Abstract
Viperin, an interferon-inducible antiviral protein, is shown to bind an iron-sulfur cluster, based on iron analysis as well as UV-Vis and electron paramagnetic resonance spectroscopic data. The reduced protein contains a [4Fe-4S]1+ cluster whose g-values are altered upon addition of S-adenosylmethionine (SAM), consistent with SAM coordination to the cluster. Incubation of reduced viperin with SAM results in reductive cleavage of SAM to produce 5′-deoxyadenosine (5′-dAdo), a reaction characteristic of the radical SAM superfamily. The 5′-dAdo cleavage product was identified by a combination of HPLC and mass spectrometry analysis.
Keywords: Viperin, Antiviral, Radical SAM, Iron-sulfur cluster, Metalloenzyme, S-Adenosylmethionine
1. Introduction
Viperin (virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible) has been shown to be strongly induced by both type I and II interferons in response to a variety of different viral infections [1–7], however the specific means by which viperin acts to carry out its antiviral function has yet to be identified. Insight into a possible mode of action for viperin was provided by the recent demonstration of its interaction with farnesyl diphosphate synthase (FPPS), an enzyme that is essential for isoprenoid biosynthesis, including squalenes and sterols [7,8]. Wang et al. provided evidence that the intracellular interaction of viperin with FPPS decreases the activity of FPPS, ultimately disrupting the formation of lipid rafts and thereby increasing the lateral mobility of the plasma membrane [7]. However, the specific biochemical interactions between viperin and FPPS, and the roles these might play in the pathways downstream of FPPS leading up to isoprenoid biosynthesis, remain to be identified.
The finding of a relationship between viperin function and lipid raft disruption was significant due to the important roles lipid rafts play in the life cycles of a number of viruses, including both HIV and the influenza virus [9]; thus lipid raft disruption could be a novel mode of antiviral activity [7]. A simplified illustration of an immune system response to viral infection is shown in Fig. 1, wherein viral infection triggers upregulation of several genes leading to production of interferon regulatory factor 3 (IRF3), which stimulates interferon (IFN) production. Secreted IFNs bind to type I IFN receptors (IFNR) and activate formation of IFN stimulated gene factor 3 (ISGF3), which then triggers the IFN stimulated response element (ISRE) of the gene promoter. In turn, genes for numerous antiviral proteins, including viperin, are upregulated. Viperin (shown as red boxes) migrates to the cytosolic face of the endoplasmic reticulum (ER) where it can interact with FPPS (shown as green circles) [7]. Viperin binding with FPPS leads to disruption of lipid raft domains thereby forming viral buds with “stalk-like” or “daisy chain” structures, which cannot be released from the plasma membrane [7,10].
Fig. 1.
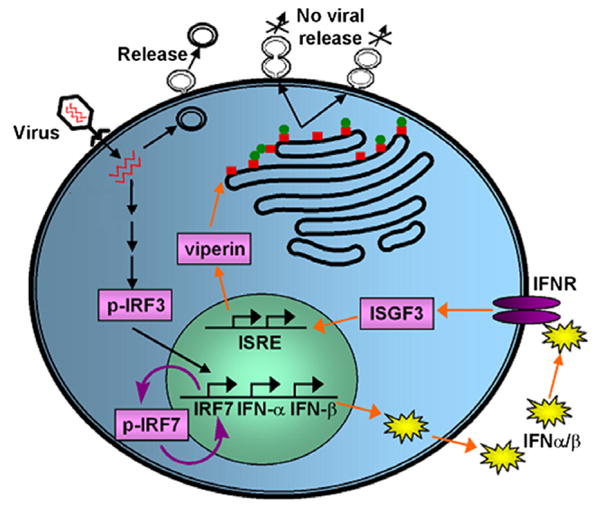
Schematic illustration of a representative immune response pathway that leads to the disruption of viral release from the plasma membrane.
Viperin has been suggested to be a member of the radical S-adenosylmethionine (SAM) superfamily due to the presence of a CX3CX2C motif, as well as some additional motifs found in other radical SAM enzymes [11]. However, the biochemical characteristics and the precise function of viperin remain unknown. Sequence analysis suggests that human viperin is a three-domain protein, with the radical SAM domain (residues 77–209) flanked by a leucine zipper domain (residues 1–76), and a C-terminal domain [12]. The leucine zipper domain may be responsible for proper folding of the protein as well as anchoring the protein to the cytoplasmic side of the ER [13].
We report here the first spectroscopic and biochemical evidence that human viperin is a radical SAM enzyme that contains the characteristic [4Fe-4S] cluster and catalyzes the reductive cleavage of SAM. The identification of viperin as a radical SAM enzyme provides a foundation for understanding its specific mode of action in the immune response to viral infection.
2. Materials and methods
2.1. Materials
S-Adenosylmethionine was synthesized from methionine and ATP using SAM synthetase and subsequently purified using cation exchange chromatography as previously described [14]. All other chemicals were obtained from commercial sources.
2.2. Cloning of viperin
The cDNA for human viperin (cig5) lacking the N-terminal sequence encoding the leucine zipper (cDNA provided in pcDNA3.1) was received as a generous gift from Peter Cresswell at Yale University; the cig5 cDNA in pLNCX2 was also received as a generous gift from Michael Beard at the University of Adelaide, Australia. Cig5 was PCR amplified from pcDNA3.1. The PCR fragments were purified, digested and ligated into a pET-14b vector (Novagen) containing an N-terminal 6x-Histidine affinity tag (His6) (see Supplementary material).
2.3. Viperin expression, purification, and reconstitution
Viperin lacking the leucine zipper domain (viperin43–340 hereafter) was overproduced in Escherichia coli Rosetta(DE3)pLysS cells (Novagen). Growth of cell lines and protein expression was performed as described in Supplementary material. Lysis, purification and reconstitution procedures were all carried out under anaerobic conditions in an anaerobic chamber (Coy Laboratories) (see Supplementary material). Protein concentrations were determined by the Bradford method using bovine serum albumin as the standard [15]. Iron content was evaluated spectrophotometrically [16].
2.4. Spectroscopic characterization
For UV–Vis absorption experiments, samples were transferred to an anaerobic cuvette within an anaerobic chamber (Mbraun) containing 1 ppm or less of O2. Room temperature UV−Vis absorption data were acquired using a Cary 6000i UV–Vis/near-IR spectrophotometer (Varian) with Spectrosil 1.4 ml anaerobic cuvettes (Starna). Low temperature X-band CW (continuous wave) electron paramagnetic resonance (EPR) spectra were recorded using a Varian E-109 spectrometer modified with a National Instruments computer interface (for data collection and field control) and equipped with an Air Products and Chemicals LTD-3-110 Heli-Tran Liquid Helium Transfer Refrigerator (Allentown, PA). EPR parameters were as follows: sample temperature 12.5 K; microwave frequency 9.24 GHz; microwave power 2 mW; time constant 0.50; each spectrum shown is the average of three scans.
2.5. Reduction of viperin
Reduction of viperin was carried out in an anaerobic chamber (Mbraun). Reaction mixtures consisted of 5 mM DTT, 50–100 mM Tris pH 7.0, 58–135 μM of as-isolated or reconstituted viperin in HEPES buffer pH 7.0, and 50–100 μM 5-deazariboflavin (DAF) or 1 mM dithionite depending on method of reduction. Mixtures were prepared by combining reagents from anoxic stock solutions in the order listed above, with DAF or dithionite always added last. Reactions using DAF were illuminated with a 300 W halogen lamp for 1 h and kept cool during illumination by immersion in an ice water bath. Reductions using dithionite were incubated on ice for 10 min. After the reduction of viperin, SAM (3 equiv to protein) was added to the samples and allowed to incubate for 5–10 min. Samples to be used for EPR analysis were placed in an EPR tube and flash frozen. Samples to be used for UV–Vis analysis were immediately transferred to an anaerobic cuvette and analyzed spectroscopically.
2.6. In vitro SAM cleavage assay
SAM cleavage samples were prepared anaerobically. The detection of the cleavage and degradation products including 5′-deoxyadenosine, S-(5′-adenosyl)-l-homocysteine and methylthioadenosine was accomplished using HPLC and confirmed by mass spectrometry (see Supplementary material).
3. Results and discussion
3.1. Purification and biochemical properties of viperin
The cloning of cig5 (minus the N-terminal region encoding the putative membrane anchor) into the pET14b vector was confirmed with restriction enzyme digestion. Synthesizing viperin43–340 from the bacterial plasmid did not result in significant overproduction (Fig. S1), even though the E. coli Rosetta(DE3)-pLysS cells, containing some of the rare codons found in mammalian genes but absent in bacteria, were used. Large scale growths generally yielded ∼25 mg of protein per 60 g of cell paste. The instability of the viperin43–340 protein led to high levels of precipitation when concentrated to levels >250 μM or during repeated freeze-thaw cycles. The observed instability of viperin43–340 could be related to the removal of the N-terminal domain during the cloning process. Hinson and Cresswell showed that the lack of the N-terminal domain could negatively affect protein dimerization [13], and it is clear that the lack of this domain prevents membrane localization; either or both of these effects could have an impact on overall protein stability. In addition, viperin contains several putative N-glycosylation sites in the radical SAM domain, and these would not be glycosylated upon expression in a bacterial host; the lack of glycosylation could also have an affect on protein stability.
3.2. Viperin binds a reducible [4Fe-4S] cluster
Purified viperin43–340 was found to contain iron (0.9 ± 0.2 mol Fe/mol viperin) and was dark yellow to light brown in color at concentrations of ∼100 μM. After reconstitution, the protein became dark brown in color at similar concentrations and contained an enhanced amount of iron (3.7 ± 0.1 mol Fe/mol viperin). The UV–Vis absorption spectra for the purified as-isolated and reconstituted viperin43–340 samples show broad overlapping ligand-to-metal charge transfer (LMCT) transitions characteristic of iron-sulfur clusters, including a sharp feature at ∼315 nm and a broad peak between ∼370 and 450 nm, both of which are typical features for proteins containing [4Fe-4S]2+ clusters (Fig. 2A) [17–19]. The reconstituted enzyme exhibits higher intensity for the LMCT features, consistent with its higher iron numbers, as well as an additional feature at ∼615 nm, perhaps indicative of the presence of [2Fe-2S]2+ clusters (Fig. 2A) [20]. Reduction of the reconstituted protein with either dithionite or photoreduced 5-deazariboflavin results in decreased intensity throughout the visible region (Fig. 2B), which is consistent with the reduction of a [4Fe-4S]2+ cluster to a [4Fe-4S]1+ cluster. Addition of SAM to the reduced enzyme results in a slight decrease in intensity, but no other significant changes in the visible absorption spectrum (Fig. 2B).
Fig. 2.
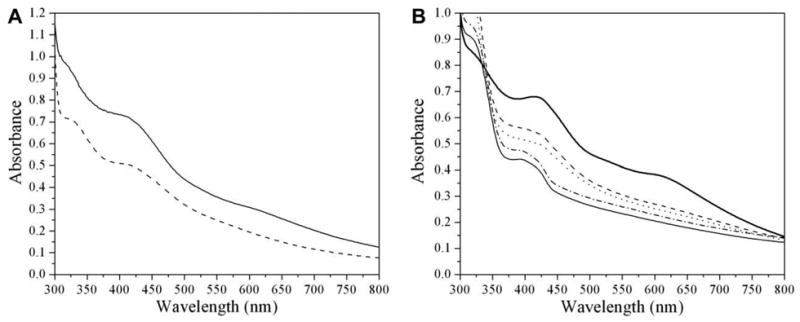
UV–Vis spectroscopic characterization of viperin. (A) As-isolated (dashed line, normalized to 100 μM, 0.7 Fe/protein) or reconstituted (solid line, 100 μM, 2.5 Fe/protein). (B) Reconstituted (normalized to 42 μM, 3.7 Fe/protein, thick solid line), reduced with dithionite (dashed line), reduced with dithionite followed by the addition of SAM (dotted line), reduced by photoillumination (dashed-dotted line), reduced by photoillumination followed by the addition of SAM (thin solid line).
Reconstituted viperin43–340 exhibits a nearly isotropic electron paramagnetic resonance (EPR) signal characteristic of a [3Fe-4S]1+ cluster and accounting for 0.09 spins/protein (Fig. 3A); the fact that this signal accounts for only ∼10% of the iron in the sample indicates that the remainder is in an EPR silent form, most likely primarily [4Fe-4S]2+ clusters. Such [3Fe-4S]1+ clusters are frequently observed for radical SAM enzymes, and are believed to result from oxidative loss of the “unique” iron of the [4Fe-4S] cluster [21–24]. Photoreduction of reconstituted viperin43–340 results in loss of the [3Fe-4S]+ signal and appearance of a nearly axial EPR signal with g-values (g = 2.02, 1.92, 1.91; 0.28 spins/protein) characteristic of [4Fe-4S]1+ clusters (Fig. 3B); this signal is observed only below ∼40 K, as would be expected for a [4Fe-4S]+ cluster. Addition of SAM alters the EPR signal (g = 2.03, 1.95 and 1.88; 0.28 spins/protein Fig. 3C), consistent with the coordination of SAM to the [4Fe-4S] cluster as observed for other radical SAM enzymes [25,26]. Together, the UV–Vis and EPR spectroscopic data provide the first clear evidence that viperin binds an iron-sulfur cluster with properties characteristic of the radical SAM enzymes.
Fig. 3.
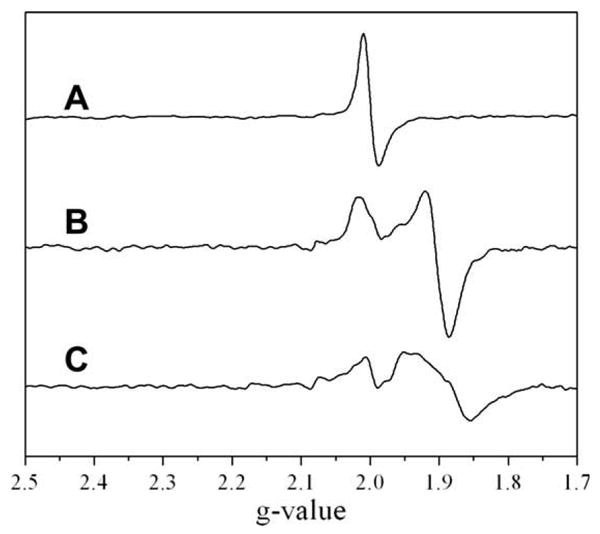
X-band EPR spectra of reconstituted viperin (96 μM). (A) As reconstituted. (B) Reconstituted and photoreduced. (C) Reconstituted and photoreduced followed by addition of SAM.
3.3. Viperin catalyzes the reductive cleavage of SAM
The most characteristic feature of the radical SAM superfamily enzymes is their ability to catalyze the reductive cleavage of SAM to yield methionine and a 5′-deoxyadenosyl radical intermediate (5′-dAdo͘). In the normal catalytic reactions, the 5′-dAdo͘ abstracts a hydrogen atom from substrate to initiate subsequent transformations. Many of these enzymes, however, exhibit uncoupled SAM cleavage, whereby even in the absence of substrate the production of methionine and 5′-deoxyadenosine (5′-dAdo) can be observed under reducing conditions. Because the substrate for viperin is currently unknown, we examined the ability of viperin43–340 to catalyze uncoupled reductive cleavage of SAM.
Fig. 4A shows that in the absence of viperin, SAM remains uncleaved with one major peak eluting at 2.4 min (SAM) along with a minor peak (11.0 min) representing a small amount of degradation of SAM into methylthioadenosine (MTA). In the presence of viperin43–340, HPLC analysis demonstrates the appearance of two new peaks eluting at 5.8 and 6.5 min, which correspond to the elution times of S-(5′-adenosyl)-l-homocysteine (SAH) and 5′-dAdo, respectively (Fig. 4B). To confirm the identity of the compounds eluting at 5.8 and 6.5 min, the peaks were collected and subjected to electrospray-ionization (ESI) mass spectrometry. MS analysis for the sample collected at 5.8 min provided evidence for the presence of both SAH [major component, mz = 384.9 (M+H)] and 5′-dAdo [minor component, mz = 249.0 (M−2H), mz = 252.1 (M+H)]. The peak eluting at 6.5 min gave similar results by MS analysis with a peak corresponding to 5′-dAdo [major component, m/z = 249.0 (M−2H), m/z = 252.2 (M+H)] as well as a peak for SAH [minor component, m/z = 385.0 (M+H)].
Fig. 4.
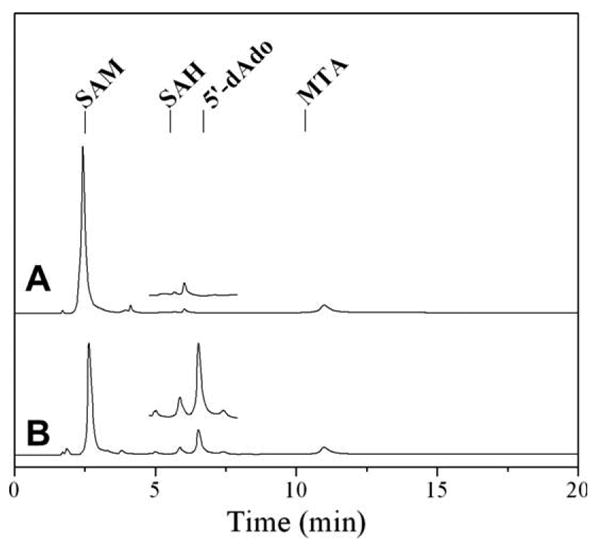
HPLC analysis of SAM cleavage assay. (A) Control assay without viperin. The elution times of standard solutions, as shown above by the labeled tick marks, are: S-adenosylmethionine (SAM) at 2.5 min, S-(5′-adenosyl)-l-homocysteine (SAH) at 5.5 min, 5′-deoxyadenosine (5′-dAdo) at 6.7 min, and methylthioadenosine (MTA) at 10.3 min. (B) Assay containing viperin. Inset: expanded region between 5 and 8 min highlighting the appearance of 5′-dAdo.
3.4. Conclusion
We demonstrate here that viperin43–340 binds a reducible [4Fe-4S] cluster and catalyzes the reductive cleavage of SAM, thus pointing directly to a role for radical SAM chemistry in the antiviral response. Shaveta et al. recently reported that viperin is a radical SAM enzyme, however the only evidence presented was a brown color and the corresponding UV–Vis spectrum after reconstituting viperin [27]. Unlike our work reported herein, Shaveta et al. did not remove excess iron and sulfide after reconstitution; as a result, the UV–Vis data indicates a huge excess of adventitious iron-sulfur clusters, as the extinction coefficient is an order of magnitude higher than typically observed for a single [4Fe-4S] cluster per protein [27]. The iron content of the reconstituted protein examined by Shaveta et al. was not reported, and thus quantitative comparisons to our results cannot be made. Regardless, the observation of binding of iron and sulfide to a protein does not constitute evidence that the protein belongs to the radical SAM superfamily. Our current results demonstrating the binding of a reducible [4Fe-4S] cluster that interacts with SAM, as evidenced by spectroscopy, and that reductively cleaves SAM, as evidenced by HPLC/MS, does provide clear evidence that viperin is a radical SAM enzyme.
The radical SAM superfamily enzymes catalyze a diverse range of reactions, all of which are initiated by a hydrogen atom abstraction from substrate [28]. Given the evidence that viperin inhibits FPPS and disrupts lipid raft formation, two particular classes of radical SAM reactions seem particularly relevant in providing insights into the mode of action of viperin. First, the glycyl radical activating enzymes provide precedent for radical SAM enzyme-catalyzed protein modification [28], which could be a basis for inactivation of FPPS by viperin. Second, radical SAM enzymes catalyzing alkane functionalization [29] support the possibility of viperin-catalyzed functionalization of a metabolite downstream of FPPS; such modification could disrupt cholesterol biosynthesis or the ability of the functionalized cholesterol to form lipid rafts. While the substrate for viperin has yet to be identified, the results presented herein provide significant new insight into the chemistry underlying the antiviral response mediated by viperin.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Peter Cresswell for providing us with cig5 cDNA. This research has been supported by the NIH (GM54608 to J.B.B. and RR-020185), and by the Montana NSF EPSCOR (EPS-0701906). The Astrobiology Biogeocatalysis Research Center is supported by the NASA Astrobiology Institute (NAI05-19).
Abbreviations
- SAM
S-adenosylmethionine
- EPR
electron paramagnetic resonance
- 5′ -dAdo
5′ -deoxyadenosine
- SAH
S-(5′-adenosyl)-l-homocysteine
- MTA
methylthioadenosine
- DAF
5-deazariboflavin
Footnotes
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.febslet.2010.02.041.
References
- 1.Boudinot P, Riffault S, Salhi S, Carrat C, Sedlik C, Mahmoudi N, Charley B, Benmansour A. Vesicular stomatitis virus and pseudoravbies virus induce a vig5/cig5 homologue in mouse dendritic cells via a different pathway. J Gen Virol. 2000;81:2675–2682. doi: 10.1099/0022-1317-81-11-2675. [DOI] [PubMed] [Google Scholar]
- 2.Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci USA. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helbig K, Lau DY, Semendric L, Harley H, Beard M. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology. 2005;42:702–710. doi: 10.1002/hep.20844. [DOI] [PubMed] [Google Scholar]
- 4.Khaiboullina S, Rizvanov A, Holbrook M, St Jeor S. Yellow fever virus strains Asibi and 17D-204 infect human umbilical cord endothelial cells and induce novel changes in gene expression. Virology. 2005;342:167–176. doi: 10.1016/j.virol.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 5.Rivieccio M, Suh HS, Zhao Y, Zhao ML, Chin KC, Lee S, Brosnan C. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J Immunol. 2006;177:4735–4741. doi: 10.4049/jimmunol.177.7.4735. [DOI] [PubMed] [Google Scholar]
- 6.Severa M, Coccia E, Fitzgerald K. Toll-like receptor-dependent and – independent viperin gene expression and counter-regulation by PRD1-binding factor-1/BLIMP1. J Biol Chem. 2006;281:26188–26195. doi: 10.1074/jbc.M604516200. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Hinson E, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Ashby M, Edwards P. Identification and regulation of a rat liver cDNA encoding farnesyl pyrophosphate synthetase. J Biol Chem. 1989;264:635–640. [PubMed] [Google Scholar]
- 9.Ono A, Freed EO. Role of lipid rafts in virus replication. In: Roy P, editor. Advances in virus research. Elsevier Academic Press; San Diego: 2005. pp. 311–358. [DOI] [PubMed] [Google Scholar]
- 10.Waheed AA, Freed EO. Influenza virus not cRAFTy enough to dodge viperin. Cell Host Microbe. 2007;2:71–72. doi: 10.1016/j.chom.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block T, Guo JT. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol. 2008;82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinson E, Cresswell P. The N-terminal amphipathic α-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J Biol Chem. 2009;284:4705–4712. doi: 10.1074/jbc.M807261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsby C, Hong W, Broderick W, Cheek J, Ortillo D, Broderick J, Hoffman B. Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active [4Fe-4S]+ cluster of pyruvate formate-lyase activating enzyme. J Am Chem Soc. 2002;124:3143–3151. doi: 10.1021/ja012034s. [DOI] [PubMed] [Google Scholar]
- 15.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Fish W. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Meth Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 17.Hagen K, Watson A, Holm R. Synthetic routes to iron sulfide (Fe2S2, Fe3S4, Fe4S4, and Fe6S9), clusters from the common precursor tetrakis(ethanethiolate)ferrate(2-) ion ([Fe(SC2H5)4]2-): structures and properties of [Fe3S4(SR)4]3- and bis(ethanethiolate)nonathioxohexaferrate-(4-) ion ([Fe6S9(SC2H5)2]4-), examples of the newest types of Fe-S-SR clusters. J Am Chem Soc. 1983;105:3905–3913. [Google Scholar]
- 18.Kulzer R, Pils T, Kappl R, Hutterman J, Knappe J. Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. J Biol Chem. 1998;273:4897–4903. doi: 10.1074/jbc.273.9.4897. [DOI] [PubMed] [Google Scholar]
- 19.Ugulava N, Frederick K, Jarrett J. Control of adenosylmethionine-dependent radical generation in biotin synthase: a kinetic and thermodynamic analysis of substrate binding to active and inactive forms of BioB. Biochemistry. 2003;42:2708–2719. doi: 10.1021/bi0261084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ugulava N, Gibney B, Jarrett J. Biotin synthase contains two distinct iron-sulfur cluster binding sites: chemical and spectroelectrochemical analysis of iron-sulfur cluster interconversions. Biochemistry. 2001;40:8343–8351. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broderick J, et al. Pyruvate formate-lyase activating enzyme: strictly anaerobic isolation yields active enzyme containing a [3Fe-4S]+ cluster. Biochem Biophys Res Commun. 2000;269:451–456. doi: 10.1006/bbrc.2000.2313. [DOI] [PubMed] [Google Scholar]
- 22.Petrovich R, Ruzicka F, Reed G, Frey P. Characterization of iron-sulfur clusters in lysine 2,3-aminomutase by electron paramagnetic resonance spectroscopy. Biochemistry. 1992;31:10774–10781. doi: 10.1021/bi00159a019. [DOI] [PubMed] [Google Scholar]
- 23.Krebs C, Broderick WE, Henshaw TF, Broderick JB, Huynh BH. Coordination of adenosylmethionine to a unique iron site of the [4Fe-4S] of pyruvate formate-lyase activating enzyme: a Mössbauer spectroscopic study. J Am Chem Soc. 2002;124:912–913. doi: 10.1021/ja017562i. [DOI] [PubMed] [Google Scholar]
- 24.Krebs C, Henshaw TF, Cheek J, Huynh BH, Broderick JB. Conversion of 3Fe-4S to 4Fe-4S clusters in native pyruvate formate lyase activating enzyme: Mössbauer characterization and implications for mechanism. J Am Chem Soc. 2000;122:12497–12506. [Google Scholar]
- 25.Liu A, Gräslund A. Electron paramagnetic resonance evidence for a novel interconversion of [3Fe-4S]+ and [4Fe-4S]+ clusters with endogenous iron and sulfide in anaerobic ribonucleotide reductase activase in vitro. J Biol Chem. 2000;275:12367–12373. doi: 10.1074/jbc.275.17.12367. [DOI] [PubMed] [Google Scholar]
- 26.Walsby C, Ortillo D, Yang J, Nnyepi M, Broderick WE, Hoffman BM, Broderick JB. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “Radical SAM” protein superfamily. Inorg Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 27.Shaveta G, Shi J, Chow VTK, Song J. Structural characterization reveals that viperin is a radical S-adenosyl-l-methionine (SAM) enzyme. Biochem Biophys Res Commun. 2010;391:1390–1395. doi: 10.1016/j.bbrc.2009.12.070. [DOI] [PubMed] [Google Scholar]
- 28.Frey P, Hegeman A, Ruzicka F. The radical SAM superfamily. Crit Rev Biochem Mol Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 29.Booker SJ. Anaerobic functionalization of unactivated C–H bonds. Curr Opin Chem Biol. 2009;13:58–73. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


