Abstract
Marine Synechococcus undergo a wide range of environmental stressors, especially high and variable irradiance, which may induce oxidative stress through the generation of reactive oxygen species (ROS). While light and ROS could act synergistically on the impairment of photosynthesis, inducing photodamage and inhibiting photosystem II repair, acclimation to high irradiance is also thought to confer resistance to other stressors. To identify the respective roles of light and ROS in the photoinhibition process and detect a possible light-driven tolerance to oxidative stress, we compared the photophysiological and transcriptomic responses of Synechococcus sp. WH7803 acclimated to low light (LL) or high light (HL) to oxidative stress, induced by hydrogen peroxide (H2O2) or methylviologen. While photosynthetic activity was much more affected in HL than in LL cells, only HL cells were able to recover growth and photosynthesis after the addition of 25 μm H2O2. Depending upon light conditions and H2O2 concentration, the latter oxidizing agent induced photosystem II inactivation through both direct damage to the reaction centers and inhibition of its repair cycle. Although the global transcriptome response appeared similar in LL and HL cells, some processes were specifically induced in HL cells that seemingly helped them withstand oxidative stress, including enhancement of photoprotection and ROS detoxification, repair of ROS-driven damage, and regulation of redox state. Detection of putative LexA binding sites allowed the identification of the putative LexA regulon, which was down-regulated in HL compared with LL cells but up-regulated by oxidative stress under both growth irradiances.
Marine cyanobacteria must cope with a wide range of fluctuating environmental factors that, by affecting the cellular redox status, may lead to oxidative stress (Asada, 1994; Hirayama et al., 1995; Latifi et al., 2005, 2009; Schwarz and Forchhammer, 2005; Takahashi and Murata, 2008). For instance, absorption of solar radiation, especially UV wavelengths, by dissolved organic matter in aquatic environments leads to the photochemical production of various reactive transients, including reactive oxygen species (ROS; Mopper and Kieber, 2000). These compounds are partially reduced forms of oxygen, between atmospheric molecular oxygen and its fully reduced form, water, and constitute powerful oxidizing agents. Environmental stress usually induces an increase of ROS intracellular production in photosynthetic organisms, particularly when photosynthetic electron transport outpaces the rate of electron consumption during CO2 fixation (Knox and Dodge, 1985; Latifi et al., 2009; Rastogi et al., 2010). This may occur when cells are exposed to excess light or UV radiation but also under nutrient and salt stress, which induce a slowing down of cell metabolism. For instance, Latifi et al. (2005) have shown that cells of Anabaena sp. PCC 7120 subjected to iron limitation exhibited a 100-fold increase in the amount of ROS compared with replete cells. Similarly, high salinity induces the release of hydrogen peroxide (H2O2) from estuarine Microcystis aeruginosa cells (Ross et al., 2006). Thus, as environmental stress is intimately linked to oxidative stress, disentangling the direct effects of environmental stressors from the indirect effects of stress-induced ROS formation is not an easy task.
In cyanobacteria and other oxygenic phototrophs, the major source of ROS is the photosynthetic apparatus, although respiratory machinery also contributes to the generation of ROS compounds (Asada, 1999; Mittler, 2002; Ledford and Niyogi, 2005; Nishiyama et al., 2006; Latifi et al., 2009). Within PSI, transfer of electrons from reduced iron-sulfur centers to oxygen leads to the formation of superoxide radicals (O2·-) that undergo further dismutation into H2O2 and hydroxyl radicals (OH·; Asada, 1999). PSII also contributes to ROS formation: at the PSII acceptor side, excessive reduction of electron acceptors, particularly during limitation of electron transport between PSII and PSI, may result in a reduction of molecular oxygen into O2·-, which subsequently results in the formation of H2O2 and OH·. At the PSII donor side, incomplete oxidation of water at the water-splitting manganese complex induces the formation of H2O2, which can be further oxidized into O2·- by manganese complex radicals or reduced into OH· by free metals released by damaged PSII (Chen et al., 1992; Vass et al., 1992; Keren et al., 1997; Pospísil, 2009). Energy transfer from photoexcited pigments to oxygen in the light-harvesting antenna also leads to the formation of singlet oxygen (Knox and Dodge, 1985; Zolla and Rinalducci, 2002).
Excess ROS may severely affect the cell by damaging proteins, cleaving nucleic acids, peroxidizing unsaturated fatty acids, and inhibiting protein synthesis; this can ultimately lead to growth inhibition and cell death (Asada, 1999; Storz and Imlay, 1999; Ledford and Niyogi, 2005; Nishiyama et al., 2006). In particular, ROS have been reported to induce deleterious effects on phytoplankton by affecting cell membranes and photosynthesis (Lesser, 2006). Thus, the photosynthetic machinery is not only a major source of ROS but also constitutes a major target for these compounds. ROS are indeed thought to be involved in PSII photoinactivation, a process that has been intensively studied in freshwater cyanobacteria (Tichý and Vermaas, 1999; Nishiyama et al., 2001, 2004, 2006; Song et al., 2006; Kanesaki et al., 2007; Murata et al., 2007). Although PSII photoinactivation continually occurs under regular and stable growth conditions, it may increase under environmental stress conditions that induce an increase in the rate of ROS formation (Long et al., 1994; Hirayama et al., 1995; Asada, 1999; Latifi et al., 2005; Nishiyama et al., 2006; Takahashi and Murata, 2008). Furthermore, photoinhibition induced by ROS seems to be higher in cyanobacteria than in other phytoplanktonic organisms (Drabkova et al., 2007).
PSII photoinactivation is thought to occur primarily at the level of the oxygen-evolving complex, immediately followed by the destruction of the PSII core protein D1 (Hakala et al., 2005; Nishiyama et al., 2006). To maintain photosynthesis, photoinactivation is countered by a rapid repair cycle that includes the proteolytic removal of photodamaged D1 protein, the coordinated high rate of de novo D1 protein synthesis, and its insertion into PSII (Park et al., 1995; Tyystjärvi and Aro, 1996; Nixon et al., 2005; Ohnishi et al., 2005; Nishiyama et al., 2006). Thus, cells must modulate PSII repair cycle activity to avoid outrunning the repair by PSII photoinactivation, which would result in net photoinhibition of photosynthesis (Aro et al., 2005; Six et al., 2008). The role of ROS in PSII photoinactivation has been much debated over the past 15 years and remains controversial (Adir et al., 2003; Edelman and Mattoo, 2008; Latifi et al., 2009; Nixon et al., 2010). ROS have been suggested to be involved in the direct inactivation of the PSII reaction center through direct D1 degradation and by increasing D1 susceptibility to proteases (Vass et al., 1992; Aro et al., 1993; Miyao et al., 1995; Okada et al., 1996; Keren et al., 1997; Lupínková and Komenda, 2004; Hideg et al., 2007). Another nonexclusive hypothesis proposed that ROS primarily affect PSII by inhibiting the synthesis of D1 and therefore impairing the repair cycle (Nishiyama et al., 2004, 2006; Takahashi and Murata, 2008). Strengthening this idea, Kojima et al. (2007) have shown that the translation elongation factor EF-G is a primary target of H2O2, thus suggesting that the synthesis of the D1 protein may be impaired at the elongation step of the protein translation machinery (Nishiyama et al., 2001, 2004, 2006).
Taken together, these data show the close interactions between light and ROS in photosynthetic organisms. On the one hand, the photosynthetic machinery constitutes a major source of ROS, with ROS generation particularly promoted when light absorption and the concomitant electron transport exceed the consumption of photochemically generated reductants. On the other hand, light and ROS seem to act synergistically in PSII photoinhibition, either through direct damage to PSII and/or by inhibiting repair. The influence of growth irradiance on the sensitivity to reactive oxygen stressors remains unclear. In particular, it is not well understood whether low light (LL)- and high light (HL)-acclimated cells, which show very different photophysiologies (Kana and Glibert, 1987a, 1987b; Kana et al., 1988; Moore et al., 1995; Six et al., 2004, 2005) and different tolerance to stress factors (MacDonald et al., 2003; Bouchard et al., 2005; Garczarek et al., 2008), are equally able to withstand oxidative stress. Cells acclimated to LL have a slow electron transport rate, presumably low rates of primary ROS generation, but would not be expected to maintain full induction of ROS detoxification paths. Cells acclimated to HL have a faster electron transport rate and presumably higher rates of primary ROS generation, but they are in parallel expected to maintain stronger induction of ROS detoxification paths. The net cellular response to additional ROS, therefore, depends upon the relative magnitudes of the underlying effects of primary cellular ROS generation and the counteracting effects of ROS detoxification.
In order to address this question, we used Synechococcus sp. WH7803, a physiologically well-characterized marine cyanobacterial isolate (Kana and Glibert, 1987a, 1987b; Toledo et al., 1999; Lindell and Post, 2001; Garczarek et al., 2008; Jia et al., 2010) for which the complete genome sequence (Dufresne et al., 2008) and a whole-genome microarray are available. We monitored PSII photophysiology and the global transcriptome response of LL- and HL-acclimated cells of this strain upon exposure to various concentrations of a direct reactive oxygen stressor, H2O2, and a cell-dependent O2·--producing agent, methylviologen (MV).
RESULTS
Effect of H2O2 and MV on PSII Function of LL and HL Cells
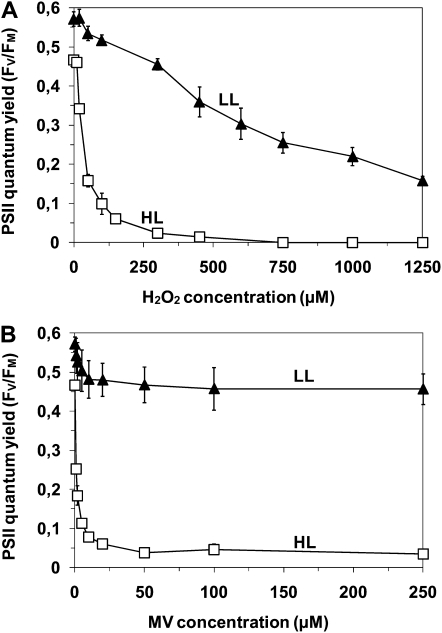
Synechococcus sp. WH7803 cells acclimated to LL (18 μmol photons m−2 s−1) and HL (250 μmol photons m−2 s−1) are clearly in different initial states. In particular, the initial PSII activity, as indicated by the PSII maximal quantum yield (Fv/Fm) values before the application of any treatment, is lower in HL-acclimated cells than in LL-acclimated ones (Fig. 1). Incubation of LL- and HL-acclimated Synechococcus in the presence of different concentrations of H2O2 and MV for 2 h under their respective growth light irradiances indicated that the higher the concentration of oxidizing agent, the stronger the decrease of PSII activity (Fv/Fm; Fig. 1). LL and HL cells, however, showed quite distinct photoinactivation patterns in response to oxidizing stress. For LL-acclimated cells, increasing concentrations up to 1,250 μm H2O2 induced a progressive PSII photoinactivation, but PSII of HL-grown cells was already completely inactivated at 250 μm H2O2 (Fig. 1A). This suggests a synergistic effect of light and H2O2 on PSII photoinactivation. Concentrations of MV higher than approximately 25 μm led to a stable PSII photoinactivation rate within 2 h, suggesting that 25 μm MV is a saturating concentration, with no additional ROS production above this level in both LL and HL cells (Fig. 1B). The saturating level of 25 μm MV induced a stress equivalent to exogenous application of about 125 μm H2O2, as at these concentrations both stressors induced a similar decrease of PSII quantum yield of about 20% for LL cells and 90% for HL cells.
Figure 1.
Effect of photoacclimation on PSII photochemistry. Fv/Fm of Synechococcus sp. WH7803 cells acclimated to LL (black triangles) or HL (white squares) was measured 2 h after the addition of a range of concentrations of H2O2 (A) or MV (B). These data are based on three to five independent experiments and are expressed as means ± sd.
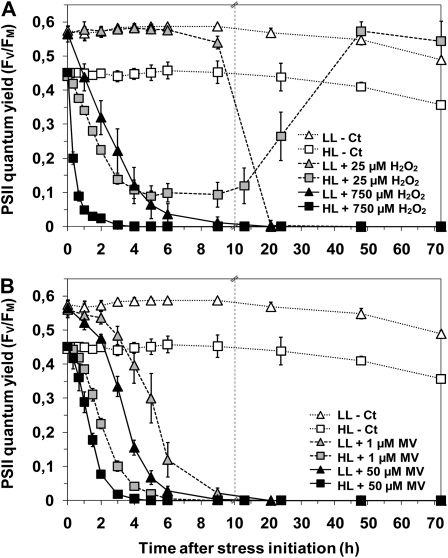
Following this first experiment, two H2O2 concentrations inducing a comparable 50% PSII photoinactivation after 2 h were selected, 750 μm H2O2 for LL cells and 25 μm H2O2 for HL cells (Fig. 1A), to monitor the effect of oxidative stress on PSII activity over 3 d (Fig. 2A). For MV, since the saturating level was reached quickly and this oxidizing agent induced a very different photoinhibition level depending on the growth irradiance at which the culture was acclimated, comparable photoinactivation levels in LL and HL cells could not be reached within 2 h. Thus, two concentrations of MV, a subsaturating one (1 μm) and a saturating one (50 μm), were chosen to monitor their kinetic effects on PSII inactivation (Fig. 2B). As expected from the previous experiment, HL cells, under their growth irradiance, exhibited much faster and stronger photoinhibition than did LL cells in response to both H2O2 and MV. At both MV concentrations tested, HL cells reached full PSII inactivation about 2.5 h earlier than LL cells. However, all cultures submitted to MV eventually died, independently of their growth irradiance. This is probably due to the inability of cells to get rid of MV, which continued to produce ROS as long as the MV redox cycle was maintained and MV·+ was continuously reoxidized (Supplemental Fig. S1; Bus and Gibson, 1984).
Figure 2.
PSII activity in response to H2O2 (A) and MV (B). The time course of changes in Fv/Fm of Synechococcus sp. WH7803 cells acclimated to LL (triangles) or HL (squares) was measured after the addition of 25 μm (gray symbols) or 750 μm (black symbols) H2O2 and 1 μm (gray symbols) or 50 μm (black symbols) MV. Control cultures without stress induction are indicated by white symbols. Broken vertical lines indicate changes of time scale on the x axis. These data are based on three to five independent experiments and are expressed as means ± sd.
The response to 25 μm H2O2 differed tremendously between HL- and LL-acclimated cultures. While LL cells did not show any change in their PSII quantum yield for more than 10 h, HL cells exhibited fast and dramatic PSII inactivation under their growth irradiance once exposed to 25 μm H2O2 (Fig. 2A). During the first 3 h, the rate of PSII inactivation was similar between LL cells incubated with 750 μm H2O2 and HL cells incubated with 25 μm H2O2. After this initial period, however, the evolution of PSII activity markedly differed between the two sets of cultures: while in LL cells subjected to 750 μm H2O2, photoinactivation led to a total shutdown of PSII activity, HL cells exposed to 25 μm H2O2 exhibited an increase of PSII function until full recovery after 3 d. Moreover, as measured by flow cytometry, the growth rate observed after this recovery was very similar to that observed before stress initiation (data not shown). Taken together, these data demonstrate the capacity of HL cells to recover after exposure to a low dose of H2O2 by completely detoxifying the cells from this oxidizing agent and restoring photosynthetic activity and most probably other affected cellular processes.
Effect of Light Acclimation and Electron Transport Rate on MV-Mediated ROS Production and PSII Inactivation
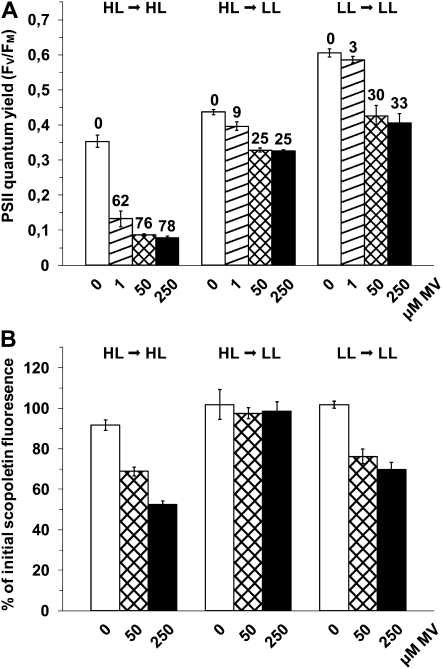
If it is correlated with photosynthetic electron flow at the PSI acceptor side, MV-mediated ROS production is expected to differ between LL- and HL-grown cells under their respective growth irradiance. To check this hypothesis, HL-acclimated cultures were either maintained under HL or shifted to LL conditions (inducing a sharp decrease in photosynthetic electron flow) 10 min before the addition of either subsaturating (1 μm) or saturating (50 or 250 μm) concentrations of MV (Fig. 3). For all MV concentrations tested, the shift to LL led to a much lower PSII inactivation after 2 h (Fig. 3A) and an interruption of H2O2 production, as assessed by the quasiconstancy of the fluorescence of scopoletin, a compound sensitive to H2O2 (Fig. 3B), compared with cells maintained under HL. This demonstrates that the MV-mediated PSII photoinactivation rate is highly dependent upon the photosynthetic electron transport rate. Surprisingly, the relative PSII photoinactivation in HL cells shifted to LL was only slightly lower than that of LL cells maintained in LL in the presence of MV (Fig. 3A), even though the H2O2 production was significantly higher in the LL cultures (Fig. 3B).
Figure 3.
Effect of photosynthetic electron transport on the MV-mediated photoinhibition and H2O2 production. A, Fv/Fm was measured 2 h after the addition of 0, 1, 50, and 250 μm MV (white, dashed, crossed, and dark bars, respectively) to Synechococcus sp. WH7803 cells acclimated to HL and maintained in HL (HL → HL), acclimated to HL but shifted to LL 10 min before the stress initiation (HL → LL), or acclimated to LL and maintained under LL (LL → LL). The percentage of photoinactivation of stressed cells compared with untreated cells for the same light treatment is indicated above the bars. B, Scopoletin fluorescence was measured 0.5 h after the addition of MV in the same conditions as in A. The decrease of fluorescence, relative to its initial value, is due to the scopoletin degradation by H2O2 from the cells. All these data are based on three independent experiments and are expressed as means ± sd.
Relation between D1 Protein Turnover and ROS-Dependent Photoinactivation of PSII
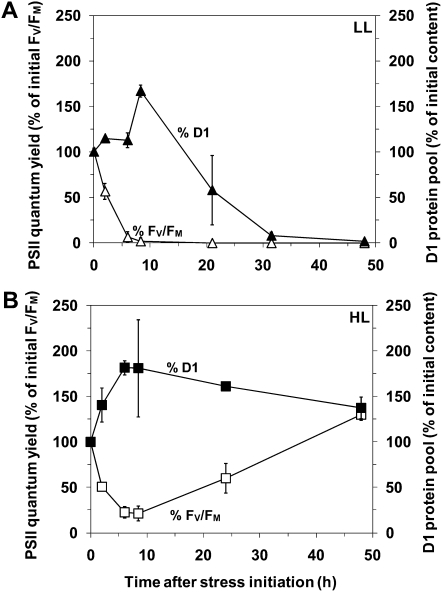
In order to compare the PSII repair capacities of LL and HL cultures exposed to 25 μm H2O2, we plotted the time-course variations of the relative D1 protein cell content (Fig. 4). While both LL and HL cells were able to increase their D1 protein content in response to oxidative stress during the first 8 h, only the HL cells sustained a high level of D1 protein over 2 d, thus enabling sufficient D1 protein turnover until complete recovery of PSII function. In contrast, PSII activity of LL cells dropped sharply and ceased after 30 h, suggesting that even a low dose of H2O2 provoked an irreversible impairment of the PSII repair cycle within about 1 d and likely subsequent cell death.
Figure 4.
D1 protein content in response to 25 μm H2O2. The D1 protein content (black symbols) in LL-acclimated (A) and HL-acclimated (B) Synechococcus sp. WH7803 cultures was measured by western-blot analysis after the addition of 25 μm H2O2 and expressed as percentage of the initial D1 content. The evolution of the PSII quantum yield after the stress initiation, expressed as percentage of initial values, is indicated by white symbols. These data are based on three independent experiments and are expressed as means ± sd.
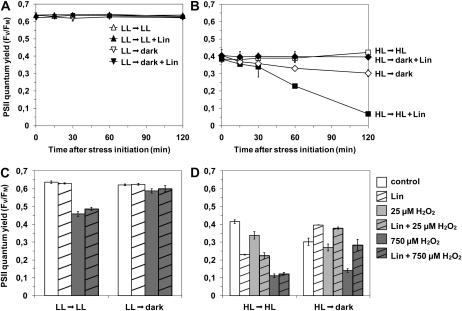
To assess the initial D1 repair capacities of HL and LL cells and the effect of H2O2 on the balance between PSII inactivation and repair, HL and LL Synechococcus cells were either maintained under their initial growth irradiance or transferred to dark and exposed to H2O2 and/or lincomycin, an inhibitor of protein synthesis. The results showed that no PSII damage was detectable after 2 h in LL-acclimated cells without the addition of any oxidant, either in the dark or under light, independently of the presence of lincomycin (Fig. 5A). In contrast, HL cells suffered a strong progressive light-dependent PSII inactivation in the presence of lincomycin (Fig. 5B), reaching 70% inactivation after 2 h compared with control (Ct) cultures without lincomycin. This demonstrates that, in contrast to LL cells, PSII complexes from HL Synechococcus undergo considerable photoinactivation under their growth irradiance, which must be counteracted by the induction of a fast PSII repair cycle at a much higher rate than in LL-acclimated cells.
Figure 5.
Effect of H2O2 on the balance between PSII damage and repair. To estimate the initial PSII repair capacity of Synechococcus sp. WH7803, cells acclimated to LL (A) and HL (B) were maintained under their initial growth irradiance (triangles and squares for LL and HL cells, respectively) or shifted to dark (upside down triangles and diamonds, respectively) in the presence (black symbols) or absence (white symbols) of lincomycin (Lin). Fv/Fm was then followed for 2 h. The effect of H2O2 on PSII damage and repair in LL (C) and HL (D) cells was estimated on cells maintained under their initial growth irradiance or shifted in the dark, by measuring the Fv/Fm 2 h after the addition of lincomycin (dashed bars) or water (open bars) and of 0 μm (white bars), 25 μm (light gray bars), or 750 μm (dark gray bars) H2O2. These data are based on three independent experiments and are expressed as means ± sd.
Second, the stronger effect on LL cells of 750 μm H2O2 plus lincomycin compared with lincomycin alone (Fig. 5C) suggested that, at this concentration and under light exposure, H2O2 induces direct damage on PSII complexes. This effect is seemingly light dependent, since no significant decrease of the Fv/Fm occurred upon the addition of H2O2 plus lincomycin in the dark. A similar photoinactivation under light with or without lincomycin indicated that the D1 repair rate was too low to be measured, either because it was initially low, due to the lack of light-dependent photodamages, and/or because of an inhibitory effect of H2O2. The absence of detectable light-driven damage in LL cells, which would necessitate an active repair (Fig. 5A), did not allow us to discriminate between these two hypotheses.
In HL cells, addition of 25 μm H2O2 and lincomycin provoked a drop in Fv/Fm equal to the HL plus lincomycin only treatment, suggesting that this amount of oxidizing agent did not induce additional direct damage to PSII above that induced by HL alone, when repair was fully blocked (Fig. 5D). In contrast, the 25 μm H2O2 treatment induced a modest decrease in PSII quantum yield compared with HL-Ct cells, suggesting some direct inhibitory effect of H2O2 on PSII repair. Notably, the inhibition was only partial at 25 μm H2O2, since Fv/Fm did not drop to the level obtained with the lincomycin treatment to fully block PSII repair. This H2O2 effect on repair was confirmed by the absence of any significant effect of 25 μm H2O2 alone in the dark, when no light-induced PSII damage occurred.
The stronger effect under light of 750 μm H2O2 plus lincomycin compared with lincomycin alone indicated that, at this concentration, the oxidant also induced direct damage to PSII (Fig. 5D). Furthermore, 750 μm H2O2 induced the same level of PSII photoinactivation with and without lincomycin, indicating that the HL cultures also underwent a complete inhibition of PSII repair at this oxidant concentration. Incubation of the culture in the dark further indicated that, in contrast to cells submitted to 25 μm H2O2, HL cultures exposed to 750 μm also underwent light-independent damage, since inactivation occurred with or without lincomycin in the dark.
Altogether, these results suggest that high concentrations of H2O2 affect photosynthetic activity by acting on PSII repair as well as by inducing both light-dependent and light-independent damage on PSII itself.
Global Transcriptomic Response of Synechococcus sp. WH7803 to Oxidative Stress
The global gene expression of Synechococcus sp. WH7803 was determined in response to H2O2 and MV doses leading to 50% PSII photoinactivation (note that the time needed to reach this state varies between treatments; Fig. 2). Microarray analyses were performed either by expressing the data with regard to the common reference hybridized on all arrays or by pairwise comparisons between nonstressed and stressed conditions (i.e. LL-Ct versus LL+MV, LL-Ct versus LL+H2O2, HL-Ct versus HL+MV, and HL-Ct versus HL+H2O2) or between HL and LL cultures (i.e. LL-Ct versus HL-Ct). Overall, 1,202 out of the 2,623 genes present on the array were significantly differentially expressed (false discovery rate [FDR] ≤ 0.05 using t test and/or LIMMA) in at least one of the pairwise comparisons. As expected, a higher number of genes were identified as differentially expressed in response to oxidative stress (450, 715, 530, and 585 genes in LL+H2O2 versus LL-Ct, LL+MV versus LL-Ct, HL+H2O2 versus HL-Ct, and HL+MV versus HL-Ct, respectively, when considering only the FDR cutoff) than between the two light acclimation conditions (121 genes). The complete set of gene expression data are available in Supplemental Table S1.
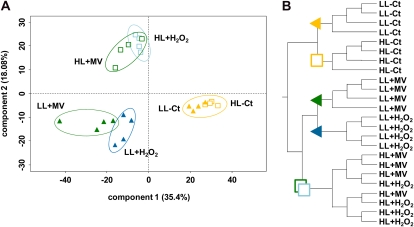
Principal component analysis (PCA; Fig. 6A) as well as hierarchical clustering dendrograms of the set of genes significantly differentially expressed in at least one of the pairwise comparisons (Fig. 6B) indicated good reproducibility among biological replicates used for microarray analyses, since all replicates grouped together except for the HL-stressed ones. This suggests a similar effect of H2O2 and MV on HL cells (Fig. 6). The most highly differentiated data sets were the oxidatively stressed cells versus Ct ones, independently of the reactive oxidizing agent used. The contribution of the stress factor seems to be dominant in PCA component 1, responsible for 35.4% of the variability of the data (Fig. 6A). The second major factor appears to be light acclimation, which segregated LL and HL data sets within both Ct and stressed conditions. This effect would be mainly represented by PCA component 2, responsible for 18.08% of the data variability. Furthermore, the greater distance between the HL and LL data sets in stressed conditions compared with Ct ones suggests that component 2 takes into account the synergistic effect of oxidative stress and HL acclimation.
Figure 6.
Multivariate analyses of microarray data. PCA was performed (A) and hierarchical clustering dendrograms were drawn (B) using the R Stats package on a subset identified as significantly differentially expressed (FDR ≤ 0.05) in at least one of the pairwise comparisons. Experiments with LL- and HL-acclimated cells are indicated by colored triangles and white squares, respectively. Treatments using 25 μm H2O2, 750 μm H2O2, and 50 μm MV are indicated by light blue, dark blue, and dark green symbols, respectively. Nontreated controls are in orange. In PCA, each replicate is indicated by one symbol, and percentages of variability accounted for by the two main components are shown near the axes.
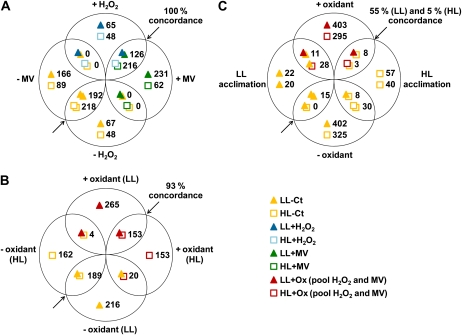
In order to identify the proportion of genes responding to the different treatments, Venn diagrams were created using the same data sets (Fig. 7). These analyses showed that MV and H2O2 induced a similar effect on both LL and HL cells (Fig. 7A). Indeed, 82% of the genes affected by H2O2 in HL cells (434 out of 530 genes) were also affected by MV, and 74% (434 out of 585) were affected the opposite way around. Very similar results were obtained for LL-acclimated cells, 71% and 44%, respectively. Furthermore, among the genes common to MV- and H2O2-induced stresses, 100% showed concordance, meaning that no gene activated by one oxidizing agent was repressed by the other. This indicates that both oxidative treatments induced similar response mechanisms.
Figure 7.
Pairwise comparisons of microarray data sets. Venn diagrams were created with genes showing a statistically significant (FDR ≤ 0.05; see “Materials and Methods”) differential expression in response to the different treatments. The number of genes differentially expressed in each data set is indicated near its representing symbol (see bottom right). The percentage concordance is related to genes that were significantly differentially expressed between the two compared conditions (represented by overlapping symbols) and that are additionally regulated in the same way (up- or down-regulation) in both conditions. A, Comparison of the transcriptomic responses to H2O2 and MV treatments for both LL- and HL-acclimated cells. B, Comparison of the pooled responses to oxidative stress (+/− oxidizing agent; i.e. genes responding to H2O2 and/or MV) between LL- and HL-acclimated cells. C, Comparison of the pooled responses to oxidative stress (+/− reactive oxygen stressors) in LL (top signs/values) and HL (bottom signs/values) cells and of the genes differentially expressed in HL- compared with LL-acclimated conditions. Concordant and discordant genes (i.e. common genes regulated in opposite ways between two conditions) from this diagram are listed in Table I.
To be able to directly compare the response of LL and HL cells, genes responding to MV and/or H2O2 were pooled per light condition, and the resulting gene sets were called LL+Ox and HL+Ox. Pairwise comparison of LL+Ox versus LL-Ct and HL+Ox versus HL-Ct confirmed that, generally speaking, LL and HL cells responded similarly to oxidative stress. Indeed, Figure 7B shows that about half of the genes responding to oxidative stress in one light condition were also differentially expressed in the other (366 genes out of 847 for LL+Ox versus LL-Ct, 366 out of 681 for HL+Ox versus HL-Ct) and that the transcriptomic response of these genes was concordant in 93% of the cases (i.e. most genes were regulated in the same way by oxidant in LL- and HL-acclimated cells).
Additionally, these pooled sets were also compared with the effect of photoacclimation (HL-Ct versus LL-Ct) to check whether acclimation to HL and oxidative stress may lead to a common response. As shown in Figure 7C, only 42 genes (corresponding to about 5% of the LL+Ox versus LL-Ct) were differentially regulated in both LL+Ox versus LL-Ct and HL-Ct versus LL-Ct comparisons. Furthermore, 50% of these genes showed an opposite transcriptomic response (11 + 8 versus 15 + 8 genes). In contrast, although 61 genes were differentially regulated in both HL-Ox versus HL-Ct and HL-Ct versus LL-Ct sets, most of them (58 out of 61) were discordant, indicating a different expression behavior between LL and HL cells in our analyses. The full list of genes showing concordant or opposite transcriptomic patterns between the effects of acclimation to HL and ROS stress is available in Table I. Among discordant genes in HL cells, the few that are also discordant in LL cells were retrieved. Noteworthy, these common genes include lexA and recA, which play key roles in the control of DNA repair (Kuzminov, 1999; Butala et al., 2009; Kolowrat et al., 2010). The expression of these two genes was activated by oxidative stress but repressed by HL acclimation. Most of the other discordant genes encode ribosomal subunits or proteins without attributed function.
Table I. List of genes showing concordant or opposite transcriptomic patterns between the effects of acclimation to HL and the effects of oxidative stress in LL- and HL-acclimated cells, as represented in Figure 7C.
Asterisks indicate genes down-regulated by HL acclimation or up-regulated by LL acclimation.
| Open Reading Frame Identifier | Product |
| Concordant genes | |
| Up-regulated by ROS in LL cells and by HL acclimation | |
| 0203 | Putative potassium channel, VIC family |
| 0343 | Putative 3-methyladenine DNA glycosylase |
| 0514 | Predicted HAD superfamily phosphatase |
| 0791 | HIT family hydrolase protein |
| 0947 | Conserved hypothetical protein |
| 1068 | Uncharacterized conserved membrane protein |
| 1501 | Hypothetical protein |
| 2354 | Conserved hypothetical membrane protein |
| Down-regulated by ROS in LL cells and by HL acclimation* | |
| 1029, 2182 | Conserved hypothetical proteins |
| 1348 | Hypothetical protein |
| 1566 | Uncharacterized conserved membrane protein |
| 1732 | Sulfate ion transporter |
| 1796 | Uncharacterized conserved secreted protein, pili subunit superfamily |
| 2068 | β-Glycosidase (chitinase-like), family 18 |
| 2167 | HlyD-like secretion protein superfamily, related to AcrA |
| galE_0242 | UDP-Glc 4-epimerase |
| mpeY_0494 | Putative phycobilin:C-PE II lyase |
| psbA_0784 | PSII protein D1 |
| rpoD_2501 | Alternative RNA polymerase σ factor, σ-70 family |
| 15_Syn_WH7803i235_s415, 208_Syn_WH7803i519_s480, 309_Syn_WH7803i1818_q350 | |
| Up-regulated by ROS in HL cells and by HL acclimation | |
| 0947 | Conserved hypothetical protein |
| metK_0513 | S-Adenosyl-Met synthetase |
| tmk_2244 | Thymidylate kinase |
| Down-regulated by ROS in HL cells and by HL acclimation* | |
| None | |
| Discordant genes | |
| Up-regulated by ROS in LL cells and down-regulated by HL acclimation* | |
| 0095, 0096, 0934, 1953 | Hypothetical proteins |
| 1500 | Conserved hypothetical protein |
| 1797, 2532 | Uncharacterized conserved secreted protein |
| 1799 | Uncharacterized conserved secreted protein, pili subunit superfamily |
| 2116 | Uncharacterized membrane protein |
| lexA_1680 | SOS-response transcriptional repressor LexA |
| recA_0439 | Recombinase A |
| Down-regulated by ROS in LL cells and up-regulated by HL acclimation | |
| 0347 | Conserved hypothetical protein |
| 1329 | 30S ribosomal protein S2 |
| 2236 | Porin, P stress induced |
| petF_1580 | Ferredoxin |
| ppiB_1251 | Peptidyl-prolyl cis-trans-isomerase |
| psb29_1213 | PSII biogenesis protein Psb29 |
| rplE_0422, rpmF_1115 | 50S ribosomal proteins L5 and L32 |
| Up-regulated by ROS in HL cells and down-regulated by HL acclimation* | |
| 0095, 0096, 0934, 1953 | Hypothetical proteins |
| 0543 | LysM-repeat protein |
| 0739, 1797 | Uncharacterized conserved secreted proteins |
| 0971 | Possible multisubunit Na+/H+ antiporter, MnhD subunit |
| 1117 | Uncharacterized conserved membrane protein |
| 0785, 0940, 1497, 1500, 2182 | Conserved hypothetical proteins |
| 1732 | Sulfate ion transporter |
| 1796, 1798, 1799 | Uncharacterized conserved secreted proteins, pili subunit superfamily |
| 2116 | Uncharacterized membrane protein |
| 2208 | Iron-sulfur oxidoreductase |
| 2329 | para-Aminobenzoate synthase component II |
| bicA_0719 | SulP-type bicarbonate transporter |
| ctaD_1872 | Cytochrome c oxidase subunit I |
| hemD_2221 | Uroporphyrinogen-III synthase |
| lexA_1680 | SOS-response transcriptional repressor LexA |
| mpeY_0494 | Putative phycobilin:C-PE II lyase |
| recA_0439 | Recombinase A |
| trpB_2299 | Trp synthase β-chain |
| Down-regulated by ROS in HL cells and up-regulated by HL acclimation | |
| 1068 | Uncharacterized conserved membrane protein |
| 1115 | 50S ribosomal protein L32 |
| 0347, 1312, 1313, 1379, 1423, 2521 | Conserved hypothetical proteins |
| 2002 | Probable 30S ribosomal protein PSRP-3 (Ycf65-like protein) |
| 2198 | Inositol monophosphatase family protein |
| 2257 | RNA-binding protein, RRM domain |
| 2494 | Conserved hypothetical protein specific to cyanobacteria |
| amt_0297 | Ammonium transporter |
| focA_2493 | FNT formate/nitrate transporter family protein |
| glnAI_1347 | Gln synthetase, Glu ammonia ligase |
| mraY_2522 | Phospho-N-acetylmuramoyl-pentapeptide-transferase |
| nusA_2077 | Transcription elongation factor NusA |
| pdxJ_1371 | Pyridoxal phosphate biosynthetic protein PdxJ |
| petF_1580 | Ferredoxin |
| ppiB_1251 | Peptidyl-prolyl cis-trans-isomerase |
| rplE_0422 | 50S ribosomal proteins L5, L15, L19, L23 |
| rpsB_1329 | 30S ribosomal proteins S2, S5, S14, S16 |
| pstSII_1045 | ABC transporter, substrate-binding protein, phosphate |
| 325_Syn_WH7803i1973_q360 | |
Identification of Functional Groups Responding to Oxidative Stress and Light Acclimation
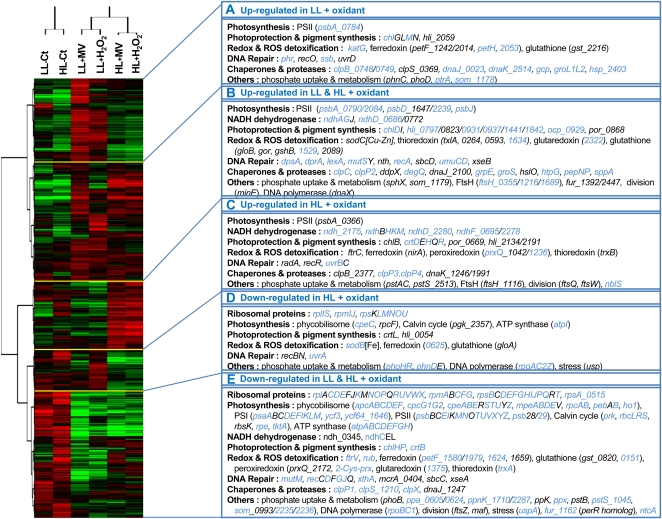
Two-dimensional hierarchical clustering analysis allowed the identification of five gene clusters behaving similarly in the different treatments (Fig. 8): two clusters corresponding to transcripts mainly activated by ROS under LL (group A) or HL (group C) conditions only, a third cluster consisting of oxidant-induced genes under both light conditions (group B), and two clusters including transcripts mainly repressed by oxidative stress in HL cells (group D) or in both LL and HL cells (group E). These five clusters highlighted the behavior of coherent functional groups. The two most obvious ones included ribosomal and photosynthesis genes that were strongly down-regulated by oxidants in both light conditions (group E), suggesting a decrease of photosynthetic and protein translation activities. Indeed, most of the genes encoding the subunits of the complexes driving these processes (ATP synthase, PSI and PSII, phycobilisomes, Calvin cycle, ribosomes) exhibited a similar response. Group E also includes numerous genes involved in phosphate uptake (pstB, pstSII, phnDE, etc.), storage (ppx, ppa), and regulation (phoB) as well as DNA repair genes such as mutM and most rec genes (except recA, located in group B). Another functional group with a characteristic behavior consists of genes involved in photoprotection, which are mainly induced by oxidative stress in LL and/or HL conditions (groups A–C). Among those, genes encoding HL-inducible proteins are particularly well represented in group B, corresponding to transcripts induced in both light conditions, which also include the ocp gene coding for the so-called orange carotenoprotein (Kerfeld et al., 2005; Wilson et al., 2006; Kirilovsky, 2007). In contrast, genes involved in the synthesis of other carotenoids (crtDEHQR genes), including zeaxanthin, are located in group C, containing genes induced by ROS in HL cells only. As expected, chaperones and proteases encoding genes (clpBCPX, dnaJK, groLS, etc.) were induced by ROS and were also mainly represented in groups A to C.
Figure 8.
Two-way cluster analysis diagram of gene expression data. This analysis is based on 1,548 genes corresponding to all genes present on the array but microarray controls, small RNA oligonucleotides, as well as unknown and hypothetical protein-encoding genes. Each row in the diagram represents a gene, and each column represents a light acclimation or a stress condition. The dendrogram at the top is similar to Figure 6, except that each column was obtained from the mean of four biological replicates performed for each condition. The color saturation represents differences in gene expression across the samples: red indicates higher than median expression (black), and green indicates lower than median expression. The color intensity indicates the degree of gene regulation. The boxes to the right represent gene clusters A to E and show some of the genes present in these clusters and belonging to specific functional categories. Genes showing a statistically significant differential expression in at least one condition (Student’s t test and/or LIMMA with FDR ≤ 0.05) are indicated in blue.
DISCUSSION
Are HL Acclimation and Oxidative Stress Cumulative?
The physiological experiments performed in this study revealed that under their growth irradiance, HL-acclimated cells were more affected than LL cells by H2O2 stress with regard to PSII photoinactivation, and this was verified across the whole range of H2O2 concentrations tested (Figs. 1 and 2). A possible hypothesis explaining this interesting observation could be that cells grown at 250 μmol photons m−2 s−1 were already facing a stress situation, so that they could not handle an additional stress. However, marine Synechococcus exhibit a large physiological flexibility that allows them to grow under a wide range of irradiance in laboratory conditions (Kana and Glibert, 1987a, 1987b; Six et al., 2004, 2005), with the maximum growth rates usually observed between 200 to 300 μmol photons m−2 s−1 (Kana and Glibert, 1987a; Moore et al., 1995; Six et al., 2004). Thus, the HL irradiance used in this study should not represent highly stressful growth conditions. Furthermore, microarray analyses revealed that only 121 genes were differentially expressed in the HL-Ct versus LL-Ct pairwise comparison, while four to six times more genes responded to the addition of oxidizing agents. None of the 121 genes seemed to be involved in ROS defense or detoxification mechanisms, except for a ferredoxin-encoding gene (petF_1580) and a peroxiredoxin-encoding gene (prxQ_1236), which were both significantly up-regulated in HL compared with LL cells. Interestingly, lexA and recA genes, which are involved in the SOS response (Butala et al., 2009; Kolowrat et al., 2010), were down-regulated in HL cells, supporting the interpretation that those cells were not under HL stress, which would require the activation of this DNA repair system.
Why Are the HL Cells More Sensitive Than LL Cells to MV-Mediated Oxidative Stress?
MV is a very low potential redox compound that primarily accepts electrons from the FA and FB iron-sulfur clusters of PSI centers at the expense of ferredoxin reduction (Supplemental Fig. S1; Fujii et al., 1990). By reacting very quickly with molecular oxygen, the reduced MV·+ radical is spontaneously reoxidized back to MV with the formation of the membrane-impermeable superoxide anion. The latter compound undergoes further dismutation to form H2O2, which can readily pass through biological membranes (Bus and Gibson, 1984; Goldstein et al., 2002). PSI electron transport includes both linear electron flow (LEF) from PSII through PSI to NADP+ and cyclic electron flow (CEF) from the PSI acceptor side back to the cytochrome b6f complex (Supplemental Fig. S1; Bendall and Manasse, 1995; Joliot and Joliot, 2002). By reacting with any electrons coming from PSI, MV inhibits CEF and diverts LEF (Yu et al., 1993; Herbert et al., 1995) toward ROS production (Jia et al., 2008; Fan et al., 2009). In the presence of MV, the ROS production in a cell is thus mostly dependent upon the absolute number of electrons passing through PSI complexes per time unit, which results from two parameters that are both strongly influenced by growth irradiance: the electron transport rate at PSI complexes and the number of PSI complexes per cell. LEF is thus a critical parameter when considering the differential response of LL and HL cells to MV-mediated oxidative stress.
At their growth irradiance, HL cells mediate much faster LEF than do LL cells, thereby producing more ROS per PSI. For PSI content, previous studies have shown that in cyanobacteria, acclimation to high irradiance is accompanied by a strong decrease of the thylakoid surface and chlorophyll a content per cell (Kana and Glibert, 1987a; Moore et al., 1995; Six et al., 2004). In cyanobacteria, chlorophyll a is almost exclusively bound to reaction centers (especially to PSI complexes); therefore, HL cells must contain fewer photosystems than LL cells. Thus, HL cells exhibit a high PSI electron transport rate favoring a large MV-mediated ROS production rate, but a lower photosystem cell content and therefore a lower number of potential ROS formation sites, than LL cells.
Addition of MV concentrations higher than 25 μm induced a saturation of PSII photoinactivation in both LL and HL cells (Fig. 1B), most likely originating from a saturation in the ROS production rate from PSI centers. Such a saturating effect has previously been observed in the freshwater Synechococcus sp. PCC 7942, albeit at a somewhat higher MV concentration (100 μm; Roncel et al., 1988). While the MV concentration leading to the saturation of PSII photoinactivation rate was similar between LL and HL cells, the remaining PSII quantum yield at the saturating MV concentration was, in contrast, almost 10-fold lower in HL cells than in LL cells (Fig. 1B). If the number of PSI complexes per cell was a major factor influencing ROS production, a severe PSII photoinactivation would have been expected in LL cells, given their high PSI cell content. Since this is not the case, this rather suggests that the LEF rate is the major factor controlling the amplitude of the MV-induced oxidative stress. Furthermore, when HL cells were suddenly shifted to LL conditions, lowering LEF without affecting the PSI cell content, the production of H2O2 almost stopped (Fig. 3B), with a consequent slowing down of PSII photoinactivation compared with the cells maintained in HL conditions (Fig. 3A). This clearly highlights the importance of LEF in the amplitude of MV-induced oxidative stress in HL cells. Still, it is worth noting that this only partially explains the higher sensitivity of HL cells to oxidative stress. First, despite the sharp reduction in MV-dependent ROS production, the HL cells shifted to LL still exhibited a 25% photoinhibition (Fig. 3). Moreover, even though this stronger PSII inactivation could, at least in part, be due to a higher rate of endogenous ROS formation in the presence of MV, this justification does not hold true for H2O2, since the same oxidant concentration, added exogenously, also produces a much stronger photoinhibition in HL cells than in LL ones (Fig. 1A). Thus, besides inducing a higher MV-dependent ROS production, HL acclimation also seems to result in a higher sensitivity to ROS-driven damage.
Effect of ROS on PSII Damage and Repair
The PSII repair cycle is a crucial process in the response of photosynthetic organisms to various stressful conditions (Park et al., 1995; Nishiyama et al., 2006; Six et al., 2007a). Several hypotheses have been proposed to explain the detrimental effect of ROS on PSII function, either through direct damage to PSII reaction centers with consequent degradation of the D1 protein (Hideg et al., 2007; Vass and Cser, 2009) or through effects on the de novo D1 protein synthesis through ROS inhibition of protein translation (Nishiyama et al., 2004; Kojima et al., 2007; Takahashi and Murata, 2008). Our results suggest that H2O2 induces both direct PSII damage and inhibition of PSII repair, with the respective importance of these processes being dependent upon both photoacclimation and oxidant concentration.
In HL cells, a treatment with lincomycin and 25 μm H2O2 induced the same level of photoinactivation as the lincomycin-only treatment. This shows that at this concentration and under this growth irradiance, H2O2 does not induce damage beyond that observed when the PSII repair cycle is inhibited (Fig. 5D). Therefore, the moderate decrease of PSII function in the presence of 25 μm H2O2 and the absence of lincomycin suggests that H2O2, at this concentration and under this growth irradiance, acts by partially inhibiting PSII repair rather than by directly damaging PSII. Despite the inhibitory effect of H2O2 in these conditions, a strong induction of D1 synthesis occurred right after stress initiation (Fig. 4). This de novo synthesis of D1 was not sufficient to maintain PSII quantum yield, indicating that most of these D1 proteins were not included in functional PSII centers (Garczarek et al., 2008).
While PSII repair cycle inhibition was partial at 25 μm, it seemed to be complete in the presence of 750 μm H2O2, since this oxidant had the same effect on HL cells with and without lincomycin (Fig. 5D). Furthermore, this H2O2 concentration also induced strong direct PSII damage, since the level of photoinactivation was about twice higher than when the repair cycle was blocked by lincomycin alone. These ROS-driven damages seemed mostly light independent, since the photoinactivation in the presence of 750 μm H2O2 alone was similar in the dark. Such damages could not be restricted to PSII only but could be attributed to a more global degradation of cellular components by ROS (Imlay, 2003), including translation machinery (Kojima et al., 2007). Thus, at high H2O2 concentrations, the decrease of PSII quantum yield in HL cells reflected both an impairment of the PSII repair cycle and direct damage to PSII complexes.
In contrast to HL cells, LL cells initially showed a nearly constant D1 pool in the presence of 25 μm H2O2, which appeared sufficient to maintain PSII quantum yield at its initial level during 6 h (Figs. 2 and 4). This suggests that 25 μm H2O2 did not provoke additional PSII damage beyond the very low photoinactivation rate at this light level, which was below detection. However, an effect leading to PSII damage was clearly noted at 750 μm H2O2 (Fig. 5C). In contrast to HL cells, these damages were light dependent, as almost no damage occurred when H2O2 was added in the dark (Fig. 5, C and D). A possible explanation for the light dependence of these damages relies on the formation of reactive OH· through oxidation by H2O2 of reduced metals, such as the iron or manganese atoms located in reaction center II (Imlay, 2003). Indeed, PSII nonheme iron, one of the prosthetic groups of the D1 protein, has been reported to be oxidized by H2O2 and is thought to be involved in a specific OH·-induced cleavage of this protein (Diner and Petrouleas, 1987; Lupínková and Komenda, 2004). Such reactions are expected to be light dependent, since the redox state of the nonheme iron depends on photosynthetic electron transfer (Ishikita and Knapp, 2005). Similarly, at the PSII donor side, the redox status of the manganese cluster, which can also be oxidized by H2O2 (Lupínková and Komenda, 2004), is also dependent on PSII electron transport. In darkness, the oxidized state of such PSII-bound metals might prevent a reaction with H2O2 and therefore the resulting formation of OH· leading to D1 protein cleavage.
Interestingly, a cumulative effect of light and oxidative stress on PSII function does not agree with the results obtained on Synechocystis sp. PCC 6803, for which oxidative stress induced by H2O2 did not accentuate light-induced damage but only affected the rate of PSII repair (Allakhverdiev and Murata, 2004). It should be noted, however, that there are major experimental differences between those results and our study. Indeed, the Synechocystis experiments were mainly performed by monitoring the recovery of PSII activity after cultures were briefly exposed to very HL conditions (250–2,000 μmol photons m−2 s−1), inducing strong PSII photoinactivation that the cells were only able to withstand for a short time. In contrast, in our study, Synechococcus sp. WH7803 cells were fully acclimated to the different growth irradiances. Furthermore, a large range of H2O2 concentrations was tested, as the dose proved to be important to differentiate between ROS-induced direct damage and inhibition of repair. For instance, the effect of H2O2 on PSII repair activity could only be detected at 25 μm and on HL cells (i.e. when some light-mediated but no ROS-mediated damages were generated). Although our approach did not allow the assessment of the PSII repair cycle when no detectable photodamage had occurred, it had the advantage of taking into account the differential damage resistance/repair capacities of these cells that cannot be appreciated through a light shift. For instance, one could expect that HL cells, which possess a higher ratio of D1:2 to D1:1 protein isoforms than LL cells (Garczarek et al., 2008), also possess a higher PSII resistance to photoinactivation (Campbell et al., 1995, 1998; Tichý et al., 2003). The fact that HL cells, in contrast, were even more sensitive to photoinhibition indicates that oxidative stress has a strong effect on direct PSII damage independently of the dominant D1 isoform present in the PSII. Altogether, these data suggest that the cumulative effect of light- and ROS-driven damage in HL cells and the resulting stronger dependence on D1 repair are probably responsible for the higher PSII sensitivity of those cells to oxidative stress. For cells shifted from HL to darkness, we observed a moderate, progressive decline in Fv/Fm (Fig. 5B). Addition of lincomycin prevented this decline, which therefore depends upon protein synthesis in the dark. This decline in Fv/Fm in the dark resulted mostly from an increase in the F0 level of fluorescence in cells shifted from HL to darkness (Supplemental Fig. S2). Similar increases in F0 occurred in the absence or presence of H2O2.
Transcriptome Response to Oxidative Stress
Effects of oxidative stress on PSII do not alone determine the survival capacities of the cells. Indeed, while HL cells were able to recover from a 25 μm H2O2 stress, LL cells eventually died, even though their photosynthetic activity seemed initially less affected (Fig. 1). We performed transcriptome analyses to observe the global response of the cells and to understand why HL cells were capable of better resistance and recovery than LL cells when facing oxidative stress.
Microarray analyses showed that H2O2 altered the expression of functional groups of genes (e.g. photosynthesis, NADPH dehydrogenase, ribosomal proteins, some chaperones) in the marine Synechococcus sp. WH7803, in much the same way as what has been observed in the freshwater Synechocystis sp. PCC 6803 in response to approximately millimolar H2O2 concentrations (Li et al., 2004; Houot et al., 2007). Moreover, global transcriptomic data clearly indicated that H2O2 and MV induced similar transcriptomic responses in both LL and HL Synechococcus sp. WH7803 cells (Figs. 6 and 7A). Pairwise comparison between pooled genes responding to H2O2 or MV resulted in 93% concordance between LL and HL cells, meaning that most genes differentially expressed in both light conditions responded similarly in HL and LL cells (Fig. 7B). Hierarchical clustering analyses (Fig. 8), however, allowed highlighting some differences associated with one or the other light condition.
The photosynthetic genes are among the most affected by oxidative stress in both LL and HL cells (Fig. 8; Supplemental Table S1), including the four psbA gene copies encoding the D1 protein of the PSII core in Synechococcus sp. WH7803. At the transcriptomic level, microarray data showed that the two transcripts making up most of the psbA mRNA pool in this strain (Garczarek et al., 2008) are inversely regulated in response to oxidative stress, indicating an exchange between D1:1 (encoded by psbA_0784) and D1:2 (encoded by psbA_0790) isoforms in both HL and LL cells. The resulting D1 pool would then be dominated by the D1:2 isoform, which is thought to provide resistance to photoinhibition (Krupa et al., 1991; Clarke et al., 1993a, 1993b; Campbell et al., 1995, 1998; Tichý et al., 2003). Accordingly, ftsH_1216, the ROS-induced direct ortholog of Synechocystis sp. PCC 6803 slr0228 gene (Li et al., 2004; Houot et al., 2007), encoding the FtsH2 protease involved in the clearance of damaged D1 proteins from inactivated PSII (Komenda et al., 2006, 2010), belongs to the cluster of genes mainly activated in response to ROS in Synechococcus sp. WH7803 (cluster B; Fig. 8). It is worth noting that, as previously described, the D1 pool in the PSII of HL cells is dominated by the D1:2 isoform (Garczarek et al., 2008). HL cells would then be better prepared to sustain ROS-driven photoinhibition not only thanks to the initial dominance of the transcript coding for the D1:2 isoform but also by completing the isoform switch in response to additional stress. Most other photosynthetic genes belong to some of the main gene clusters inhibited by oxidative stress (clusters D and E; Fig. 8). This includes most of the ATP synthase, PSI and PSII subunits, as well as the genes involved in phycobilisome synthesis and the Calvin cycle, suggesting a decrease in energy supply in response to stress.
Although most of the photosynthetic genes were not significantly differentially affected by oxidative stress in LL and HL cells, some differences in gene induction gave us hints to explain the survival of HL cells in oxidative stress conditions that were lethal for LL cells. This includes a number of genes involved in photoprotection mechanisms, such as the ocp gene (Kerfeld et al., 2005; Wilson et al., 2006; Kirilovsky, 2007), whose induction was significantly higher in HL cells than in LL cells (Supplemental Table S1). In addition, the crtDEHQR genes involved in carotenoid synthesis (Masamoto et al., 1998; Klassen, 2010) mainly belong to the cluster C genes more activated in HL cells than in LL cells (Fig. 8), with crtR, a gene involved in the conversion of β-carotene into zeaxanthin, being clearly the most up-regulated. Carotenoids are thought to dissipate energy from photosensitized chlorophyll or from singlet oxygen and may have intrinsic antioxidant properties (Edge et al., 1997). In particular, zeaxanthin-deficient mutants of Synechocystis sp. PCC 6803 and Synechococcus sp. PCC 7002 are more sensitive than their respective wild types to HL and oxidative stress, and it has been shown that such mutants indeed accumulate ROS (Schäfer et al., 2005; Zhu et al., 2010). The genes differentially regulated by oxidative stress also include the high-light-inducible proteins encoding genes, three of which are induced at higher levels in HL cells than in LL cells (hli_0937, hli_0797, hli_1441). HL-inducible proteins have been shown to be involved in protecting cells against photodamage, either by direct or indirect dissipation of excess absorbed light energy (Montané and Kloppstech, 2000; He et al., 2001; Havaux et al., 2003) or by binding and storage of free chlorophylls (Funk and Vermaas, 1999; Vavilin et al., 2007; Kufryk et al., 2008).
Among the genes clearly induced by oxidants in HL cells (cluster C; Fig. 8), those encoding the NADPH dehydrogenase subunits are highly represented. This complex is involved in a variety of cellular processes, including respiratory electron flow, CEF around PSI, and indirect control of the redox state of the plastoquinone pool, as well as in CO2 uptake (Cooley and Vermaas, 2001; Ogawa and Mi, 2007; Battchikova et al., 2010). The active induction of ndh genes would allow maintaining energy production of HL cells under oxidative stress. Notably, a direct effect of NDH-dependent CEF in alleviating photooxidative stress has also been suggested in tobacco (Nicotiana tabacum) chloroplast by compensating the stromal overreduction that induces the formation of ROS (Wang et al., 2006).
Tolerance of the deleterious effects of ROS may result from the activity of a number of mechanisms that maintain ROS at a harmless level or that actively repair ROS-induced damage. ROS-scavenging enzymes such as catalases, peroxidases, and superoxide dismutases, as well as antioxidants such as glutathione, tocopherol, and ascorbic acid, are involved in the maintenance of low levels of ROS (Noctor and Foyer, 1998; Storz and Imlay, 1999; Mittler, 2002; Masip et al., 2006). Such antioxidant activities involved in ROS scavenging and redox homeostasis have been reported in freshwater cyanobacteria such as Synechocystis sp. PCC 6803 and Synechococcus sp. PCC 7942 (Regelsberger et al., 2002; Yousef et al., 2003; Kobayashi et al., 2004; Hosoya-Matsuda et al., 2005; Kanesaki et al., 2007). Most of the genes involved in antioxidant activities have also been reported in the genome of Synechococcus sp. WH7803, suggesting that a similar detoxification systems occurs in this marine cyanobacterium (Scanlan et al., 2009). Among these genes, some, like the catalase/peroxidase-encoding gene katG, were induced in response to oxidative stress in both LL and HL cells, but others showed an obvious difference between HL and LL. While the expression of the cytosolic (iron [Fe]) superoxide dismutase (Fe-SOD)-encoding gene sodB was down-regulated in both LL and HL cells submitted to oxidative stress, the response of the periplasmic (copper/zinc [Cu/Zn]) superoxide dismutase (Cu/Zn-SOD) gene sodC was up-regulated in HL cells only (Supplemental Table S1; Langford et al., 2002; Culotta et al., 2006). In contrast, in Synechocystis sp. PCC 6803, which only carries the sodB gene, the latter seemed to be induced in response to H2O2 (Li et al., 2004; Houot et al., 2007). In Synechococcus sp. PCC 7942, the cytosolic Fe-SOD does not protect the cells from ROS generated within the thylakoid membrane (Thomas et al., 1998), suggesting that other SODs might confer such a protection. Thus, the increase in periplasmic Cu/Zn-SOD activity would confer a resistance to the light-driven ROS production in HL cells. Another gene involved in ROS detoxification, dpsA, encoding a DNA-binding protein, was also up-regulated in response to oxidative stress in HL cells only. DpsA has been shown to be involved in the detoxification of Fe2+ ions and H2O2 and to more specifically protect DNA from oxidative damage (Chiancone and Ceci, 2010). In Synechococcus sp. PCC 7942, while an insoluble DpsA fraction would bind DNA, a soluble faction, localized into the thylakoid membranes, would allow maintaining metal homeostasis of the photosynthetic apparatus (Durham and Bullerjahn, 2002). Thus, as the Cu/Zn-SOD, DpsA would confer a better resistance of HL cells to the ROS generated within the thylakoid membrane. The genes ftrC and ftrV, encoding subunits of the ferredoxin-thioredoxin reductase (FTR) complex, are clearly differentially regulated in response to oxidative stress between LL and HL cells: both genes are down-regulated in LL cells but not in HL cells, for which ftrC is up-regulated. The FTR complex links PSI redox status with the regulation of numerous photosynthetic processes and is required for the growth of plants that are submitted to oxidative stress (Buchanan and Balmer, 2005; Schürmann and Buchanan, 2008). Thus, the activation of the FTR system in HL cells, while it is inhibited in LL cells, may partially prevent HL cells from cellular damage due to reactive oxygen stressors by means of a better retrocontrol of the redox status of the cells.
A number of chaperone- and protease-encoding genes were also affected by ROS in only one of the light acclimation conditions. Among these, the three genes encoding the GroEL/GroES system (groS, groL1, groL2) and htpG were particularly activated in LL cells. These two major chaperone systems in bacteria are known to be activated in response to cold stress, high irradiance, and MV in Synechococcus sp. PCC 7942 (Hossain and Nakamoto, 2002, 2003; Sato et al., 2010). In contrast, clpP genes mostly responded to oxidative stress in HL cells, with three being activated (clpP2, clpP4, clpP3), while the clpP1 gene was repressed in both LL and HL cells (Fig. 8; Supplemental Table S1). The expression of these genes may also be related to clpX, which encodes a regulatory ATPase/chaperone interacting with ClpP and was surprisingly inhibited by ROS. Indeed, despite the fact that clpX and clpP are typically induced by heat, high salt, oxidation, and Glc deprivation in bacteria, such an inhibitory effect by H2O2 on clpX expression has already been observed in Synechococcus sp. PCC 7942 (Schelin et al., 2002) and may be linked to a possible exchange of Clp proteolytic core complexes in response to stress (Stanne et al., 2007; A. Clarke, personal communication). Finally, degQ, which encodes a periplasmic protease known to confer some stress resistance (Barker et al., 2006), was also somewhat more induced in HL cells than in LL cells.
An element that could contribute to the better ROS resistance of HL cells is the capacity to induce an SOS response, which is involved in protecting the cells from the damaging effects of increased mutation rates. Although some authors have suggested that cyanobacteria could lack an SOS response similar to that of Escherichia coli (Domain et al., 2004; Michel, 2005; Patterson-Fortin et al., 2006), it seems that Synechococcus sp. WH7803, like Prochlorococcus (Kolowrat et al., 2010), could well possess such an inducible pathway involved in DNA repair. The SOS response is under the control of two regulatory components, RecA and LexA, the latter being a transcriptional regulator of SOS genes, including itself. The consensus sequence motif for the LexA-binding site in cyanobacteria has been proposed to be RGTACNNNDGTWCB (Mazón et al., 2004). Among the 43 genes belonging to the LexA regulon in E. coli (Courcelle et al., 2001), 10 seem to be present in the Synechococcus sp. WH7803 genome, namely lexA, recAN, umuCD, uvrAD, ssb, and ruvAB, but only lexA, recA, and umuD show potential LexA-binding sites in their regulatory regions, independently on their orientation (Table II). In order to identify other putative SOS genes, Synechococcus sp. WH7803 intergenic regions were screened with the consensus motif RRTACNNNDGTWYB, which integrates one degenerated position from the LexA box of the lexA gene. Six other putative LexA-binding sites have been detected downstream of the wcaG, dprA (or smf), WH7803_0096, WH7803_0814, WH7803_0843, and WH7803_1500 genes (Table II). These findings differ slightly from those of Li et al. (2010), who, by using another cyanobacterial consensus, failed to detect significant sites in the regulatory regions of lexA and umuCD, leading them to conclude that the SOS response regulation might be attenuated in Synechococcus sp. WH7803. According to our analysis, all genes from the LexA regulon (i.e. that carry a putative LexA box in their regulatory region) were very similarly regulated in response to HL acclimation and to oxidative stress, strengthening the hypothesis that LexA may indeed regulate these genes and that the gene products with unknown functions might be involved in the SOS response. In contrast, genes that are thought to be part of the LexA regulon in E. coli but that do not have a LexA-binding motif in their promoter region in Synechococcus sp. WH7803 are not similarly regulated and are most probably not under the control of LexA. Using the WebLogo tool (Crooks et al., 2004), the nine putative binding sites found in Synechococcus sp. WH7803 allowed us to define the consensus of the LexA box in this strain, which seems to be degenerated from the palindromic TACAN2TGTA consensus (Supplemental Fig. S3). Interestingly, the expression of lexA was more induced in HL cells than in LL cells, suggesting that the SOS response may indeed play a role in the better resistance of HL cells to oxidative stress. Although it might appear surprising that both repressor-encoding (lexA) and derepressor-encoding (recA) genes are simultaneously induced, the constant production of LexA would allow its reaccumulation to repress SOS genes once DNA damage has been repaired (Michel, 2005). At last, the expression of uvrA, which was clearly differently regulated compared with all other SOS genes, was strongly repressed in response to oxidative stress in HL cells, and that may be due to the fact that uvrA is only expressed in very early stages of the SOS response (Kuzminov, 1999; Michel, 2005).
Table II. Putative LexA regulon and DNA-binding sites in Synechococcus sp. WH7803.
| Gene | Putative LexA-Binding Site | Position from ATGa | Log2 Fold Change from Microarray Datab |
Gene Clusterc | Gene Product | ||||
| HL-Ct/LL-Ct | LL+H2O2/LL-Ct | LL+MV/LL-Ct | HL+H2O2/HL-Ct | HL+MV/HL-Ct | |||||
| Genes with a putative LexA-binding site in their upstream intergenic region | |||||||||
| lexA | GGTACACATGTATT | −21 | −0.1 | 0.9 | (0.2) | 1.4 | 1.4 | B | SOS-response transcriptional repressor |
| recA | AGTACAGATGTACG | −137 | −1.1 | 1.9 | 1.0 | 1.1 | 1.4 | B | Recombinase A |
| umuD umuCd | AGTACAGATGTTCT | −40 | (−0.3) (−0.3) | 2.6 1.6 | 1.2 0.6 | 1.2 (0.4) | 1.3 0.6 | B B | Subunits of DNA polymerase for DNA repair |
| dprA | AGTACACATGTATT | +1 | −0.5 | 0.6 | (0.2) | 0.6 | 0.8 | B | Putative DNA-protecting protein |
| wcaG | AGTACAAACGTATT | −8 | (−0.3) | (−0.1) | 0.5 | 0.7 | 0.9 | B | Nucleoside-diphosphate-sugar epimerase |
| 0096e | AATACAGTTGTATT | −88 | −2.1 | 2.2 | (0.6) | 2.9 | 3.2 | ND | Hypothetical protein |
| 0814e | GGTACGCCTGTTCT | −96 | (−0.6) | 1.7 | 1.4 | 1.9 | 2.3 | ND | Conserved hypothetical protein |
| 0843e | AGTACAGATGTACT | −20 | −0.7 | 1.6 | 1.8 | 1.4 | 1.5 | ND | Conserved hypothetical protein |
| 1500e | AGTACAGATGTACT | −13 | −1.5 | 1.5 | (0.4) | 2.2 | 2.0 | ND | Conserved hypothetical protein |
| Consensus | |||||||||
| RRTACRNNYGTWYK | |||||||||
| Genes with no LexA-binding site found but whose orthologs are thought to be part of the E. coli LexA regulon | |||||||||
| recN | (−0.4) | (−0.2) | (−0.5) | (0.1) | (−0.3) | D | ATPase involved in DNA repair | ||
| ruvB | (−0.3) | (−0.2) | (0.7) | 0.9 | (0.4) | B | Holliday ATP-dependent DNA helicase | ||
| ssb | −0.8 | (−0.1) | (0.5) | 1.0 | 1.0 | A | Single-stranded DNA-binding protein | ||
| uvrA | (0.1) | (0.1) | −0.7 | −1.2 | −1.2 | D | Excinuclease ATPase subunit | ||
| uvrD | (−0.4) | (−0.4) | (−0.1) | (−0.4) | (−0.2) | A | DNA-dependent ATPase I and helicase II | ||
Positions have been calculated between the first nucleotide of the start codon and the middle of the putative binding site.
Values in parentheses are not statistically significant (i.e. with FDR > 0.05).
See hierachical clustering analysis in Figure 8. ND, Not determined (hypothetical proteins were excluded from the analysis).
umuD and umuC genes are likely organized in an operon.
The complete open reading frame number is SynWH7803_XXXX.
CONCLUSION
Our data show that the susceptibility of Synechococcus cells to oxidative stress varies depending on culture light history. HL cells are more immediately affected by oxidant exposure than are LL cells, but HL cells then show better capacity to ultimately acclimate and recover from low concentrations of H2O2. By testing a range of oxidant concentrations and treatment durations on LL and HL cultures, we show that, depending upon the oxidant concentration, PSII damage observed in oxidant-stressed HL cells results both from cumulative effects of direct ROS damage to PSII and ROS inhibition of the rapid PSII repair needed to counter the high photoinactivation rate. These findings largely reconcile earlier findings that ROS directly inactivates PSII with other studies showing ROS inhibition of PSII repair. Even though the active PSII repair cycle of HL cells is initially inhibited by oxidative stress, it nonetheless allowed them to fully recover when H2O2 exposure did not exceed 25 μm. Although the oxidative stress induced a similar global transcriptomic response in HL and LL cells, in HL cells we detected the induction of other cellular processes, including more efficient photoprotection mechanisms, some ROS detoxification, and a better redox/energy homeostasis, that most likely helped prevent the death of HL cells. Thus, in HL cells, physiological changes induced by ROS were reversible when the intracellular level of ROS decreased, while in LL cells, similar ROS treatment led to cell death. More generally, identification of the factors that set the threshold at which a cell makes the transition from successful acclimation/resistance to oxidative stress-induced cell death will be of major interest to understanding the cumulative effect of environmental stress conditions.
MATERIALS AND METHODS
Culture and Oxidative Stress Conditions
Synechococcus sp. WH7803 (Roscoff Culture Collection strain RCC752) was grown at 22°C in 0.2 μm filtered PCR-S11 medium (Rippka et al., 2000) supplemented with 1 mm NaNO3. Cultures were acclimated during many generations to continuous LL (18 μmol photons m−2 s−1) and HL (250 μmol photons m−2 s−1) provided by Sylvania Daylight 58W/154 fluorescent bulbs. Growth rates for this strain grown at several irradiances have been published elsewhere (Kana and Glibert, 1987a; Garczarek et al., 2008). For all stress experiments performed in this study, exponentially growing cultures (1–3 × 107 cells mL−1) were split into subcultures and incubated in the presence of different concentrations (see “Results”) of oxidizing agent under various light conditions: the subcultures were either set back to their growth light conditions, or shifted to lower light to investigate the influence of photosynthetic electron flux on ROS production, or shifted to dark to differentiate direct ROS-induced damage from light-driven damage. The oxidative stress induced by H2O2 was compared with that provoked by the herbicide MV (Sigma Aldrich). H2O2 stock solutions were titrated before each experiment by A240 (Nanodrop ND-1000; NanoDrop Technologies) using a molar extinction coefficient of 43.6 m−1 cm−1 (Hildebraunt and Roots, 1975) and adjusted at 100 mm.
Synechococcus sp. WH7803 growth was monitored by flow cytometry using a FACS Canto flow cytometer (Becton Dickinson Biosciences) as described previously (Marie et al., 1997, 1999) and by phycoerythrin fluorescence emission maximum (excitation at 530 nm) using an LS-50B spectrofluorometer (Perkin-Elmer).
PSII Quantum Yield and Repair
PSII fluorescence quantum yield (Fv/Fm) was measured using a pulse amplitude-modulated fluorometer (PhytoPAM; Walz) as described previously (Six et al., 2007b; Garczarek et al., 2008).
In order to measure PSII damage and repair, 500 μg mL−1 lincomycin (Sigma-Aldrich), an inhibitor of protein synthesis, was added or not to the culture immediately before submitting the cells to oxidative stress, following a procedure similar to the one described by Six et al. (2007a). When cultures were shifted from one light condition to another, cells were shifted to the new light condition 10 min prior to the addition of oxidizing agent, immediately after the lincomycin.
Measurement of H2O2 Production during MV Stress
H2O2 production by cells upon MV-induced oxidative stress was monitored by the decrease of scopoletin fluorescence (Patterson and Myers, 1973; Tichý and Vermaas, 1999), as this molecule is degraded by H2O2 in the presence of horseradish peroxidase. Cells (0.5–1 × 108) from exponentially growing cultures were pelleted (6,000g for 10 min; Eppendorf 5424R), resuspended in 3 mL of 25 mm HEPES (pH 7.0), and incubated in a cuvette for 30 min under their initial HL or LL acclimation conditions. After the addition of 1.33 μm scopoletin (Sigma-Aldrich) and 25 units mL−1 horseradish peroxidase (Sigma-Aldrich), the cells where incubated for 10 min under their initial irradiance or shifted to LL irradiance, and 0, 50, or 250 μm MV was then added. After 30 min, the scopoletin fluorescence emission was monitored at 460 nm (excitation at 350 nm) using an LS-50B spectrofluorometer (Perkin-Elmer). After subtracting the dye-free background signal, fluorescence was expressed as a percentage of the initial value.
D1 Protein Quantification by Immunoblotting
A culture volume of 50 mL was centrifuged in the presence of the nonionic surfactant Pluronic F-68 (10 mg L−1; Sigma-Aldrich) to avoid cell adhesion at the surface of the tubes, and the pellet was flash frozen in liquid nitrogen and stored at −80°C until analysis. Total protein assays, PAGE, protein transfer onto a polyvinylidene fluoride membrane, immunoreaction, and chemoluminescence detection were performed as described by Garczarek et al. (2008). D1 protein amounts were expressed as a percentage of the initial conditions.
RNA Sampling and Extraction
For the transcriptomic analyses, Synechococcus sp. WH7803 cultures were submitted to oxidative stress by the addition of H2O2 or MV and harvested when PSII quantum yield fell to half of the initial value. For H2O2 experiments, this level of PSII photoinactivation was reached 2 h after submitting LL and HL cultures to 750 μm and 25 μm, respectively (Fig. 2). Because of the large divergence in dose and kinetics responses to MV between LL and HL cells, it was not possible to find MV concentrations leading to 50% decrease of quantum yield at the same time for both light acclimations. Thus, array analyses for MV were performed on HL and LL cultures incubated at the same MV concentration (50 μm) but harvested once PSII quantum yield was halved (i.e. after 1 and 3.5 h of stress, respectively; Fig. 2).
For every experiment, a 300-mL volume of stressed culture was harvested by centrifugation (10,000g for 7 min; Eppendorf 5417R) in the presence of the nonionic surfactant Pluronic F-68 (10 mg L−1; Sigma-Aldrich). Pellets were then flash frozen in liquid nitrogen and stored at −80°C until analysis. Four independent biological replicates of the experiments were carried out.
Total RNA was extracted with acid pH-guanidinium thiocyanate/phenol/chloroform (TRIzol reagent; Invitrogen) following a modified procedure from Millican and Bird (1998) and purified on a column (Qiagen RNeasy Mini Kit). Briefly, frozen cells were resuspended in 500 μL of TRIzol (Invitrogen) and submitted to three cycles of 2 min of heating at 65°C and flash freezing in liquid nitrogen. Following 10 min of incubation at 65°C with vortexing, the clear lysate was vigorously mixed with 100 μL of chloroform and incubated at room temperature for 5 min in microtubes containing an interphase gel barrier (Phase Lock Gel Heavy; 5 PRIME). After centrifugation at 9,000g for 15 min, the upper aqueous fraction was collected and the total RNA was purified following the Qiagen RNeasy Mini Kit procedure. Two consecutive DNase treatments (Qiagen RNase-Free DNase Set) were performed on the RNeasy column during the purification step, as recommended by the manufacturer. RNA was retrieved by two consecutive elutions with 30 μL of RNA-free water (Ambion), and its quality was checked by capillary electrophoresis on a Bioanalyzer 2100 using the Procayote total RNA nano chips (Agilent).
Microarray Analysis
Array Design, cDNA Labeling, and Hybridization
Microarray experiments were performed using a homemade array targeting 2,497 protein-coding genes out of the 2,586 genes identified so far in the Synechococcus sp. WH7803 genome (Dufresne et al., 2008) as well as 126 potential small RNAs (W.R. Hess, personal communication). The 60-mer oligonucleotides were designed and synthesized by Eurogentec, resuspended at a final concentration of 20 μm into a 1× spotting buffer (Schott Nexterion), and spotted in duplicate on Schott Nexterion slides using facilities of the Rennes transcriptomic platform.
cDNA synthesis from 5 μg of total RNA and indirect CyDye cDNA labeling were performed using the ChipShot Indirect Labeling and Clean-Up System (Promega) following the manufacturer’s procedure. After resuspension, labeled cDNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific) and vacuum concentrated to use equal amounts of Cy3- and Cy5-labeled probes (20–100 pmol) for hybridization. All hybridizations were performed on four independent biological replicates. A pool of RNA from all samples investigated in this study was used as a reference sample. This allowed us to maximize the number of spots with a significant signal. Furthermore, in order to minimize bias due to differential dye bleaching or unequal incorporation of the Cy3 and Cy5 dyes during reverse transcription reactions, two out of the four replicate samples for each condition were hybridized in dye-swap experiments.
Printed slides were rinsed and blocked by 5 min of incubation in 0.1% Triton X-100 followed by 2 min in 0.0037% HCl, 10 min in 100 mm KCl, and finally 15 min at 55°C in a blocking solution (0.1% SDS, 0.1 m Tris, pH 9, and 0.03% ethanolamine). The slides were then prehybridized during 60 min at 55°C in prehybridization buffer (150 μm bovine serum albumin, 3.5% SSC, and 0.7% SDS). Labeled cDNA was denatured at 90°C for 3 min and immediately spun down and kept at 55°C until loading onto the slide. The slides were then covered with a coverslip (Corning), placed in a hybridization chamber (Telechem), and hybridized for 17 h in a water bath at 55°C. The washing consisted in steps of 5 min at 55°C in 4× SSC, 5 min in the same buffer at room temperature, 1 min in 0.2× SSC at room temperature, and finally 1 min in 0.1× SSC at room temperature. Immediately after washing, slides were briefly rinsed with distilled water before being dried by centrifugation at 300g for 90 s. Scanning was performed immediately after this step.
Image Acquisition and Statistical Analysis
Scanning of the arrays was performed using a Genepix 4000A scanner (Molecular Devices). Photomultiplier gain at 570 nm (Cy3) and 660 nm (Cy5) was adjusted automatically using a threshold saturation of 0.001%, as implemented in Genepix 6.0 (Molecular Devices). Addressing and segmentation of spots were automatically detected and manually corrected, and intensities were quantified using the Genepix 6.0 software. All microarray experiments were MIAME compliant, and raw data were deposited under accession number E-MTAB-681 at the ArrayExpress database of the EMBL-European Bioinformatics Institute (http://www.ebi.ac.uk/microarray-as/ae/).
Data treatments were done using custom-designed scripts written under an R environment (R Development Core Team, 2009). Normalization of the Cy3 and Cy5 signal intensities within arrays was performed by Loess normalization (Yang and Thorne, 2003) followed by quantile normalization between arrays, as implemented in the Bioconductor package LIMMA (Smyth and Speed, 2003; Gentleman et al., 2004). The value of each sampling time point is an average of the technical replicates on a slide. Student’s t test, linear modeling features, and empirical Bayes test statistics of the LIMMA package (Smyth, 2004) were used to perform pairwise comparison of the stress induced by either H2O2 or MV on both LL and HL cultures (i.e. LL-Ct versus LL+MV, LL-Ct versus LL+H2O2, HL-Ct versus HL+MV, and HL-Ct versus HL+H2O2) as well as comparing the steady-state acclimation to different light conditions (i.e. LL-Ct versus HL-Ct). To take into account multiple testing, P values were adjusted using the Benjamini and Hochberg algorithm (Thissen et al., 2002). We particularly focused our attention on genes that were statistically significant using Student’s t test and/or LIMMA with FDR lower than or equal to 0.05, also taking into account a fold change (FC) cutoff between two conditions [log2(FC) < −1 or log2(FC) > 1].
Finally, hierarchical clustering analyses (Bolstad et al., 2004) and PCA (Raychaudhuri et al., 2000) were performed to investigate the technical and biological reproducibility of our results and highlight the main groups of genes, excluding Ct oligonucleotides, that shared similar expression patterns over multiple conditions. The clustering was performed using the hclust function from the R Stats package (R Development Core Team, 2009), using the clustering method “ward” and a Pearson correlation, and PCA analysis was carried out using the FactoMineR package (Husson et al., 2008). Analyses were performed on a subset of genes corresponding to the genes significantly differentially expressed (FDR ≤ 0.05 using t test and/or LIMMA) in at least one of the pairwise comparisons (1,202 genes left), except for two-dimensional (genes versus arrays) hierarchical clustering analyses, where no FDR restriction had been applied but where oligonucleotides targeting unknown proteins, hypothetical proteins, and small RNAs were removed (1,548 genes left).
Validation by Real-Time Quantitative PCR
To validate microarray data, real-time quantitative PCR (qPCR) was performed on 10 Synechococcus sp. WH7803 genes. This set includes genes that were either differentially expressed in microarray analyses or representative of key processes, including ROS detoxification processes and PSII turnover. Gene-specific primers for both reverse transcription and qPCR were designed using PrimerExpress software version 2.0 (Applied Biosystems; Supplemental Table S2), and the linearity of the cDNA content to CT (for cycle at threshold) ratio was checked for every set of primers within the dilution range used. Reverse transcription was carried out on 100 ng of RNA using SuperScriptII reverse transcriptase (Gibco-BRL) as described previously (Six et al., 2007b). qPCR was performed on the 1:100 diluted cDNA obtained, using the DNA Engine/Chromo4 Real Time PCR-Detector (Bio-Rad) and using absolute SYBR Green ROX Mix (Abgene), as described previously (Garczarek et al., 2008), in the presence of 200 nm primers, except for the sodB primers, which were used at 500 nm. The relative expression of each gene between two conditions was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001), using the rnpB gene as an internal standard (Mary and Vaulot, 2003). All genes and pairwise comparisons tested showed a similar response (up- or down-regulation) in qPCR and microarray experiments (Supplemental Table S2; Pearson correlation coefficient of 0.85).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NC_009481.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of MV on ROS production through photosynthetic electron transport.
Supplemental Figure S2. Effect of dark adaptation on PSII basal fluorescence in HL cells.
Supplemental Figure S3. Graphical representation of the predicted LexA motif identified in Synechococcus sp. WH7803.
Supplemental Table S1. Complete set of gene expression data as measured by microarray analyses.
Supplemental Table S2. Validation of microarray data using qPCR analyses on selected genes from Synechococcus sp. WH7803.
Acknowledgments
Nathalie Babic and Amandine Etcheverry are warmly thanked for their help with acquiring microarray data. The health genomic platform Biogenouest (Rennes, France) is acknowledged for allowing us access to its facilities. We also thank Frédéric Partensky and Amanda M. Cockshutt for very useful discussions on Synechococcus photophysiology as well as Emeric Dubois, Catherine Chevalier, and Rémi Houlgatte for discussions on array analyses.
References
- Adir N, Zer H, Shochat S, Ohad I. (2003) Photoinhibition: a historical perspective. Photosynth Res 76: 343–370 [DOI] [PubMed] [Google Scholar]
- Allakhverdiev SI, Murata N. (2004) Environmental stress inhibits the synthesis de novo of proteins involved in the photodamage-repair cycle of photosystem II in Synechocystis sp. PCC 6803. Biochim Biophys Acta 1657: 23–32 [DOI] [PubMed] [Google Scholar]
- Aro EM, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A, Battchikova N, Rintamäki E. (2005) Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot 56: 347–356 [DOI] [PubMed] [Google Scholar]
- Aro E-M, Virgin I, Andersson B. (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Asada K. (1994) Production and action of active oxygen species in photosynthetic tissues. Foyer CH, Mullineaux PM, , Causes of Photo-Oxidative Stress and Amelioration of Defense Systems in Plants. CRC Press, Boca Raton, FL, pp 77–104 [Google Scholar]
- Asada K. (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Barker M, de Vries R, Nield J, Komenda J, Nixon PJ. (2006) The deg proteases protect Synechocystis sp. PCC 6803 during heat and light stresses but are not essential for removal of damaged D1 protein during the photosystem two repair cycle. J Biol Chem 281: 30347–30355 [DOI] [PubMed] [Google Scholar]
- Battchikova N, Vainonen JP, Vorontsova N, Keranen M, Carmel D, Aro EM. (2010) Dynamic changes in the proteome of Synechocystis 6803 in response to CO(2) limitation revealed by quantitative proteomics. J Proteome Res 9: 5896–5912 [DOI] [PubMed] [Google Scholar]
- Bendall DS, Manasse RS. (1995) Cyclic photophosphorylation and electron-transport. Biochim Biophys Acta 1229: 23–38 [Google Scholar]
- Bolstad BM, Collin F, Simpson KM, Irizarry RA, Speed TP. (2004) Experimental design and low-level analysis of microarray data. Int Rev Neurobiol 60: 25–58 [DOI] [PubMed] [Google Scholar]
- Bouchard JN, Campbell DA, Roy S. (2005) Effects of UV-B radiation on the D1 protein repair cycle of natural phytoplankton communities from three latitudes (Canada, Brazil, and Argentina). J Phycol 41: 273–286 [Google Scholar]
- Buchanan BB, Balmer Y. (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Bus JS, Gibson JE. (1984) Paraquat: model for oxidant-initiated toxicity. Environ Health Perspect 55: 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butala M, Zgur-Bertok D, Busby SJW. (2009) The bacterial LexA transcriptional repressor. Cell Mol Life Sci 66: 82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D, Eriksson MJ, Oquist G, Gustafsson P, Clarke AK. (1998) The cyanobacterium Synechococcus resists UV-B by exchanging photosystem II reaction-center D1 proteins. Proc Natl Acad Sci USA 95: 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D, Zhou GQ, Gustafsson P, Oquist G, Clarke AK. (1995) Electron transport regulates exchange of two forms of photosystem II D1 protein in the cyanobacterium Synechococcus. EMBO J 14: 5457–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GX, Kazimir J, Cheniae GM. (1992) Photoinhibition of hydroxylamine-extracted photosystem II membranes: studies of the mechanism. Biochemistry 31: 11072–11083 [DOI] [PubMed] [Google Scholar]
- Chiancone E, Ceci P. (2010) The multifaceted capacity of Dps proteins to combat bacterial stress conditions: detoxification of iron and hydrogen peroxide and DNA binding. Biochim Biophys Acta 1800: 798–805 [DOI] [PubMed] [Google Scholar]
- Clarke AK, Hurry VM, Gustafsson P, Oquist G. (1993a) Two functionally distinct forms of the photosystem II reaction-center protein D1 in the cyanobacterium Synechococcus sp. PCC 7942. Proc Natl Acad Sci USA 90: 11985–11989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AK, Soitamo A, Gustafsson P, Oquist G. (1993b) Rapid interchange between two distinct forms of cyanobacterial photosystem II reaction-center protein D1 in response to photoinhibition. Proc Natl Acad Sci USA 90: 9973–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley JW, Vermaas WFJ. (2001) Succinate dehydrogenase and other respiratory pathways in thylakoid membranes of Synechocystis sp. strain PCC 6803: capacity comparisons and physiological function. J Bacteriol 183: 4251–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. (2001) Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158: 41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotta VC, Yang M, O’Halloran TV. (2006) Activation of superoxide dismutases: putting the metal to the pedal. Biochim Biophys Acta 1763: 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner BA, Petrouleas V. (1987) Light-induced oxidation of the acceptor-side Fe(II) of photosystem-II by exogenous quinines acting through the QB binding-site. 2. Blockage by inhibitors and their effects on the Fe(III) electron-paramagnetic-res spectra. Biochim Biophys Acta 893: 138–148 [Google Scholar]
- Domain F, Houot L, Chauvat F, Cassier-Chauvat C. (2004) Function and regulation of the cyanobacterial genes lexA, recA and ruvB: lexA is critical to the survival of cells facing inorganic carbon starvation. Mol Microbiol 53: 65–80 [DOI] [PubMed] [Google Scholar]
- Drabkova M, Matthijs HCP, Admiraal W, Marsalek B. (2007) Selective effects of H2O2 on cyanobacterial photosynthesis. Photosynthetica 45: 363–369 [Google Scholar]
- Dufresne A, Ostrowski M, Scanlan DJ, Garczarek L, Mazard S, Palenik BP, Paulsen IT, de Marsac NT, Wincker P, Dossat C, et al. (2008) Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol 9: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham KA, Bullerjahn GS. (2002) Immunocytochemical localization of the stress-induced DpsA protein in the cyanobacterium Synechococcus sp. strain PCC 7942. J Basic Microbiol 42: 367–372 [DOI] [PubMed] [Google Scholar]
- Edelman M, Mattoo AK. (2008) D1-protein dynamics in photosystem II: the lingering enigma. Photosynth Res 98: 609–620 [DOI] [PubMed] [Google Scholar]
- Edge R, McGarvey DJ, Truscott TG. (1997) The carotenoids as anti-oxidants: a review. J Photochem Photobiol B 41: 189–200 [DOI] [PubMed] [Google Scholar]
- Fan DY, Jia HS, Barber J, Chow WS. (2009) Novel effects of methyl viologen on photosystem II function in spinach leaves. Eur Biophys J 39: 191–199 [DOI] [PubMed] [Google Scholar]
- Fujii T, Yokoyama E, Inoue K, Sakurai H. (1990) The sites of electron donation of photosystem-I to methyl viologen. Biochim Biophys Acta 1015: 41–48 [Google Scholar]
- Funk C, Vermaas W. (1999) A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38: 9397–9404 [DOI] [PubMed] [Google Scholar]
- Garczarek L, Dufresne A, Blot N, Cockshutt AM, Peyrat A, Campbell DA, Joubin L, Six C. (2008) Function and evolution of the psbA gene family in marine Synechococcus: Synechococcus sp. WH7803 as a case study. ISME J 2: 937–953 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein S, Samuni A, Aronovitch Y, Godinger D, Russo A, Mitchell JB. (2002) Kinetics of paraquat and copper reactions with nitroxides: the effects of nitroxides on the aerobic and anoxic toxicity of paraquat. Chem Res Toxicol 15: 686–691 [DOI] [PubMed] [Google Scholar]
- Hakala M, Tuominen I, Keränen M, Tyystjärvi T, Tyystjärvi E. (2005) Evidence for the role of the oxygen-evolving manganese complex in photoinhibition of photosystem II. Biochim Biophys Acta 1706: 68–80 [DOI] [PubMed] [Google Scholar]
- Havaux M, Guedeney G, He Q, Grossman AR. (2003) Elimination of high-light-inducible polypeptides related to eukaryotic chlorophyll a/b-binding proteins results in aberrant photoacclimation in Synechocystis PCC6803. Biochim Biophys Acta 1557: 21–33 [DOI] [PubMed] [Google Scholar]
- He QF, Dolganov N, Bjorkman O, Grossman AR. (2001) The high light-inducible polypeptides in Synechocystis PCC6803: expression and function in high light. J Biol Chem 276: 306–314 [DOI] [PubMed] [Google Scholar]
- Herbert SK, Martin RE, Fork DC. (1995) Light adaptation of cyclic electron transport through photosystem I in the cyanobacterium Synechococcus sp. PCC 7942. Photosynth Res 46: 277–285 [DOI] [PubMed] [Google Scholar]
- Hideg E, Kós PB, Vass I. (2007) Photosystem II damage induced by chemically generated singlet oxygen in tobacco leaves. Physiol Plant 131: 33–40 [DOI] [PubMed] [Google Scholar]
- Hildebraunt AG, Roots I. (1975) Reduced nicotinamide adenine dinucleotide phosphate (NADPH)-dependent formation and breakdown of hydrogen peroxide during mixed function oxidation reactions in liver microsomes. Arch Biochem Biophys 171: 385–397 [DOI] [PubMed] [Google Scholar]
- Hirayama S, Ueda R, Sugata K. (1995) Effect of hydroxyl radical on intact microalgal photosynthesis. Energy Convers Manage 36: 685–688 [Google Scholar]
- Hosoya-Matsuda N, Motohashi K, Yoshimura H, Nozaki A, Inoue K, Ohmori M, Hisabori T. (2005) Anti-oxidative stress system in cyanobacteria: significance of type II peroxiredoxin and the role of 1-Cys peroxiredoxin in Synechocystis sp. strain PCC 6803. J Biol Chem 280: 840–846 [DOI] [PubMed] [Google Scholar]
- Hossain MM, Nakamoto H. (2002) HtpG plays a role in cold acclimation in cyanobacteria. Curr Microbiol 44: 291–296 [DOI] [PubMed] [Google Scholar]
- Hossain MM, Nakamoto H. (2003) Role for the cyanobacterial HtpG in protection from oxidative stress. Curr Microbiol 46: 70–76 [DOI] [PubMed] [Google Scholar]
- Houot L, Floutier M, Marteyn B, Michaut M, Picciocchi A, Legrain P, Aude JC, Cassier-Chauvat C, Chauvat F. (2007) Cadmium triggers an integrated reprogramming of the metabolism of Synechocystis PCC6803, under the control of the Slr1738 regulator. BMC Genomics 8: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson F, Josse J, Lê S. (2008) FactoMineR: an R package for multivariate analysis. J Stat Softw 25: 1–18 [Google Scholar]
- Imlay JA. (2003) Pathways of oxidative damage. Annu Rev Microbiol 57: 395–418 [DOI] [PubMed] [Google Scholar]
- Ishikita H, Knapp EW. (2005) Oxidation of the non-heme iron complex in photosystem II. Biochemistry 44: 14772–14783 [DOI] [PubMed] [Google Scholar]
- Jia H, Oguchi R, Hope AB, Barber J, Chow WS. (2008) Differential effects of severe water stress on linear and cyclic electron fluxes through photosystem I in spinach leaf discs in CO2-enriched air. Planta 228: 803–812 [DOI] [PubMed] [Google Scholar]
- Jia Y, Shan JY, Millard A, Clokie MRJ, Mann NH. (2010) Light-dependent adsorption of photosynthetic cyanophages to Synechococcus sp. WH7803. FEMS Microbiol Lett 310: 120–126 [DOI] [PubMed] [Google Scholar]
- Joliot P, Joliot A. (2002) Cyclic electron transfer in plant leaf. Proc Natl Acad Sci USA 99: 10209–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana TM, Glibert PM. (1987a) Effect of irradiances up to 2000 μE m−2s−1 on marine Synechococcus WH7803-I: growth, pigmentation, and cell composition. Deep Sea Research Part A Oceanographic Research Papers 34: 479–495 [Google Scholar]
- Kana TM, Glibert PM. (1987b) Effect of irradiances up to 2000 μE m−2s−1 on marine Synechococcus WH7803-II: photosynthetic responses and mechanisms. Deep Sea Research Part A Oceanographic Research Papers 34: 497–516 [Google Scholar]
- Kana TM, Glibert PM, Goericke R, Welschmeyer NA. (1988) Zeaxanthin and beta-carotene in Synechococcus WH7803 respond differently to irradiance. Limnol Oceanogr 33: 1623–1627 [Google Scholar]
- Kanesaki Y, Yamamoto H, Paithoonrangsarid K, Shoumskaya M, Suzuki I, Hayashi H, Murata N. (2007) Histidine kinases play important roles in the perception and signal transduction of hydrogen peroxide in the cyanobacterium, Synechocystis sp. PCC 6803. Plant J 49: 313–324 [DOI] [PubMed] [Google Scholar]
- Keren N, Berg A, van Kan PJM, Levanon H, Ohad I. (1997) Mechanism of photosystem II photoinactivation and D1 protein degradation at low light: the role of back electron flow. Proc Natl Acad Sci USA 94: 1579–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. (2005) Protein structures forming the shell of primitive bacterial organelles. Science 309: 936–938 [DOI] [PubMed] [Google Scholar]
- Kirilovsky D. (2007) Photoprotection in cyanobacteria: the orange carotenoid protein (OCP)-related non-photochemical-quenching mechanism. Photosynth Res 93: 7–16 [DOI] [PubMed] [Google Scholar]
- Klassen JL. (2010) Phylogenetic and evolutionary patterns in microbial carotenoid biosynthesis are revealed by comparative genomics. PLoS ONE 5: e11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP, Dodge AD. (1985) Singlet oxygen and plants. Phytochemistry 24: 889–896 [Google Scholar]
- Kobayashi M, Ishizuka T, Katayama M, Kanehisa M, Bhattacharyya-Pakrasi M, Pakrasi HB, Ikeuchi M. (2004) Response to oxidative stress involves a novel peroxiredoxin gene in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 45: 290–299 [DOI] [PubMed] [Google Scholar]
- Kojima K, Oshita M, Nanjo Y, Kasai K, Tozawa Y, Hayashi H, Nishiyama Y. (2007) Oxidation of elongation factor G inhibits the synthesis of the D1 protein of photosystem II. Mol Microbiol 65: 936–947 [DOI] [PubMed] [Google Scholar]
- Kolowrat C, Partensky F, Mella-Flores D, Le Corguillé G, Boutte C, Blot N, Ratin M, Ferréol M, Lecomte X, Gourvil P, et al. (2010) Ultraviolet stress delays chromosome replication in light/dark synchronized cells of the marine cyanobacterium Prochlorococcus marinus PCC9511. BMC Microbiol 10: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J, Barker M, Kuviková S, de Vries R, Mullineaux CW, Tichý M, Nixon PJ. (2006) The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J Biol Chem 281: 1145–1151 [DOI] [PubMed] [Google Scholar]
- Komenda J, Knoppova J, Krynicka V, Nixon PJ, Tichý M. (2010) Role of FtsH2 in the repair of photosystem II in mutants of the cyanobacterium Synechocystis PCC 6803 with impaired assembly or stability of the CaMn4 cluster. Biochim Biophys Acta 1797: 566–575 [DOI] [PubMed] [Google Scholar]
- Krupa Z, Oquist G, Gustafsson P. (1991) Photoinhibition of photosynthesis and growth-responses at different light levels in psbA gene mutants of the cyanobacterium Synechococcus. Physiol Plant 82: 1–8 [Google Scholar]
- Kufryk G, Hernandez-Prieto MA, Kieselbach T, Miranda H, Vermaas W, Funk C. (2008) Association of small CAB-like proteins (SCPs) of Synechocystis sp. PCC 6803 with photosystem II. Photosynth Res 95: 135–145 [DOI] [PubMed] [Google Scholar]
- Kuzminov A. (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev 63: 751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford PR, Sansone A, Valenti P, Battistoni A, Kroll JS. (2002) Bacterial superoxide dismutase and virulence. Methods Enzymol 349: 155–166 [DOI] [PubMed] [Google Scholar]
- Latifi A, Jeanjean R, Lemeille S, Havaux M, Zhang CC. (2005) Iron starvation leads to oxidative stress in Anabaena sp. strain PCC 7120. J Bacteriol 187: 6596–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A, Ruiz M, Zhang CC. (2009) Oxidative stress in cyanobacteria. FEMS Microbiol Rev 33: 258–278 [DOI] [PubMed] [Google Scholar]
- Ledford HK, Niyogi KK. (2005) Singlet oxygen and photo-oxidative stress management in plants and algae. Plant Cell Environ 28: 1037–1045 [Google Scholar]
- Lesser MP. (2006) Oxidative stress in marine environments: biochemistry and physiological ecology. Annu Rev Physiol 68: 253–278 [DOI] [PubMed] [Google Scholar]
- Li H, Singh AK, McIntyre LM, Sherman LA. (2004) Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J Bacteriol 186: 3331–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xu M, Su Z. (2010) Computational analysis of LexA regulons in cyanobacteria. BMC Genomics 11: 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D, Post AF. (2001) Ecological aspects of ntcA gene expression and its use as an indicator of the nitrogen status of marine Synechococcus spp. Appl Environ Microbiol 67: 3340–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Long SP, Humphries S, Falkowski PG. (1994) Photoinhibition of photosynthesis in nature. Annu Rev Plant Physiol Plant Mol Biol 45: 633–662 [Google Scholar]
- Lupínková L, Komenda J. (2004) Oxidative modifications of the photosystem II D1 protein by reactive oxygen species: from isolated protein to cyanobacterial cells. Photochem Photobiol 79: 152–162 [DOI] [PubMed] [Google Scholar]
- MacDonald TM, Dubois L, Smith LC, Campbell DA. (2003) Sensitivity of cyanobacterial antenna, reaction center and CO2 assimilation transcripts and proteins to moderate UVB: light acclimation potentiates resistance to UVB. Photochem Photobiol 77: 405–412 [DOI] [PubMed] [Google Scholar]
- Marie D, Brussaard CPD, Thyrhaug R, Bratbak G, Vaulot D. (1999) Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl Environ Microbiol 65: 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D, Partensky F, Jacquet S, Vaulot D. (1997) Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl Environ Microbiol 63: 186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary I, Vaulot D. (2003) Two-component systems in Prochlorococcus MED4: genomic analysis and differential expression under stress. FEMS Microbiol Lett 226: 135–144 [DOI] [PubMed] [Google Scholar]
- Masamoto K, Misawa N, Kaneko T, Kikuno R, Toh H. (1998) Beta-carotene hydroxylase gene from the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol 39: 560–564 [DOI] [PubMed] [Google Scholar]
- Masip L, Veeravalli K, Georgiou G. (2006) The many faces of glutathione in bacteria. Antioxid Redox Signal 8: 753–762 [DOI] [PubMed] [Google Scholar]
- Mazón G, Lucena JM, Campoy S, Fernández de Henestrosa AR, Candau P, Barbé J. (2004) LexA-binding sequences in Gram-positive and cyanobacteria are closely related. Mol Genet Genomics 271: 40–49 [DOI] [PubMed] [Google Scholar]
- Michel B. (2005) After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol 3: e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millican DS, Bird IM. (1998) Extraction of cellular and tissue RNA. Methods Mol Biol 105: 315–324 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Miyao M, Ikeuchi M, Yamamoto N, Ono T. (1995) Specific degradation of the D1 protein of photosystem II by treatment with hydrogen peroxide in darkness: implications for the mechanism of degradation of the D1 protein under illumination. Biochemistry 34: 10019–10026 [DOI] [PubMed] [Google Scholar]
- Montané MH, Kloppstech K. (2000) The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): was the harvesting of light their primary function? Gene 258: 1–8 [DOI] [PubMed] [Google Scholar]
- Moore LR, Goericke R, Chisholm SW. (1995) Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar Ecol Prog Ser 116: 259–275 [Google Scholar]
- Mopper K, Kieber DJ. (2000) Marine photochemistry and its impact on carbon cycling. Demers S, de Mora S, Vernet M, , Effects of UV Radiation on Marine Ecosystems. Cambridge University Press, Cambridge, UK, pp 101–129 [Google Scholar]
- Murata N, Takahashi S, Nishiyama Y, Allakhverdiev SI. (2007) Photoinhibition of photosystem II under environmental stress. Biochim Biophys Acta 1767: 414–421 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Allakhverdiev SI, Murata N. (2006) A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim Biophys Acta 1757: 742–749 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Allakhverdiev SI, Yamamoto H, Hayashi H, Murata N. (2004) Singlet oxygen inhibits the repair of photosystem II by suppressing the translation elongation of the D1 protein in Synechocystis sp. PCC 6803. Biochemistry 43: 11321–11330 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N. (2001) Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J 20: 5587–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu JF, Boehm M, Komenda J. (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot (Lond) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PJ, Sarcina M, Diner BA. (2005) Protein constituents of photosystem II: the D1 and D2 core proteins. Wydrzynski T, Satoh K, , Photosystem II: The Water/Plastoquinone Oxido-Reductase In Photosynthesis, Vol 22. Kluwer Academic Publications, Dordrecht, The Netherlands, pp 71–94 [Google Scholar]
- Noctor G, Foyer CH. (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Mi H. (2007) Cyanobacterial NADPH dehydrogenase complexes. Photosynth Res 93: 69–77 [DOI] [PubMed] [Google Scholar]
- Ohnishi N, Allakhverdiev SI, Takahashi S, Higashi S, Watanabe M, Nishiyama Y, Murata N. (2005) Two-step mechanism of photodamage to photosystem II: step 1 occurs at the oxygen-evolving complex and step 2 occurs at the photochemical reaction center. Biochemistry 44: 8494–8499 [DOI] [PubMed] [Google Scholar]
- Okada K, Ikeuchi M, Yamamoto N, Ono TA, Miyao M. (1996) Selective and specific cleavage of the D1 and D2 proteins of photosystem II by exposure to singlet oxygen: factors responsible for the susceptibility to cleavage of the proteins. Biochim Biophys Acta 1274: 73–798765097 [Google Scholar]
- Park YI, Chow WS, Anderson JM. (1995) Light inactivation of functional photosystem-II in leaves of peas grown in moderate light depends on photon exposure. Planta 196: 401–411 [Google Scholar]
- Patterson CO, Myers J. (1973) Photosynthetic production of hydrogen peroxide by Anacystis nidulans. Plant Physiol 51: 104–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson-Fortin LM, Colvin KR, Owttrim GW. (2006) A LexA-related protein regulates redox-sensitive expression of the cyanobacterial RNA helicase, crhR. Nucleic Acids Res 34: 3446–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospísil P. (2009) Production of reactive oxygen species by photosystem II. Biochim Biophys Acta 1787: 1151–1160 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2009) R: a language and environment for statistical computing. http://www.R-project.org (February 1, 2011)
- Rastogi RP, Singh SP, Häder DP, Sinha RP. (2010) Detection of reactive oxygen species (ROS) by the oxidant-sensing probe 2′,7′-dichlorodihydrofluorescein diacetate in the cyanobacterium Anabaena variabilis PCC 7937. Biochem Biophys Res Commun 397: 603–607 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Stuart JM, Altman RB. (2000) Principal components analysis to summarize microarray experiments: application to sporulation time series. Pac Symp Biocomput 5: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regelsberger G, Jakopitsch C, Plasser L, Schwaiger H, Furtmuller PG, Peschek GA, Zamocky M, Obinger C. (2002) Occurrence and biochemistry of hydroperoxidases in oxygenic phototrophic prokaryotes (cyanobacteria). Plant Physiol Biochem 40: 479–490 [Google Scholar]
- Rippka R, Coursin T, Hess W, Lichtlé C, Scanlan DJ, Palinska KA, Iteman I, Partensky F, Houmard J, Herdman M. (2000) Prochlorococcus marinus Chisholm et al. 1992 subsp. pastoris subsp. nov. strain PCC 9511, the first axenic chlorophyll a2/b2-containing cyanobacterium (Oxyphotobacteria). Int J Syst Evol Microbiol 50: 1833–1847 [DOI] [PubMed] [Google Scholar]
- Roncel M, Navarro JA, Delarosa MA. (1988) Hydrogen-peroxide photoproduction sensitized with rose-bengal with semicarbazide as the electron source. J Photochem Photobiol Chem 45: 341–353 [Google Scholar]
- Ross C, Santiago-Vázquez L, Paul V. (2006) Toxin release in response to oxidative stress and programmed cell death in the cyanobacterium Microcystis aeruginosa. Aquat Toxicol 78: 66–73 [DOI] [PubMed] [Google Scholar]
- Sato T, Minagawa S, Kojima E, Okamoto N, Nakamoto H. (2010) HtpG, the prokaryotic homologue of Hsp90, stabilizes a phycobilisome protein in the cyanobacterium Synechococcus elongatus PCC 7942. Mol Microbiol 76: 576–589 [DOI] [PubMed] [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. (2009) Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer L, Vioque A, Sandmann G. (2005) Functional in situ evaluation of photosynthesis-protecting carotenoids in mutants of the cyanobacterium Synechocystis PCC6803. J Photochem Photobiol B 78: 195–201 [DOI] [PubMed] [Google Scholar]
- Schelin J, Lindmark F, Clarke AK. (2002) The clpP multigene family for the ATP-dependent Clp protease in the cyanobacterium Synechococcus. Microbiology 148: 2255–2265 [DOI] [PubMed] [Google Scholar]
- Schürmann P, Buchanan BB. (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal 10: 1235–1274 [DOI] [PubMed] [Google Scholar]
- Schwarz R, Forchhammer K. (2005) Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology 151: 2503–2514 [DOI] [PubMed] [Google Scholar]
- Six C, Finkel ZV, Irwin AJ, Campbell DA. (2007a) Light variability illuminates niche-partitioning among marine picocyanobacteria. PLoS ONE 2: e1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six C, Finkel ZV, Rodriguez F, Marie D, Partensky F, Campbell DA. (2008) Contrasting photoacclimation costs in ecotypes of the marine eukaryotic picoplankter Ostreococcus. Limnol Oceanogr 53: 255–265 [Google Scholar]
- Six C, Joubin L, Partensky F, Holtzendorff J, Garczarek L. (2007b) UV-induced phycobilisome dismantling in the marine picocyanobacterium Synechococcus sp. WH8102. Photosynth Res 92: 75–86 [DOI] [PubMed] [Google Scholar]
- Six C, Thomas JC, Brahamsha B, Lemoine Y, Partensky F. (2004) Photophysiology of the marine cyanobacterium Synechococcus sp. WH8102, a new model organism. Aquat Microb Ecol 35: 17–29 [Google Scholar]
- Six C, Thomas JC, Thion L, Lemoine Y, Zal F, Partensky F. (2005) Two novel phycoerythrin-associated linker proteins in the marine cyanobacterium Synechococcus sp. strain WH8102. J Bacteriol 187: 1685–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. (2003) Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Song YG, Liu B, Wang LF, Li MH, Liu Y. (2006) Damage to the oxygen-evolving complex by superoxide anion, hydrogen peroxide, and hydroxyl radical in photoinhibition of photosystem II. Photosynth Res 90: 67–78 [DOI] [PubMed] [Google Scholar]
- Stanne TM, Pojidaeva E, Andersson FI, Clarke AK. (2007) Distinctive types of ATP-dependent Clp proteases in cyanobacteria. J Biol Chem 282: 14394–14402 [DOI] [PubMed] [Google Scholar]
- Storz G, Imlay JA. (1999) Oxidative stress. Curr Opin Microbiol 2: 188–194 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Murata N. (2008) How do environmental stresses accelerate photoinhibition? Trends Plant Sci 13: 178–182 [DOI] [PubMed] [Google Scholar]
- Thissen D, Steinberg L, Kuang D. (2002) Quick and easy implementation of the Benjamini-Hochberg procedure for controlling the false positive rate in multiple comparisons. J Educ Behav Stat 27: 77–83 [Google Scholar]
- Thomas DJ, Avenson TJ, Thomas JB, Herbert SK. (1998) A cyanobacterium lacking iron superoxide dismutase is sensitized to oxidative stress induced with methyl viologen but is not sensitized to oxidative stress induced with norflurazon. Plant Physiol 116: 1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichý M, Lupínková L, Sicora C, Vass I, Kuviková S, Prásil O, Komenda J. (2003) Synechocystis 6803 mutants expressing distinct forms of the photosystem II D1 protein from Synechococcus 7942: relationship between the psbA coding region and sensitivity to visible and UV-B radiation. Biochim Biophys Acta 1605: 55–66 [DOI] [PubMed] [Google Scholar]
- Tichý M, Vermaas W. (1999) In vivo role of catalase-peroxidase in Synechocystis sp. strain PCC 6803. J Bacteriol 181: 1875–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo G, Palenik B, Brahamsha B. (1999) Swimming marine Synechococcus strains with widely different photosynthetic pigment ratios form a monophyletic group. Appl Environ Microbiol 65: 5247–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyystjärvi E, Aro EM. (1996) The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc Natl Acad Sci USA 93: 2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass I, Cser K. (2009) Janus-faced charge recombinations in photosystem II photoinhibition. Trends Plant Sci 14: 200–205 [DOI] [PubMed] [Google Scholar]
- Vass I, Styring S, Hundal T, Koivuniemi A, Aro EM, Andersson B. (1992) Reversible and irreversible intermediates during photoinhibition of photosystem II: stable reduced QA species promote chlorophyll triplet formation. Proc Natl Acad Sci USA 89: 1408–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavilin D, Yao D, Vermaas W. (2007) Small Cab-like proteins retard degradation of photosystem II-associated chlorophyll in Synechocystis sp. PCC 6803: kinetic analysis of pigment labeling with 15N and 13C. J Biol Chem 282: 37660–37668 [DOI] [PubMed] [Google Scholar]
- Wang P, Duan W, Takabayashi A, Endo T, Shikanai T, Ye JY, Mi HL. (2006) Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol 141: 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Ajlani G, Verbavatz JM, Vass I, Kerfeld CA, Kirilovsky D. (2006) A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell 18: 992–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Thorne NP. (2003) Normalization for two-color cDNA microarray data. Lecture Notes Monogr Ser 40: 403–418 [Google Scholar]
- Yousef N, Pistorius EK, Michel KP. (2003) Comparative analysis of idiA and isiA transcription under iron starvation and oxidative stress in Synechococcus elongatus PCC 7942 wild-type and selected mutants. Arch Microbiol 180: 471–483 [DOI] [PubMed] [Google Scholar]
- Yu L, Zhao JD, Muhlenhoff U, Bryant DA, Golbeck JH. (1993) psaE is required for in-vivo cyclic electron flow around photosystem I in the cyanobacterium Synechococcus sp PCC 7002. Plant Physiol 103: 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YH, Graham JE, Ludwig M, Xiong W, Alvey RM, Shen GZ, Bryant DA. (2010) Roles of xanthophyll carotenoids in protection against photoinhibition and oxidative stress in the cyanobacterium Synechococcus sp. strain PCC 7002. Arch Biochem Biophys 504: 86–99 [DOI] [PubMed] [Google Scholar]
- Zolla L, Rinalducci S. (2002) Involvement of active oxygen species in degradation of light-harvesting proteins under light stresses. Biochemistry 41: 14391–14402 [DOI] [PubMed] [Google Scholar]