Abstract
Conventional optical microscopes image cells by use of objective lenses that work together with other lenses and optical components. While quite effective, this classical approach has certain limitations for miniaturization of the imaging platform to make it compatible with the advanced state of the art in microfluidics. In this report, we introduce experimental details of a lensless on-chip imaging concept termed LUCAS (Lensless Ultra-wide field-of-view Cell monitoring Array platform based on Shadow imaging) that does not require any microscope objectives or other bulky optical components to image a heterogeneous cell solution over an ultra-wide field of view that can span as large as ~18 cm2. Moreover, unlike conventional microscopes, LUCAS can image a heterogeneous cell solution of interest over a depth-of-field of ~5 mm without the need for refocusing which corresponds to up to ~9 mL sample volume. This imaging platform records the shadows (i.e., lensless digital holograms) of each cell of interest within its field of view, and automated digital processing of these cell shadows can determine the type, the count and the relative positions of cells within the solution. Because it does not require any bulky optical components or mechanical scanning stages it offers a significantly miniaturized platform that at the same time reduces the cost, which is quite important for especially point of care diagnostic tools. Furthermore, the imaging throughput of this platform is orders of magnitude better than conventional optical microscopes, which could be exceedingly valuable for high-throughput cell-biology experiments.
Protocol
Here we discuss the experimental procedures that are involved in LUCAS [1-3]. To illustrate the proof of concept of LUCAS we will describe the imaging process for a whole blood sample.
A. Imaging Set-up
The LUCAS Imaging platform exhibits significant advantages to provide a cost-effective and compact alternative to existing point-of-care cytometry and medical diagnostic tools, especially for resource-limited settings. Rather than detecting the image of the cells, LUCAS instead captures digital holograms of the cells that are created by the interference of the scattered light from each cell with the background light. Careful control of the partial spatial coherence of the illumination is critical to enable holographic recording.
1. Digital Sensor Array
The LUCAS platform utilizes an optoelectronic sensor array to digitally record individual cell holograms. For this purpose, charged couple devices (CCD; Sample Models: KAI-11002, KAF-39000, from Kodak) or complementary metal-oxide-semiconductor chips (CMOS, Sample Model: MT9P031, Micron) can be used. Pixel sizes for the Kodak charged couple devices, KAI-11002, KAF-39000, and Micron CMOS image sensors are 9 μm, 6.8 μm and 2.2 μm, with an active FOV of 10 cm2, 18 cm2, and 24.4 mm2, respectively. [1-2].
2. Light Source
Unlike most other microscopic imaging modalities, LUCAS does not require a laser and therefore even a simple light emitting diode (LED) can be used for illumination. In order to enable tunable wavelength illumination, we can also utilize a monochromator with a Xenon Lamp (Cornerstone T260, Newport Corp.) along with a standard grade fused silica fiber which consists of a bundle of fibers (77564, Newport) and pinhole of ~100 μm diameter located at ~5-10 cm above the sensor surface. This tunable wavelength illumination configuration provides a flexible platform where the holographic signatures of the cells can be adjusted, and hybrid digital signatures can be synthesized to improve the signal to noise ratio for better characterization accuracy and specificity. [3]
B. Sample Preparation and Imaging
A proof of concept of LUCAS based on-chip imaging will be demonstrated using a heterogeneous solution as described below. A similar protocol could be applied for various other cell types [1-3].
1. Whole blood dilution and preparation of the heterogeneous solution
To begin the process, obtain a whole blood sample and polystyrene microbeads of various diameters (5, 10, and 20 μm. Duke Scientific), and bring all of them to room temperature.
Add 2 mL of RPMI 1640 classic liquid media (Fisher Scientific) as a diluent into a sterile 5 mL polypropylene tube.
After incubating the whole blood sample for 30 minutes, pipette up 10 μL of sedimented erythrocyte specimen and transfer it into the RPMI solution.
Add a total volume of 20 μL polystyrene microbeads into erythrocyte RPMI dilution, and agitate the cell solution by gently pipetting or by using magnetic stirrer with a stir bar.
Place a total volume of 5-15 μL heterogeneous solution onto a cover slip and place a second, identical cover slip over the solution by using forceps. The sample should be sandwiched between the two cover slips evenly.
Then, using a vacuum pen (NT57-636, Edmund Optics) load the sample onto the active region of the sensor array for imaging.
2. Whole Blood Staining
Starting with preparation of a mixture of 0.1 % buffered Eosin Y and diluted New Methylene Solution by using power type pure Eosin Y (MW=691.85, Acros Organics), zinc-free pure New Methylene Blue dye (MW=347.90, Acros Organics), and Potassium Oxalate Monohydrate (99.0% Reagent ACS, Acros Organics); each solution is purified with a 0.45 μm pore size Syringless Filter (Whatman) for aqueous white blood cell staining.
Then, gently stir a dissolved aqueous staining reagent with the whole blood specimen at a volume ratio of 1:1 in a polypropylene beaker by using a magnetic stirrer for 2 minutes and incubate for 10 minutes.
Transfer a stained blood solution of 50 μL into a 5 mL polypropylene tube and dilute with RPMI 1640 classic liquid solution (Fisher Scientific) of 1.95 mL to acquire a volume ratio of 1:200. Then, vortex the dilution for 30 seconds. A different dilution rate can also be introduced at this stage.
Pipette a stained cell solution of 10-100 μL between cover slips or within a microfluidic reservoir. Then, using a vacuum pen (NT57-636, Edmund Optics), place the specimen onto the active region of sensor array.
REPRESENTATIVE RESULTS
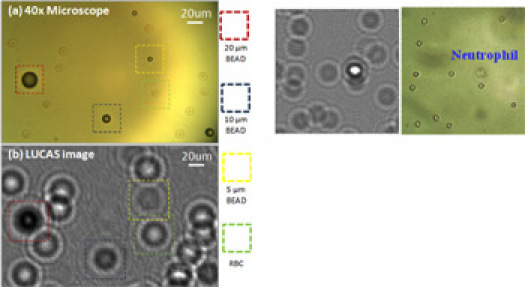 Figure 1: LUCAS enables accurate counting and digital differentiation of various cells and micro-objects based on their 2D holographic signatures without using any lenses, lasers or other bulky optical components. Characteristic LUCAS signatures of various micro-objects are illustrated in this figure and are compared against a conventional microscope image obtained with a 40X objective lens. Please click here to see a larger version of figure 1.
Figure 1: LUCAS enables accurate counting and digital differentiation of various cells and micro-objects based on their 2D holographic signatures without using any lenses, lasers or other bulky optical components. Characteristic LUCAS signatures of various micro-objects are illustrated in this figure and are compared against a conventional microscope image obtained with a 40X objective lens. Please click here to see a larger version of figure 1.
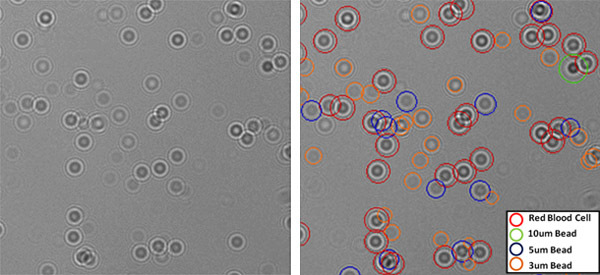 Figure 2: (Left) Raw image of a heterogeneous mixture containing red blood cells, 10um, 5um and 3 um particles. (Right) Fully-automated LUCAS characterization results for the same field of view are illustrated. Note that the decision algorithm is robust in characterizing high density regions as well as particles with low signal to noise ratio such as the 3um beads.
Figure 2: (Left) Raw image of a heterogeneous mixture containing red blood cells, 10um, 5um and 3 um particles. (Right) Fully-automated LUCAS characterization results for the same field of view are illustrated. Note that the decision algorithm is robust in characterizing high density regions as well as particles with low signal to noise ratio such as the 3um beads.
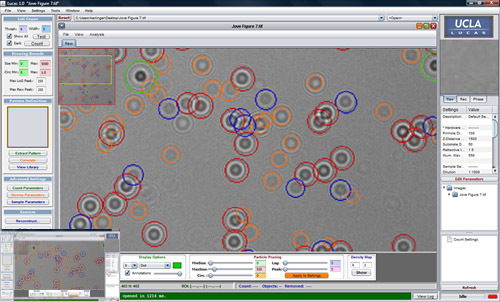 Figure 3: The LUCAS custom interface is illustrated. Java based LUCAS software allows inputs for various experimental conditions such as the sensor pixel size or the wavelength of light. Selection of a specific field of view on the image can also be made and the target cell patterns can be defined by the user to build a statistical cell shadow library. The acquired LUCAS image can then be characterized based on this training data (i.e., the cell shadow library) and the marked (counted) image is displayed to the user. Statistics of the count results are also stored as an XML file for further analysis.
Figure 3: The LUCAS custom interface is illustrated. Java based LUCAS software allows inputs for various experimental conditions such as the sensor pixel size or the wavelength of light. Selection of a specific field of view on the image can also be made and the target cell patterns can be defined by the user to build a statistical cell shadow library. The acquired LUCAS image can then be characterized based on this training data (i.e., the cell shadow library) and the marked (counted) image is displayed to the user. Statistics of the count results are also stored as an XML file for further analysis.
Discussion
We have illustrated that the LUCAS platform can accurately count and identify various micro-objects/cells on a chip based on their holographic signatures, and provides a promising tool for point-of-care medical diagnostics and high-throughput cell-biology. In order to accurately process the recorded holographic patterns, we implemented a custom-developed LUCAS decision software. This algorithm, which uses a statistical diffraction image database created by training of LUCAS images, identifies various features of cells within a heterogeneous solution; classifies the type and the relative position of individual cells and counts the desired cell type after applying thresholding to discard unwanted particle/cell patterns [1-3].
In summary, the LUCAS platform should provide a cost-effective and compact solution for point-of-care cytometry and medical diagnostics. For this end, LUCAS will especially be useful for monitoring of HIV patients in resource limited settings as well as for other global health related problems such as malaria and tuberculosis.
References
- Seo S, Su T, Tseng DK, Erlinger A, Ozcan A. "Lensfree Holographic Imaging for On-Chip Cytometry and Diagnostics". Lab Chip. 2009 doi: 10.1039/b813943a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Seo S, Erlinger A, Ozcan A. "High-throughput Imaging and Characterization of a Heterogeneous Cell Solution on a Chip" . Biotechnology and Bioengineering. 2008 doi: 10.1002/bit.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Seo S, Erlinger A, Ozcan A. "Multi-color LUCAS: Lensfree on-chip cytometry using tunable monochromatic illumination and digital noise reduction". Cellular and Molecular Bioengineering. 2008 [Google Scholar]


