Abstract
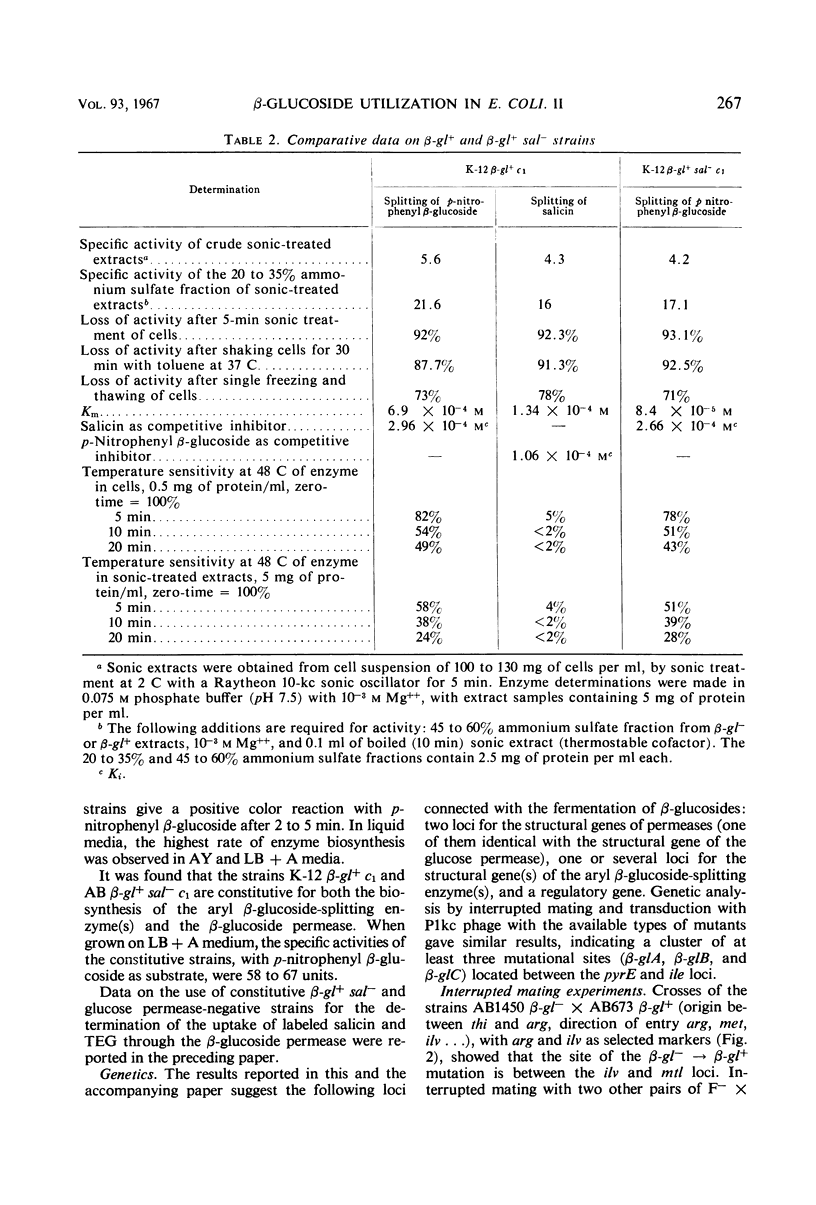
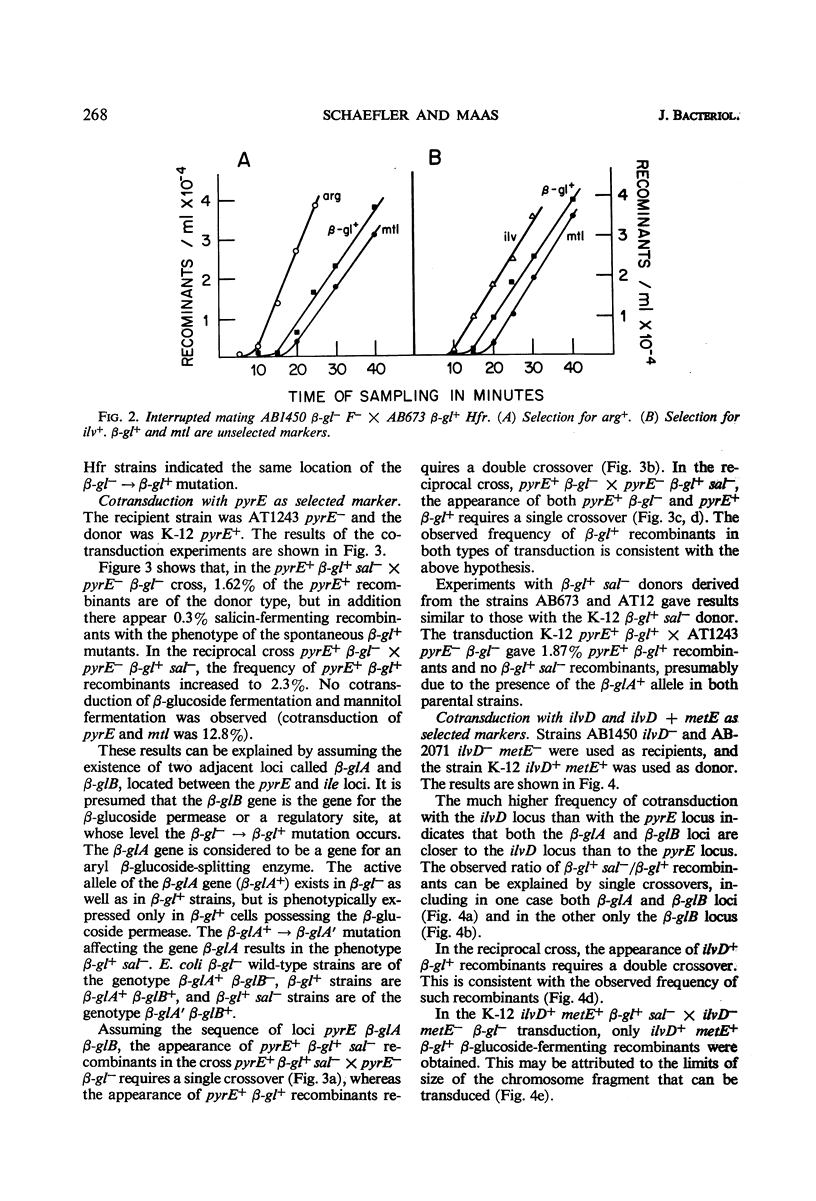
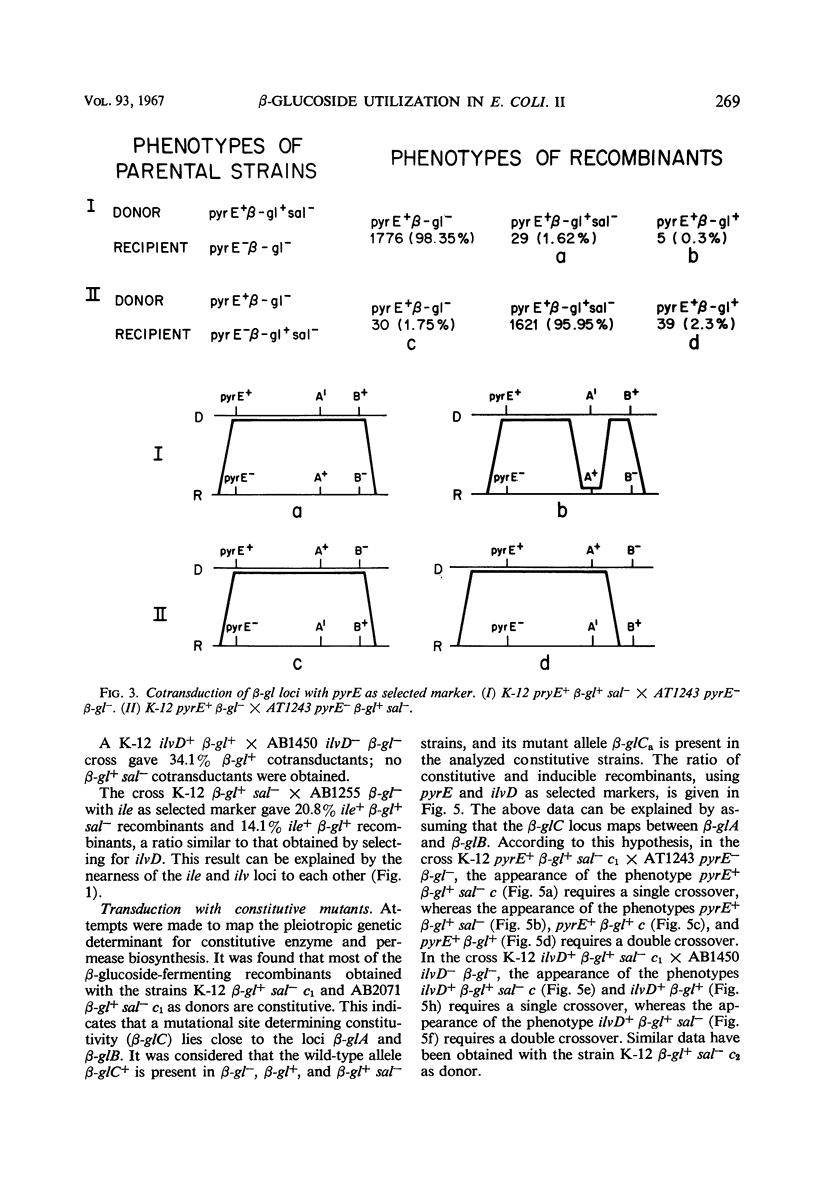
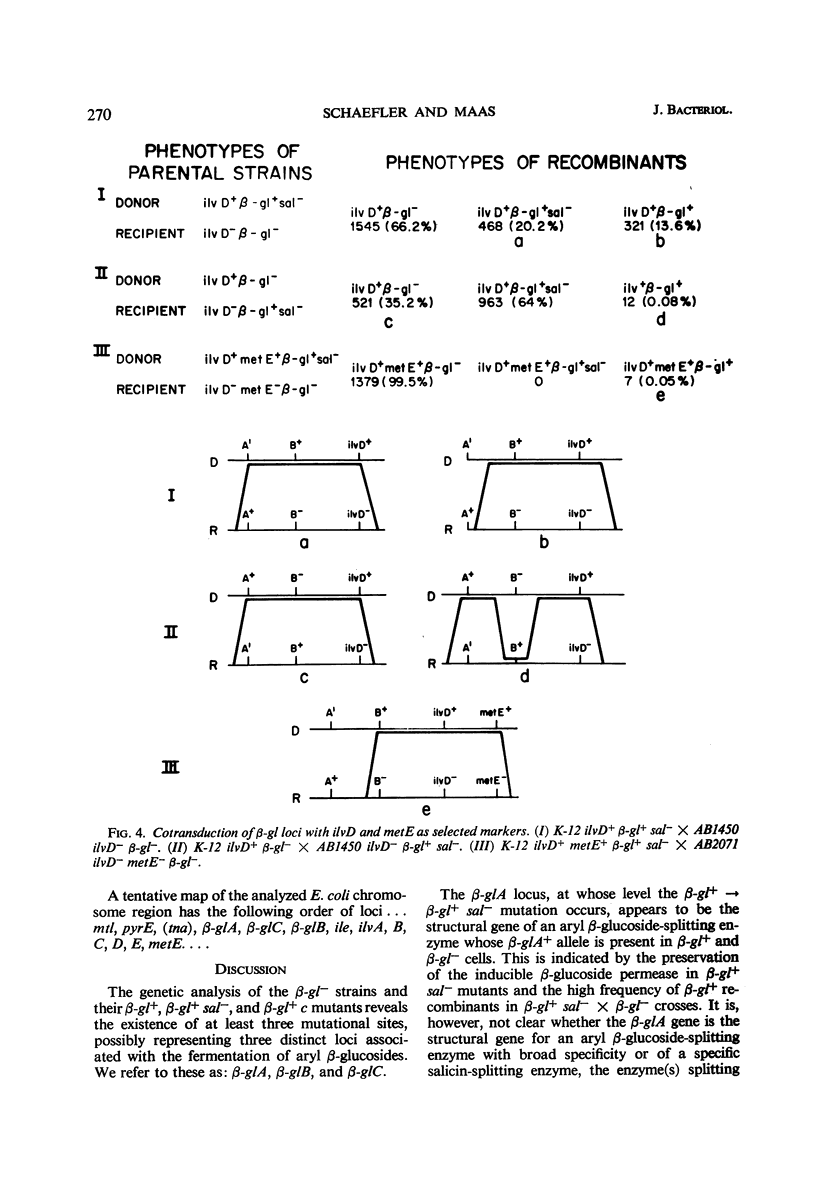
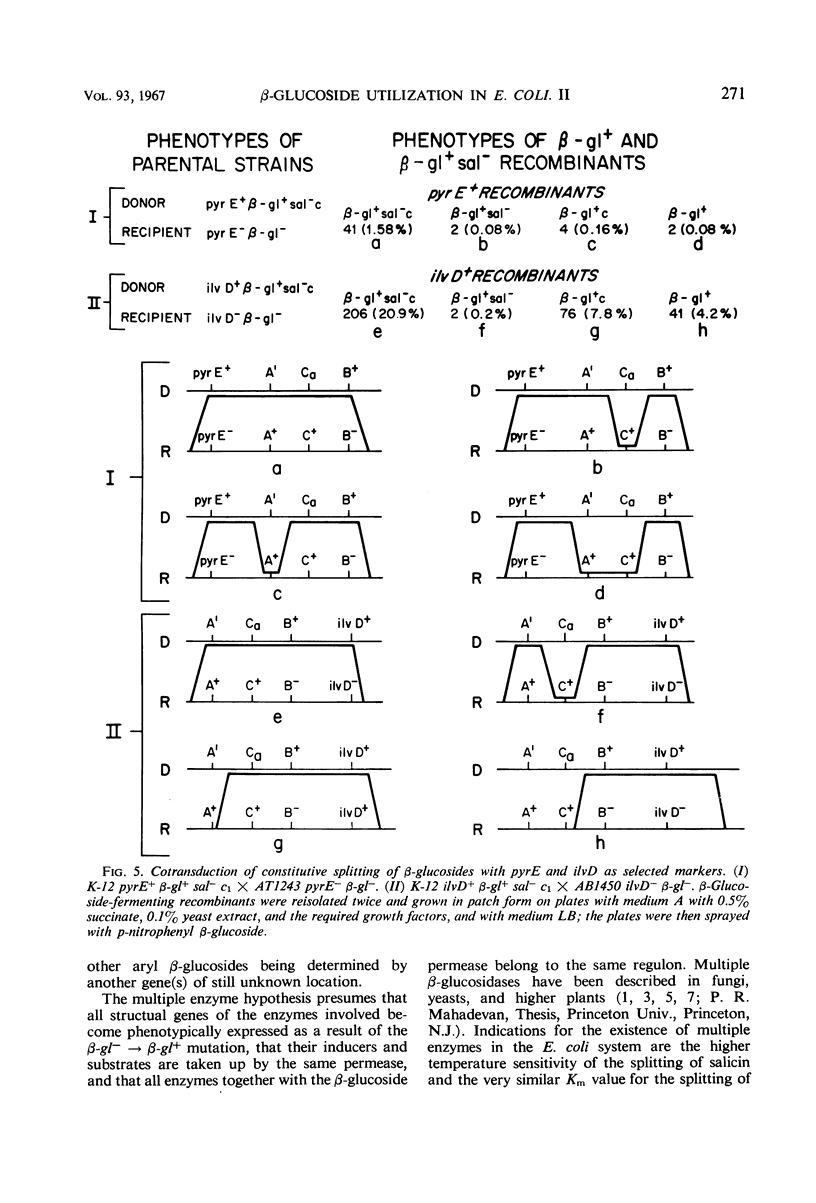
Two types of mutants obtained by treating β-gl+ cells with nitrosoguanidine are described. One type, β-gl+c, is constitutive for the biosynthesis of the aryl β-glucoside splitting enzyme(s) and for the β-glucoside permease; the other (β-gl+sal−) has lost the capacity to ferment salicin, but has retained the capacity to ferment arbutin and other aryl β-glucosides. By two successive mutational steps, β-gl+sal−c double mutants can be obtained. Determinations of the enzymatic splitting of salicin and p-nitrophenyl β-glucoside by β-gl+sal− cells and extracts showed that these mutants have lost the capacity to split salicin but do split p-nitrophenyl β-glucoside; they possess the β-glucoside permease, and in them salicin is a gratuitous inducer for enzyme and permease biosynthesis. Studies on a β-gl+ strain, which splits salicin as well as p-nitrophenyl β-glucoside, have shown that the splitting of salicin is more temperature-sensitive than that of p-nitrophenyl β-glucoside and other β-glucosides. Other properties of the two activities are similar. Interrupted mating experiments and cotransduction with P1kc phage showed that the genetic determinants of the β-glucoside system map between the pyrE and ile loci. Three distinct mutational sites were found and are presumed to have the following functions: β-glA, a structural gene for an aryl β-glucoside splitting enzyme; β-glB, either the structural gene for the β-glucoside-permease or a regulatory gene; and β-glC, a regulatory gene (or site). Escherichia coli wild-type strains are of the genotype A+ B− C+. The β-gl+ mutation determining the ability to ferment β-glucosides is considered to be a permease or regulatory mutation, and the resulting genotype is A+ B+ C+. The β-gl+sal− phenotype results from a mutation in the β-glA gene (genotype A′ B+ C+), and the constitutive phenotype results from a mutation in the β-glC gene, the genotypes A+ B+Ca and A′ B+Ca corresponding to the phenotypes β-gl+c and β-gl+sal−c.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BARNETT J. A., INGRAM M., SWAIN T. The use of beta-glucosides in classifying yeasts. J Gen Microbiol. 1956 Dec;15(3):529–555. doi: 10.1099/00221287-15-3-529. [DOI] [PubMed] [Google Scholar]
- Englesberg E., Irr J., Power J., Lee N. Positive control of enzyme synthesis by gene C in the L-arabinose system. J Bacteriol. 1965 Oct;90(4):946–957. doi: 10.1128/jb.90.4.946-957.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HELFERICH B., KLEINSCHMIDT T. ZUR KENNTNIS DES SUESSMANDELEMULSINS. Hoppe Seylers Z Physiol Chem. 1965;340:31–45. doi: 10.1515/bchm2.1965.340.1-2.31. [DOI] [PubMed] [Google Scholar]
- KAPLAN J. G. AN INDUCIBLE SYSTEM FOR THE HYDROLYSIS AND TRANSPORT OF BETA-GLUCOSIDES IN YEAST. I. CHARACTERISTICS OF THE BETA-GLUCOSIDASE ACTIVITY OF INTACT AND OF LYSED CELLS. J Gen Physiol. 1965 May;48:873–886. doi: 10.1085/jgp.48.5.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LI L. H., KING K. W. Fractionation of beta-glucosidases and related extracellular enzymes from Aspergillus niger. Appl Microbiol. 1963 Jul;11:320–325. doi: 10.1128/am.11.4.320-325.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN E. C., LERNER S. A., JORGENSEN S. E. A method for isolating constitutive mutants for carbohydrate-catabolizing enzymes. Biochim Biophys Acta. 1962 Jul 2;60:422–424. doi: 10.1016/0006-3002(62)90423-7. [DOI] [PubMed] [Google Scholar]
- MANDELL J. D., GREENBERG J. A new chemical mutagen for bacteria, 1-methyl-3-nitro-1-nitrosoguanidine. Biochem Biophys Res Commun. 1960 Dec;3:575–577. doi: 10.1016/0006-291x(60)90064-4. [DOI] [PubMed] [Google Scholar]
- Schaefler S. Inducible system for the utilization of beta-glucosides in Escherichia coli. I. Active transport and utilization of beta-glucosides. J Bacteriol. 1967 Jan;93(1):254–263. doi: 10.1128/jb.93.1.254-263.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAYLOR A. L., THOMAN M. S. THE GENETIC MAP OF ESCHERICHIA COLI K-12. Genetics. 1964 Oct;50:659–677. doi: 10.1093/genetics/50.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLSON C., PERRIN D., COHN M., JACOB F., MONOD J. NON-INDUCIBLE MUTANTS OF THE REGULATOR GENE IN THE "LACTOSE" SYSTEM OF ESCHERICHIA COLI. J Mol Biol. 1964 Apr;8:582–592. doi: 10.1016/s0022-2836(64)80013-9. [DOI] [PubMed] [Google Scholar]



