Abstract
Motivation: Increasing use of structural modeling for understanding structure–function relationships in proteins has led to the need to ensure that the protein models being used are of acceptable quality. Quality of a given protein structure can be assessed by comparing various intrinsic structural properties of the protein to those observed in high-resolution protein structures.
Results: In this study, we present tools to compare a given structure to high-resolution crystal structures. We assess packing by calculating the total void volume, the percentage of unsatisfied hydrogen bonds, the number of steric clashes and the scaling of the accessible surface area. We assess covalent geometry by determining bond lengths, angles, dihedrals and rotamers. The statistical parameters for the above measures, obtained from high-resolution crystal structures enable us to provide a quality-score that points to specific areas where a given protein structural model needs improvement.
Availability and Implementation: We provide these tools that appraise protein structures in the form of a web server Gaia (http://chiron.dokhlab.org). Gaia evaluates the packing and covalent geometry of a given protein structure and provides quantitative comparison of the given structure to high-resolution crystal structures.
Contact: dokh@unc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Modeling protein structure has become an integral part of biological studies via providing experimentally testable predictions of molecular interactions. Similarly, comparative modeling plays an important role in expanding the structural landscape of proteins. However, important quality control steps are essential to ensure that any given protein structural model conforms to known properties of proteins starting from its covalent geometry to ideal atomic packing. Several studies have measured properties like bond lengths, bond angles and bond torsions allowable in protein structure (Laskowski et al., 1993; Ramachandran et al., 1963). Other studies have also benchmarked properties like steric clashes, hydrogen bonding and rotamer outliers (Davis et al., 2007; Hooft et al., 1996; Vriend and Sander, 1993). In this study, we systematically determine the distribution of these structural parameters as well as other properties such as solvent accessibility and void volume, using high-resolution structures. Based on statistical analysis, we introduce Gaia, a new web-based tool with filters that systematically report on protein structure quality.
We select structural filters that represent some of the important features in stable protein folds: (i) close packing in the buried core of the protein, which minimizes the void volume in the protein core; (ii) minimal number of free polar/charged residues in the buried core of the protein; and (iii) minimal number of sterically overlapping atoms. These factors are incorporated into the energy functions of most protein force fields to recapitulate the structure of the folded state of a protein. However, due to approximations used in all force fields and insufficient sampling, there can be predicted low energy decoy structures possessing nonphysical structural features. Measures that compare theoretical structures with high-resolution structures on the basis of known characteristics of folded proteins can be used to ‘filter’ out the nonphysical models from a pool of predicted models. Additionally, these filters will also point to regions in a given structure that need to be refined to ensure quality of a structural model closer to that of the native structure.
In order to report on the packing quality of a given protein structure, Gaia computes its total void volume, percentage of unsatisfied hydrogen bond donors/acceptors, and extent of steric clashes. In addition, to report on the quality of a model structure's covalent geometry, Gaia also determines deviant bond lengths, angles, torsions, side-chain rotamers and the scaling of accessible surface area with protein length. The users are also provided with an option to resolve clashes using Chiron (http://chiron.dokhlab.org), our recently developed rapid clash minimization routine (Ramachandran et al., 2011) and anomalous side chain orientations using our force field, Medusa (Ding and Dokholyan, 2006).
2 METHODS
2.1 High-resolution dataset used in determining benchmark distributions
To construct our high-resolution dataset, we obtained high-resolution (<2.5 Å resolution) protein structures determined only using X-ray crystallography from the protein data bank (PDB) that were at least 25 residues long and did not contain nonprotein biomolecules (such as ligands/DNA/RNA). We also excluded structures that contained modified residues except selenomethionine (which we replaced with methionine using Medusa). Since we are interested in the properties of globular chains and not interfaces, we split the structures into individual peptide chains. We performed clustering of all structures based on similarity of sequences they represent. We considered only one chain to represent each cluster of sequences that are at least 80% similar to one another. To filter out structures that were extremely nonglobular, we used an Rg scaling cutoff of 0.66. We filtered out structures whose Rg scaled by more than L0.66, where L is the number of amino acids in the protein. For well-packed globular proteins, we found the scaling factor to be 0.36. The dataset consists of 3928 individual chains. Joosten et al. (2009) recently re-refined available high resolution X-ray structures to obtain optimized structure models featuring better fit to deposited experimental data and improved geometric quality. Of the selected 3928 chains, we retrieved the structures for those chains that were available in PDB_REDO database, thereby creating a final dataset with 2163 unique chains. We used Medusa (Ding and Dokholyan, 2006; Yin, et al., 2007) to accurately place any missing side-chain atoms in these structures. Of the 2163 structures used, 969 are enzymes as determined using GO annotation (catalytic activity) and/or the presence of an EC number in the header of the PDB file. The final list of PDB and chain IDs are provided as a table in Supplementary Material.
2.2 Steric clashes
We define a steric clash in a protein as any atomic overlap resulting in a Van der Waals repulsion energy greater than 0.5 kBT, except (i) when the atoms are bonded, (ii) when the atoms form a disulfide bond or a hydrogen bond, and (iii) when the atoms involved are backbone atoms and are separated by two residues. We compute a clash-score for the input protein, which is defined as the sum of Van der Waals repulsion energies of all clashes normalized by the number of contacts screened. A detailed description of computation of clash-score and minimization of clashes is provided elsewhere (Ramachandran et al., 2011).
2.3 Generation of surface dots
We use our implementation of the algorithm originally proposed by Le Grand and Merz (Le Grand and Merz, 1993) to compute solvent accessible and molecular surface area of proteins. The algorithm represents each atom as a set of dots placed on the surface of the atom. For improved accuracy, we used 4096 dots to represent the surface of each atom compared to 256 in the original implementation. For scale-up, we represent the dots as pairs of spherical angles θ and ϕ. In our convention, θ=[0, π] and ϕ=[0, 2π). We first generated dots on the surface of a unit sphere by randomly choosing θ and ϕ values within their respective domains. We then performed Monte Carlo-based simulated annealing to minimize the following cost function using the Metropolis criterion,
| (1) |
where N is the number of dots on the surface and dij is the Euclidian distance between the dots i and j.
2.4 Solvent accessible surface area
We define the solvent accessible surface area (SASA) of a protein as the area covered by the center of a solvent sphere, as it rolls over the protein surface. Considering the radius of the solvent sphere to be 1.4 Å (radius of one water molecule), we obtain SASA by calculating the surface area of the protein, when the radii of all its atoms are increased by 1.4 Å. We use our implementation of the algorithm proposed by LeGrand and Merz (Le Grand and Merz, 1993) for calculating SASA, where surface of each atom is represented by 4096 dots and boolean masks are used to delineate buried and exposed dots on each atom. The reported SASA of a protein therefore includes the surface area of the voids (if any) in the protein. We have modified the algorithm to ensure uniform distribution of masks on the surface of a unit sphere. We define and use a metric hij given by,
| (2) |
for generating masks instead of,
| (3) |
as proposed by LeGrand and Merz (Le Grand and Merz, 1993). We identify the dot closest to the point D on the line joining the centers of two atoms i and j and retrieve the appropriate mask to determine the fraction of surface of atom i not buried by atom j (Supplementary Fig. S1). We repeat this process for all atom pairs to determine the exposed surface of each atom. The surface area of the protein can then be computed by summing up the fractional surface areas contributed by individual atoms.
2.5 Molecular surface area
We define the molecular surface area (MSA) of a protein as the area covered by the edge of a solvent sphere, as it rolls over the protein surface. MSA is represented as a sum of three components—contact, toric and reentrant surfaces (Connolly, 1983).
Contact surface area: we compute the contact surface area using the same algorithm that we use for computing SASA, but without increasing the radii of atoms by the radius of the solvent. The contact surface area of the protein can be formally defined as:
| (4) |
where ni and ri are the number of exposed dots, and the radius of atom i, respectively, and D is the total number of dots on the atom, set to be 4096. Ac includes the contact surface area of voids (if any) in the protein core.
Toric surface area: we analytically calculate the toric surface area covered by the solvent probe on a pair of atoms i and j, using the following equation:
| (5) |
where τij is the fraction of the torus around the overlapping atoms i and j that is accessible to the solvent probe, ri and rw represent the radii of the atom i and the probe respectively, and θij is the angle subtended by the atom j at the center of atom i. We compute τij using edge masks as described by Bystroff (Bystroff, 2002). The solvent probe may roll over itself causing singularities in the toric surface. We treat such cases by computing the toric surface area by atoms i and j using the following equation when (ri+rw)sinθij<rw:
| (6) |
We use this algorithm to compute the toric surface area of all atom pairs including those forming voids (if any) in the protein core. Further details on the mathematical formulation are reported elsewhere (Bystroff, 2002).
Reentrant surface area: we apply the Gauss–Bonnet theorem to calculate the total reentrant curvature of the protein. Gauss–Bonnet theorem states that the total Gaussian curvature integrated over a closed manifold equals 2π times the Euler characteristic of the manifold. This theorem is applicable only if a normal can be generated unambiguously at every point on the surface of the manifold (orientable surface). Protein surfaces are orientable and hence the Gauss–Bonnet theorem can be used to calculate the Gaussian curvature of a protein. Since the Euler characteristic is geometrically invariant, the Gaussian curvature integral of a closed 3D surface, i.e. the Connolly molecular surface of a protein, is the same as that of a sphere and is equal to 4π. The total Gaussian curvature of the protein can be denoted as a sum of contact, toric and reentrant curvatures. However, proteins may contain voids, which are isolated continuous surfaces in the protein core. Each such void, if present, must be considered as an independent orientable manifold. Therefore, the reentrant curvature of the protein is given as
| (7) |
where Kr is the total reentrant curvature integral and Kc, Kt represent the total contact and toric curvature integrals respectively. m represents the total number of manifolds in the system including the solvent accessible surface and all the voids. The total contact curvature integral, Kc, can be obtained using,
| (8) |
and total toric curvature integral Kt can be calculated by,
| (9) |
Here, the curvature integral of toric surface is negative and we do not need to consider the overlapping of toric surfaces. Kr can be derived accordingly, which corresponds to the total reentrant curvature of the protein and voids since the contact and toric curvatures already take the corresponding curvatures from voids in the protein into account. We cluster all the exposed dots on the surface of all exposed atoms to obtain the number of independent manifolds within the protein. Our approach is different from that employed by MASKER (Bystroff, 2002) in that we derive the total reentrant surface analytically using the global Gauss–Bonnet theorem instead of computing individual reentrant surfaces, which have additional sources of errors. The reentrant surface area of the protein is then given by
| (10) |
2.6 Void volume
We define voids as those internal cavities in the protein core that are inaccessible to the bulk solvent, but feature a volume greater than or equal to at least one solvent molecule. We define void volume as the volume of such internal cavities inaccessible to the bulk solvent. To compute void volume, we first use our modified implementation of the algorithm proposed by LeGrand and Merz, to obtain all the dots on the surface of each atom that is not buried by other atoms (depicted in Supplementary Fig. S2). These exposed dots could either belong to the surface or internal voids in the protein. We identify voids by performing single-linkage clustering on these exposed dots using the distance between them as the clustering criterion. This process yields one large cluster corresponding to the solvent accessible surface and zero or more small clusters each corresponding to an internal void (Supplementary Fig. S2). Since we increase the radius of each atom by the radius of a water molecule (1.4 Å) before void identification, the minimum volume of the identified voids is equal to the molecular volume of water. We delineate the volume of each identified void into (i) solvent excluded volume—the region from the surface of the atoms to the surface traced out by the center of the solvent sphere as it rolls on the atoms lining the void and (ii) solvent accessible volume—which is accessible to the solvent, should a solvent molecule be able to approach this space within the protein (Supplementary Fig. S3).
(i) Solvent excluded volume: the solvent excluded volume is composed of three components: the contact volume (fractional volume accessible to the solvent probe touching only one atom), the toric volume (fractional volume inaccessible to the probe touching two atoms at a time) and the reentrant volume (fractional volume inaccessible to the probe when it touches three atoms simultaneously). The fractional volume accessible to the probe touching only one atom can be mathematically computed using,
| (11) |
v where Vc is the total contact volume, D represents the number of dots on the atom i, ni is the number of exposed dots facing the void, N is the number of atoms lining the void, ri is the radius of atom i and rw is the radius of the probe. To calculate the toric volume of the void, we performed analytical integration to arrive at the following equation:
| (12) |
where the terms represent the same quantities as in At. To account for singularities, we used different lower limits for integration when (ri+rw)sinθij<rw, generating the following equation.
 |
(13) |
We compute the total reentrant curvature for each identified void as described above. The reentrant volume can then be computed using
| (14) |
where Kr is the total reentrant curvature for the void. Since the void is a single orientable manifold, we do not perform manifold correction in calculation of void volume, as we perform while calculating the molecular surface area.
(ii) Accessible void volume: we calculate the accessible void volume by numerical integration: we iteratively increment the radii of all the atoms (starting from atom radius plus solvent radius) forming the void by 0.01 Å and sum up the surface area of these voids by 0.01 times at each increment till the area converges to zero. The total void volume is then obtained by summation of the independent components of solvent excluded volume and accessible void volume.
2.7 Unsatisfied hydrogen bond donor/acceptor
We define a polar nitrogen/oxygen atom as an unsatisfied hydrogen bond donor/acceptor if it is buried from the solvent and is not involved in a hydrogen bond. If a polar atom belongs to a residue whose total SASA is zero, it is marked as buried. On the other hand, if the polar atom itself is buried, but the residue it belongs to features a nonzero SASA, rotamer changes/side chain dynamics could expose the polar atom, and thus, the polar atom is classified as being in the shell: an intermediate layer between buried and solvent accessible regions of the protein. We first build all hydrogen bonds in a given protein structure using Medusa's directional hydrogen-bond potential (Ding and Dokholyan, 2006; Yin, et al., 2007), and then list all the buried/shell polar atoms that do not form hydrogen bonds.
2.8 Bond lengths, angles, torsions and side chain rotamers
To ensure the robustness of the covalent geometry of the input protein structure, we also calculate bond lengths, angles, backbone torsions and side chain rotamers to detect outliers. For side chain integrity, the nearest rotamer in the Dunbrack library (Dunbrack and Cohen, 1997) for a given side chain is determined, and then, the P-values of each of the applicable chi-angles of the given side chain with respect to the identified standard rotamer is calculated. A P-value <0.05 is reported as an outlier and presented in the output for a protein structure on the web server. The bond lengths for all standard bonds were calculated from our high-resolution dataset, and the mean and standard deviation from the resulting distributions were used in determining P-values for bond lengths of the input structure. For bonds with SD <2.5% of the mean (as calculated from the standard distribution), we reset the SD as 2.5% of the mean. We reset the SD because the force constants for the bonded term in the MD force fields allow between 2.5% and 4% deviations in bond lengths at 300K. Thus, to report realistic outliers in modeled structures, we require the SD to be at least 2.5% of the mean. Similar analysis was performed for angles and the omega dihedral of the protein backbone. For the ϕ−ψ dihedrals, a two-dimensional histogram with bin width of 2° was constructed combining all amino acid types excluding proline and glycine. A separate histogram was constructed for proline. In the input structure, residues whose ϕ−ψ values belong to a lowly populated bin (roughly <2.5% of the population) are designated as outliers. The outliers in terms of ϕ−ψ dihedrals in an input structure are plotted on top of the heat map of the two-dimensional histogram.
3 RESULTS AND DISCUSSION
For setting benchmarks to qualify a given structure, we first generated distributions of our structural filters from the high-resolution dataset. We analyze these distributions to understand the behavior of our filters with respect to high-resolution structures.
3.1 Protein surface area (SASA/MSA)
To ensure accuracy of our algorithm in calculating MSA, we first determined the different components of MSA for simple two and three sphere systems, where our algorithm accurately reproduced the analytical solution. To determine the accuracy of our method in calculating MSA of proteins, we compared on our whole dataset, different components of MSA determined using our method to that determined using MSMS (Sanner et al., 1996), one of the standard programs used by the modeling community. For most structures, we observe that the contact area computed using our algorithm is within 0.3% of the value obtained using MSMS (Supplementary Fig. S4). Additionally, for most structures, toric and reentrant surface areas obtained using our algorithm are within 2% of the value obtained using MSMS, while the total MSA is within 1% difference. Since SASA is directly proportional to contact area, the differences in SASA between our algorithm and MSMS are identical to that of contact area, <0.3%.
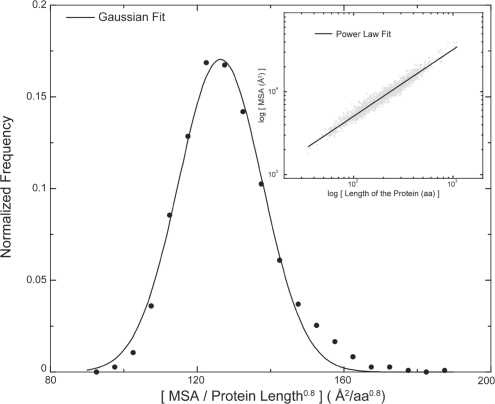
From the distribution of MSA from high-resolution structures, we observe that MSA scales as a function of chain length (N, the number of residues in the protein) with a scaling exponent of 0.8. The scaling exponent is higher than the scaling exponent of the surface of a globular object in 3D (0.667), suggesting that the protein surface is fractal-like and more rugged than the surface of a globular object in 3D. Upon normalization by the factor (chain length)−0.8, we observe that MSA of high resolution structures fits well to a Gaussian distribution (Fig. 1). If the P-value of the normalized MSA of a protein based on the distribution of high-resolution structures is greater than 0.05, the protein structure features MSA scaling characteristic of high-resolution structures. Featuring normalized MSA much greater than that observed in the standard distribution indicates excessive exposure of protein residues in a globular protein, which is unfavorable or corresponds to a nonglobular structure. Having a smaller scaled MSA than observed for high-resolution structures however is not unfavorable; it merely indicates a more compact fold than observed in high-resolution protein structures. Surprisingly, scaling of SASA as a function of chain length features a smaller scaling exponent of 0.74 (>0.667) compared to MSA (Supplementary Fig. S5).
Fig. 1.
Scaling of MSA as a function of protein size. MSA scales with size of the protein as (length)0.8. Upon normalization of MSA by (length)0.8, MSA from all structures fits to a Gaussian distribution. The raw plot of MSA versus the protein length is shown as gray points and the power-law fit is shown as a black line (inset).
3.2 Void volume
We define void volume as the volume of the free space inside a protein that is enclosed by the Connolly surface but is not accessible to bulk solvent. Voids in proteins are considered thermodynamically unfavorable, with early studies indicating destabilization upon introduction of voids (Eriksson et al., 1992). In this study, we identified individual voids in proteins of our dataset and computed their volume. We first computed the total void volume of a protein, which is a sum of the volumes of all voids in a protein. To avoid bias due to size of the protein, we divided the total void volume by the chain length (number of amino acids). Even though voids have been analyzed in proteins before (Busa et al., 2010; Cuff and Martin, 2004; Kleywegt and Jones, 1994; Liang et al., 1998), none of the studies employed such a large dataset of structures to build distributions of total void volumes.
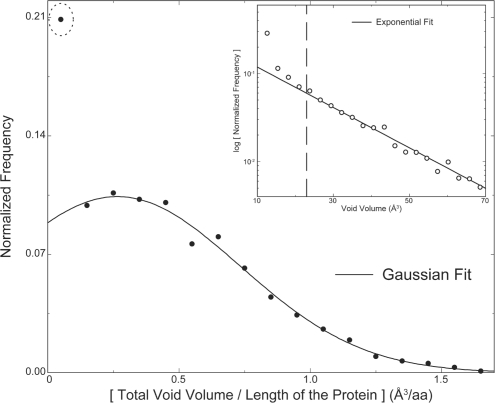
The minimum size of a void we consider is the size of a water molecule. Ignoring smaller voids may not be detrimental, since they could arise as structural deformations due to thermal vibrations of atoms. In contrast, atomic sized voids that we identify are not explained by protein dynamics, and in the absence of buried ligand (or water), these voids can lead to significant structural destabilization of the protein. To find the extent of voids tolerated by proteins, we computed the distribution of normalized total void volume of proteins in our high-resolution dataset. We observe that the normalized total void volume of proteins from our dataset feature a Gaussian distribution (Fig. 2) with a mean of 0.26 ± 0.66 Å3. Thus, a 100-residue protein would on average have a total void volume of ~26 Å3, equivalent to the molecular volume of two water molecules. However, outliers in this distribution include proteins featuring zero and nearly zero normalized total void volume (marked with a dotted circle). Around 17% of proteins in our dataset feature no voids, contributing to the significant outlier in the distribution. We find that 100% of small proteins (<50 amino acids) feature no voids, while 57% of proteins between 50 and 100 amino acids feature no voids, indicating voids as unfavorable in small and medium sized proteins (Supplementary Fig. S6). In striking contrast, majority of proteins having more than 100 amino acids feature at least one void (Supplementary Fig. S6). Using our large dataset, one can reasonably estimate the probability of voids expected in a given structure. To examine possible bias due to buried active sites in enzymes, we performed similar analyses, but with the dataset divided into enzymes and nonenzymes (Supplementary Fig. S7). The distribution for enzymes is modestly right shifted, indicating that enzymes on average feature larger total void volume. Interestingly, the outlier positioned near zero void volume for enzymes has much lower value than that observed in the distribution of nonenzymes.
Fig. 2.
Distribution of voids in proteins. The distribution of total void volume of proteins per residue fits well to a Gaussian distribution with the exception of a total void volume corresponding to values close to zero (dotted circle). Distribution of the volume of individual voids from all structures in the dataset reveal an exponential decay for larger voids, while the decay for smaller voids is much steeper than an exponential. The crossover between smaller and larger voids occurs around the volume of two water molecules (23 Å3) (inset).
From the distribution of the volume of all individual voids detected in our dataset, we can observe the range of voids that occur in proteins (Fig. 2, inset). We observe that the distribution of individual voids can be fit well with a negative exponential function if the first four points are excluded (Fig. 2, inset). The first four points feature a much steeper decay compared to the rest of the points with the crossover to a conventional exponential decay occurring around 23 Å3. Remarkably, this crossover point is very close to the volume of two water molecules, indicating that the bigger voids occur at much higher probability than expected from the distribution of smaller voids. This behavior could be explained by the fact that these bigger voids are more likely to accommodate ligands. Interestingly, the largest probability is observed for voids that can fit exactly one water molecule, indicating a prevalence of singly occurring buried water molecules.
3.3 Unsatisfied hydrogen bond partners
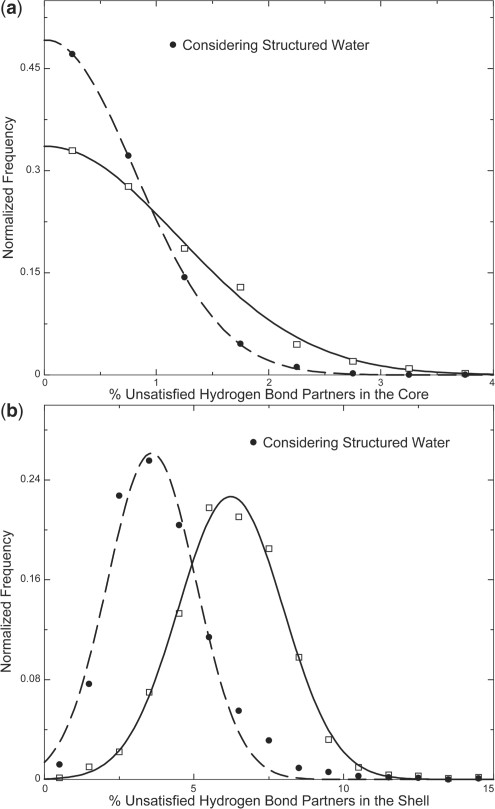
Hydrogen bonds are essential anchors that stabilize a folded protein (Fleming and Rose, 2005). Even though the exact balance of the energetics of hydrogen bonds between polar atoms in proteins compared to the hydrogen bonds between polar atoms and solvent is debated, in the absence of the solvent in the protein core, any polar atom that does not form a hydrogen bond results in destabilization. Absence of secondary structural elements or the presence of polar side chains in the core leads to unsatisfied hydrogen bonding partners (considering that the surface polar atoms form hydrogen bonds with bulk solvent). We measure unsatisfied hydrogen bonds as the percentage of total polar atoms that do not form hydrogen bonds in the buried region of the protein and the shell region of the protein (the buried and shell regions of a protein are defined in Section 2). We observe that the percentage of unsatisfied hydrogen bonding partners in the buried region of a protein across our dataset fits to a Gaussian distribution centered at 0.01% with a SD of 1.7 (Fig. 3a). Similarly, in the shell region, we observe 6.2 ± 2.5 % of the total polar atoms that do not form hydrogen bonds (Fig. 3b).
Fig. 3.
Distribution of the percentage of unsatisfied hydrogen bond partners. The distribution of unsatisfied hydrogen bond partners that are completely buried is plotted with open squares, with the corresponding Gaussian fit shown as a solid line (a). The percentage decreases when hydrogen bonds with buried structural waters are considered (solid circles, Gaussian fit is plotted as dashed lines) (a). Similar plots for hydrogen-bonding partners in the shell region of a protein (b).
In the absence of hydrogen bonding partners, polar atoms in the buried regions can form hydrogen bonds with buried structural waters. Since we use high-resolution structures in our datasets, most of structural waters in these structures are expected to be resolved. Hence we ask the question: what is the influence of buried waters in lowering the number of polar unsatisfied atoms, thus stabilizing the protein? After discounting the buried/shell unsatisfied polar atoms that are within 3.5 Å of a structural water molecule, we observe that the SD of the distribution of unsatisfied buried polar atoms decreases from 1.7% to 1.1%. Similarly, the mean percentage of polar atoms in the shell that do not form hydrogen bonds decreases from 6.2±2.5 to 3.6±2.1. Thus, structural waters make a significant contribution in forming hydrogen bonds in the shell and buried regions of the protein.
3.4 Gaia web server
We combine all the filters described here, in the web server Gaia. Gaia calculates the P-value for various filters of any protein structure model that is submitted. The P-value for a given filter is calculated based on the distribution of that filter for high-resolution structures. We have developed the web interface using PHP and Java script. The first step in the evaluation entails either uploading a PDB file or providing a PDB ID. The input structure is checked for completeness of backbone atoms and any missing side-chain atoms are reconstructed using Medusa. Subsequently, the values of all the filters are calculated for the input structure and the results are stored in a MySQL database. The calculated value of each structural filter is then plotted along with the distribution of values obtained from high-resolution structures. Filters with P-values less than 0.05 are indicated with a warning. A dossier for the input protein structure, with details of each filter, including a plot of the filter value with respect to the benchmark distribution and the P-values is made available for download as a portable document format (PDF) document.
4 CONCLUSIONS
In this study, we choose steric clashes, SASA/MSA, void volume and percentage of unsatisfied hydrogen-bond donor/acceptors as metrics reporting on the quality of packing of the protein core and the formation of proper contacts in the core and shell. We measure the quality of any given structure in terms of these metrics by comparing against the benchmarks from high-resolution crystal structures.
Servers like MolProbity and WHAT IF (Davis et al., 2007; Hooft et al., 1996; Vriend and Sander, 1993) have revolutionized structural biology by providing accurate assessment of the quality of a structure through evaluation of clashes, hydrogen bonds and protein covalent geometry. Through Gaia, we seek to complement already existing servers. Gaia is unique in several aspects of protein structure quality assessment. For example, Gaia provides a unique way to define clashes using energetics compared to fixed overlap distance cutoffs (0.4 Å) used by other servers. Furthermore, while other servers provide a list of hydrogen bonds, Gaia also evaluates unsatisfied hydrogen bond partners in a protein. Finally, Gaia uses a novel method to compute void volume in a protein and provides a statistical score for the total void volume of a protein. Thus, Gaia provides a systematic, multi-faceted evaluation of the quality of a protein structure model, including clashes, voids, hydrogen bonds and molecular surface, in addition to the local, covalent geometry of individual residues and peptide bonds.
In addition to introducing tools for quantifying the quality of protein structures, our benchmarks revealed interesting properties of protein cores. Our finding that majority of small and medium sized proteins feature no voids, suggests that single domains are well packed in proteins, while folding of multiple domains may introduce voids. Further analysis is required to substantiate this hypothesis. The distribution of the percent buried polar atoms forming no hydrogen bonds peaks at zero reiterating the strong penalty for the burial of unsatisfied polar atoms. We also observe a high probability for voids that can fit just one water molecule, implying a prevalence of singly occurring buried water that may be important for structural stability. The possible structural role of buried waters is further supported by the observation of substantial number of contacts between buried polar atoms and crystallographic waters in the structures in our dataset. This discovery is important in modeling protein structure given that most current methods for ab initio protein structure prediction do not consider buried structural waters.
The filters established in this study combined with calculation of covalent geometry of proteins are available for use by protein-structural biology community as a web server (Gaia—http://chiron.dokhlab.org). For a given input protein, Gaia calculates all the above properties and provides P-values by comparing the input protein parameters to the distributions obtained from high-resolution crystal structures. By providing a detailed report on each of these properties within minutes, Gaia serves as a final filter for estimating the quality of a given protein structural model.
Funding: American Heart Association Predoctoral Fellowship (to S.R. 09PRE2090068); the University of North Carolina Research Council (to F.D.); the National Institutes of Health (to N.V.D. grant number R01GM080742 and ARRA supplements GM080742-03S1 and GM066940-06S1).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Busa J., et al. CAVE: a package for detection and quantitative analysis of internal cavities in a system of overlapping balls: application to proteins. Comput. Phys. Commun. 2010;181:2116–2125. [Google Scholar]
- Bystroff C. MASKER: improved solvent-excluded molecular surface area estimations using Boolean masks. Protein Eng. 2002;15:959–965. doi: 10.1093/protein/15.12.959. [DOI] [PubMed] [Google Scholar]
- Connolly M. Analytical molecular surface calculation. J. Appl. Crystallogr. 1983;16:548–558. [Google Scholar]
- Cuff A.L., Martin A.C. Analysis of void volumes in proteins and application to stability of the p53 tumour suppressor protein. J. Mol. Biol. 2004;344:1199–1209. doi: 10.1016/j.jmb.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Davis I.W., et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F., Dokholyan N.V. Emergence of protein fold families through rational design. PLoS Comput. Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbrack R.L., Jr., Cohen F.E. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson A.E., et al. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- Fleming P.J., Rose G.D. Do all backbone polar groups in proteins form hydrogen bonds? Protein Sci. 2005;14:1911–1917. doi: 10.1110/ps.051454805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft R.W.W., et al. Errors in protein structures. Nature. 1996;381:272–272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Joosten R.P., et al. PDB_REDO: automated re-refinement of X-ray structure models in the PDB. J. Appl. Crystallogr. 2009;42:376–384. doi: 10.1107/S0021889809008784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt G.J., Jones T.A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta. Crystallogr. D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., et al. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Le Grand S.M., Merz K.M.J. Rapid approximation to molecular surface area via the use of Boolean logic and look-up tables. J. Comput. Chem. 1993;14:349–352. [Google Scholar]
- Liang J., et al. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran G.N., et al. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Ramachandran S., et al. Automated minimization of steric clashes in protein structures. Proteins: Struct. Funct. Bioinf. 2011;79:261–270. doi: 10.1002/prot.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner M.F., et al. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Vriend G., Sander C. Quality control of protein models: directional atomic contact analysis. J. Appl. Cryst. 1993;26:47–60. [Google Scholar]
- Yin S., et al. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15:1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.