One hundred twenty root sectors were counted in 20 root segments scanned across an imaginary grid line of the microscope ocular with a 200× magnification. Total percentage colonization comprised root intercepts containing hyphae, vesicles/spores, arbuscules, or combinations of these fungal structures. Differences between means of variables were examined by ANOVA. Statistical significance for all analyses was accepted at P ≤ 0.05. Statistical analyses were performed using SPSS 10.1. Imaging using a Leica MZ FLIII stereomicroscope with appropriate filters was done in order to select transgenic hairy roots expressing the fluorescent protein DsRED1. Optical views of stained roots were obtained using a Leica DM1000 LED microscope. Images where then processed in silico using Adobe Photoshop CS3.
Samples, RNA Isolation, and Quantitative Reverse Transcription-PCR
For autumnal senescence studies, 20 leaves were sampled from an adult poplar growing on the campus of University Henri Poincaré in Vandoeuvre-lès-Nancy (for details, see Couturier et al., 2007) at 2:00 pm on December 5, 2005. The senescing stage of leaves was confirmed by measuring the transcript level of a Cys protease gene (Bhalerao et al., 2003; Andersson et al., 2004; Couturier et al., 2007). Male and female inflorescences were harvested on the same trees on March 29 (for details, see Couturier et al., 2007). For other tissues and Pi starvation analysis, samples were collected on poplar plantlets growing in the greenhouse with a Pi-free or full-Pi nutrient solution. Bud development and comparative leaf emergence stages were determined by assigning each sample a number between 0 and 5 as follows: 0, closed bud; 1, bud swelling; 2, breaking up of the scales; 3, scales and bud open; 4, emergence of the first leaf; 5, fully expanded leaf (young leaves). Mature leaves were harvested 1 month after young leaves. Adventitious nonmycorrhizal roots (lateral roots developing from preformed primordia along the length of the cutting) were collected after 2 months of cultivation. About 40 EM root tips (selected under binocular 40×) and 100 mg of AM root tips were collected 4 months after inoculation of well-rooted P. tremula × P. alba plantlets. All samples were immediately frozen in liquid nitrogen and stored at −80°C for further analysis.
The procedure used for RNA extraction and cDNA synthesis was as described by Courty et al. (2009) and in Supplemental Materials and Methods S1. Primers used as controls or for analysis had an efficiency of greater than 95% (Supplemental Table S4). Considering the high sequence similarity (98%) of PtPT4 and PtPT7, it was not possible to design specific PCR primers for the amplification of corresponding cDNAs.
Computational Identification and Characterization of Poplar Pht1 Transporters
Selection of the Pht1 models was based on EST support and homology to a curated set of proteins. Additionally, searches were performed with the use of a range of Pht1 sequences available from plants at the National Center for Biotechnology Information GenBank and UNIPROT (http://expasy.org/) to probe the P. trichocarpa genome database using BLASTN, TBLASTN, and BLASTP algorithms at the Joint Genome Institute P. trichocarpa BLAST server. The Pht1 sequences were also used in a TBLASTN query against the poplar sequence assemblies (Tuskan et al., 2006). The putative homologs that were detected were characterized based on conserved domains, identities, and E-values in comparison with known proteins. Poplar Pht1 gene models were edited when needed. The final set of Pht1 proteins is given in Supplemental Table S1. Prediction of putative TM segments for Pht1 proteins was performed using several programs (TMHMM, TM pred, Top Pred, and prodiv-TMHMM) available on the EXPASY Web site (http://expasy.org/tools/). For phylogenetic analysis, the Pht1 amino acid sequences were aligned (745 amino acids) with additional GenBank sequences with ClustalX (multiple alignment parameters are given in Supplemental Materials and Methods S1). Neighbor-joining trees were constructed using the Jones-Taylor-Thornton substitution rate matrix in MEGA version 4.1. Bootstrap analysis was carried out with 500 replicates. Promoter regulatory regions were analyzed using FOOTPRINTER 2.0 and the PLACE program (Higo et al., 1999).
Yeast Complementation with Poplar Pht1 cDNA
PtPT1, PtPT2, PtPT4, PtPT6, PtPT9, and PtPT10 Pi transporter cDNAs were amplified by PCR using primers designed to introduce unique restriction sites at the 5′ and 3′ ends of each gene. The yeast strains used in this study were BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and Δpho84 (BY4741; YML123c::kanMX4). While the wild-type strain could normally grow on both normal and low-Pi media, the Δpho84 strain exhibited a growth phenotype under the low-Pi condition. Yeast strains were obtained from Euroscarf (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/). The full detailed procedure is given in Supplemental Materials and Methods S1.
The Δpho84 strain was transformed with either the empty vector pFL61 (Δpho84) or pFL61 harboring the full-length cDNA of PtPT1, PtPT2, PtPT4, PtPT6, PtPT9, or PtPT10. The wild-type strain was also transformed with the empty vector pFL61 and was used as a positive control. Yeast transformants were grown in liquid Synthetic Define medium with uracil to achieve a final cell suspension of optical density at 600 nm = 1.1, and then 5-μL aliquots of serial dilutions were spotted on Synthetic Define medium (pH 5.8) containing either 6 mm (sufficient Pi) or 100 μm (low Pi) inorganic Pi. The plates were incubated at 30°C for 5 d.
Immunolocalization of PtPT10
Antibodies specific for the PtPT10-derived peptides 266-[5′-H2N-QESDKLSEIRASNNYE-CONH2-3′]-282 and 328-[5′-H2N-QAADVSALEEVYETSR-CONH2-3′]-344, corresponding to a nonconserved region in the protein, were raised in rabbits. The antisera and the peptides were obtained from the custom peptide antibody production program (Eurogentec). Immunolocalization was performed essentially as described by Blancaflor et al. (2001) and Harrison et al. (2002) with some modifications (Martin et al., 2008; Supplemental Materials and Methods S1) on poplar roots mycorrhizal or not with G. intraradices. Control images in which the mycorrhizal root sections were treated by replacing primary anti-PtPT10 antibodies with preimmune immunoglobulin at the same concentration showed no specific fluorescence.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene distribution of poplar Pht1 genes.
Supplemental Figure S2. Yeast complementation of different Pht1s.
Supplemental Figure S3. Histochemical localization of GUS activity in P. tremula × P. tremuloides roots expressing reporter gene GUS under the control of the LeExt1.1 promoter from tomato.
Supplemental Figure S4. Colonization patterns of mycorrhizal hairy roots of potato and poplar (harvested after 2 and 8 months of cocultivation with D. carota hairy roots colonized with G. intraradices).
Supplemental Table S1. Characteristics of the poplar Pht1 gene family.
Supplemental Table S2. Additional Pht1 genes in the poplar genome assembly (version 2.0; http://www.phytozome.net).
Supplemental Table S3. Identity/similarity pairwise matrix of the poplar Pht1 protein sequences.
Supplemental Table S4. Primers used for quantitative reverse transcription (qRT)-PCR analysis.
Supplemental Materials and Methods S1. Detailed procedures.
Acknowledgments
We thank Dr. Sarah Wegmüller for great support of phylogenetic footprinting, Dr. Marc Buée for assistance in mycorrhizal synthesis, Dr. Jérémy Couturier for providing RNA extracts from poplar tissues, Dr. Barbara Montanini for assistance in the yeast complementation assay, Dr. Catherine Bastien for kindly providing Populus cuttings, Dr. Valérie Legué and Joëlle Gérard for technical support on confocal microscopy, Prof. Dr. Thomas Boller and Prof. Dr. Andres Wiemken for valuable discussions, and Patrice Vion for managing the greenhouse experiments.
References
- Andersson A, Keskitalo J, Sjödin A, Bhalerao R, Sterky F, Wissel K, Tandre K, Aspeborg H, Moyle R, Ohmiya Y, et al. (2004) A transcriptional timetable of autumn senescence. Genome Biol 5: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini R, Gómez-Ariza J, Lanfranco L, Bonfante P. (2007) Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Mol Plant Microbe Interact 20: 1055–1062 [DOI] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, Erlandsson R, Björkbacka H, Birve SJ, Karlsson J, Gardeström P, Gustafsson P, Lundeberg J, et al. (2003) Gene expression in autumn leaves. Plant Physiol 131: 430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancaflor EB, Zhao L, Harrison MJ. (2001) Microtubule organization in root cells of Medicago truncatula during development of an arbuscular mycorrhizal symbiosis with Glomus versiforme. Protoplasma 217: 154–165 [DOI] [PubMed] [Google Scholar]
- Brundrett M, Kendrick B. (1990) The roots and mycorrhizas of herbaceous woodland plants. I. Quantitative aspects of morphology. New Phytol 114: 457–468 [DOI] [PubMed] [Google Scholar]
- Brundrett MC, Piche Y, Peterson RL. (1984) A new method for observing the morphology of vesicular-arbuscular mycorrhizae. Can J Bot 62: 2128–2134 [Google Scholar]
- Bucher M. (2007) Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol 173: 11–26 [DOI] [PubMed] [Google Scholar]
- Bucher M, Brunner S, Zimmermann P, Zardi GI, Amrhein N, Willmitzer L, Riesmeier JW. (2002) The expression of an extensin-like protein correlates with cellular tip growth in tomato. Plant Physiol 128: 911–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Gu M, Sun S, Zhu L, Hong S, Xu G. (2011) Identification of two conserved cis-acting elements, MYCS and P1BS, involved in the regulation of mycorrhiza-activated phosphate transporters in eudicot species. New Phytol 189: 1157–1169 [DOI] [PubMed] [Google Scholar]
- Chen A, Hu J, Sun S, Xu G. (2007) Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytol 173: 817–831 [DOI] [PubMed] [Google Scholar]
- Courty PE, Hoegger PJ, Kilaru S, Kohler A, Buée M, Garbaye J, Martin F, Kües U. (2009) Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytol 182: 736–750 [DOI] [PubMed] [Google Scholar]
- Courty PE, Labbé J, Kohler A, Marçais B, Bastien C, Churin JL, Garbaye J, Le Tacon F. (2011) Effect of poplar genotypes on mycorrhizal infection and secreted enzyme activities in mycorrhizal and non-mycorrhizal roots. J Exp Bot 62: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Montanini B, Martin F, Brun A, Blaudez D, Chalot M. (2007) The expanded family of ammonium transporters in the perennial poplar plant. New Phytol 174: 137–150 [DOI] [PubMed] [Google Scholar]
- Dunlop J, Phung H, Meeking R, White D. (1997) The kinetics associated with phosphate absorption by Arabidopsis and its regulation by phosphorus status. Aust J Plant Physiol 24: 623–629 [Google Scholar]
- Frey-Klett P, Pierrat JC, Garbaye J. (1997) Location and survival of mycorrhiza helper Pseudomonas fluorescens during establishment of ectomycorrhizal symbiosis between Laccaria bicolor and Douglas fir. Appl Environ Microbiol 63: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güimil S, Chang HS, Zhu T, Sesma A, Osbourn A, Roux C, Ioannidis V, Oakeley EJ, Docquier M, Descombes P, et al. (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA 102: 8066–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Jin Y, Wussler C, Blancaflor EB, Motes CM, Versaw WK. (2008) Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol 177: 889–898 [DOI] [PubMed] [Google Scholar]
- Hampp R, Ecke M, Schaeffer C, Wallenda T, Wingler A, Kottke I, Sundberg B. (1996) Axenic mycorrhization of wildtype and transgenic hybrid aspen expressing T-DNA indoleacetic acid-biosynthetic genes. Trees (Berl) 11: 59–64 [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J. (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau E, Amasino RM. (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158: 1317–1323 [Google Scholar]
- Javot H, Pumplin N, Harrison MJ. (2007) Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ 30: 310–322 [DOI] [PubMed] [Google Scholar]
- Jensen LT, Ajua-Alemanji M, Culotta VC. (2003) The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J Biol Chem 278: 42036–42040 [DOI] [PubMed] [Google Scholar]
- Jentschke G, Winter S, Godbold DL. (1999) Ectomycorrhizas and cadmium toxicity in Norway spruce seedlings. Tree Physiol 19: 23–30 [DOI] [PubMed] [Google Scholar]
- Kara–ov V, Bucher M. (2005) Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci 10: 22–29 [DOI] [PubMed] [Google Scholar]
- Kara–ov V, Nagy R, Wegmüller S, Amrhein N, Bucher M. (2004) Evolutionary conservation of a phosphate transporter in the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 101: 6285–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda D, Ashida K, Iguchi K, Chechetka SA, Hijikata A, Okusako Y, Deguchi Y, Izui K, Hata S. (2006) Knockdown of an arbuscular mycorrhiza-inducible phosphate transporter gene of Lotus japonicus suppresses mutualistic symbiosis. Plant Cell Physiol 47: 807–817 [DOI] [PubMed] [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants. Academic Press, San Diego [Google Scholar]
- Martin F, Aerts A, Ahren D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, et al. (2008) The genome of Laccaria bicolor provides insights into the mycorrhizal symbiosis. [DOI] [PubMed] [Google Scholar]
- Martin F, Nehls U. (2009) Harnessing ectomycorrhizal genomics for ecological insights. Curr Opin Plant Biol 12: 508–515 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Shen Q, Xu G. (2009) Freeways in the plant: transporters for N, P and S and their regulation. Curr Opin Plant Biol 12: 284–290 [DOI] [PubMed] [Google Scholar]
- Mirabella R, Franken C, van der Krogt GNM, Bisseling T, Geurts R. (2004) Use of the fluorescent timer DsRED-E5 as reporter to monitor dynamics of gene activity in plants. Plant Physiol 135: 1879–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L. (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nagy R, Kara–ov V, Chague V, Kalinkevich K, Tamasloukht M, Xu G, Jakobsen I, Levy AA, Amrhein N, Bucher M. (2005) The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J 42: 236–250 [DOI] [PubMed] [Google Scholar]
- Nagy R, Vasconcelos MJV, Zhao S, McElver J, Bruce W, Amrhein N, Raghothama KG, Bucher M. (2006) Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol (Stuttg) 8: 186–197 [DOI] [PubMed] [Google Scholar]
- Pao SS, Paulsen IT, Saier MH., Jr (1998) Major facilitator superfamily. Microbiol Mol Biol Rev 62: 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP. (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassard C, Dell B. (2010) Phosphorus nutrition of mycorrhizal trees. Tree Physiol 30: 1129–1139 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Bucher M. (2002) Phosphate transport and homeostasis in Arabidopsis. The Arabidopsis Book; 1: e0024, doi/10.1193/tab.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss CP, Huang CY, Gilliham M, Tyerman SD. (2010) Channel-like characteristics of the low-affinity barley phosphate transporter PHT1;6 when expressed in Xenopus oocytes. Plant Physiol 152: 1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae AL, Cybinski DH, Jarmey JM, Smith FW. (2003) Characterization of two phosphate transporters from barley: evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol Biol 53: 27–36 [DOI] [PubMed] [Google Scholar]
- Rausch C, Daram P, Brunner S, Jansa J, Laloi M, Leggewie G, Amrhein N, Bucher M. (2001) A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414: 462–470 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schünmann PH, Richardson AE, Vickers CE, Delhaize E. (2004) Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol 136: 4205–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Dickson S, Smith FA. (2001) Nutrient transfer in arbuscular mycorrhizas: how are fungal and plant processes integrated? Aust J Plant Physiol 28: 685–696 [Google Scholar]
- Smith SE, Read DJ. (2008) Mycorrhizal Symbiosis, Ed 3. Academic Press, San Diego [Google Scholar]
- Tatry MV, El Kassis E, Lambilliotte R, Corratgé C, van Aarle I, Amenc LK, Alary R, Zimmermann S, Sentenac H, Plassard C. (2009) Two differentially regulated phosphate transporters from the symbiotic fungus Hebeloma cylindrosporum and phosphorus acquisition by ectomycorrhizal Pinus pinaster. Plant J 57: 1092–1102 [DOI] [PubMed] [Google Scholar]
- Tuominen H, Sitbon F, Jacobsson C, Sandberg G, Olsson O, Sundberg B. (1995) Altered growth and wood characteristics in transgenic hybrid aspen expressing Agrobacterium tumefaciens T-DNA indoleacetic acid-biosynthetic genes. Plant Physiol 109: 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Versaw WK, Harrison MJ. (2002) A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14: 1751–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y. (2004) Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol 135: 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Zardi G, Lehmann M, Zeder C, Amrhein N, Frossard E, Bucher M. (2003) Engineering the root-soil interface via targeted expression of a synthetic phytase gene in trichoblasts. Plant Biotechnol J 1: 353–360 [DOI] [PubMed] [Google Scholar]