Abstract
Motivation: Integrative mathematical and statistical models of cardiac anatomy and physiology can play a vital role in understanding cardiac disease phenotype and planning therapeutic strategies. However, the accuracy and predictive power of such models is dependent upon the breadth and depth of noninvasive imaging datasets. The Cardiac Atlas Project (CAP) has established a large-scale database of cardiac imaging examinations and associated clinical data in order to develop a shareable, web-accessible, structural and functional atlas of the normal and pathological heart for clinical, research and educational purposes. A goal of CAP is to facilitate collaborative statistical analysis of regional heart shape and wall motion and characterize cardiac function among and within population groups.
Results: Three main open-source software components were developed: (i) a database with web-interface; (ii) a modeling client for 3D + time visualization and parametric description of shape and motion; and (iii) open data formats for semantic characterization of models and annotations. The database was implemented using a three-tier architecture utilizing MySQL, JBoss and Dcm4chee, in compliance with the DICOM standard to provide compatibility with existing clinical networks and devices. Parts of Dcm4chee were extended to access image specific attributes as search parameters. To date, approximately 3000 de-identified cardiac imaging examinations are available in the database. All software components developed by the CAP are open source and are freely available under the Mozilla Public License Version 1.1 (http://www.mozilla.org/MPL/MPL-1.1.txt).
Availability: http://www.cardiacatlas.org
Contact: a.young@auckland.ac.nz
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
In both the clinical and research settings, a variety of techniques can be used to quantify cardiac performance at various structural and functional levels. Electrocardiography allows monitoring of the electrical activity of the heart to diagnose pathology such as rhythm disturbances, myocardial infarction and hypertrophy. Blood pressure measurements enable detection of hypertension. Cardiovascular imaging studies provide quantification of cardiac mass and volumes, as well as assessment of regional heart wall motion. The ability to integrate these multi-source, multivariate data has enormous implications for the diagnosis and clinical care of patients (Kohane, 2009). Mathematical and computational models can be used to integrate data in a standardized way, providing both a ‘big picture’ population map of the various factors that determine cardiac function as well as highly detailed information which can be used to characterize function in an individual patient (patient specific models). These models can elucidate the complex interaction of electrical, anatomical and functional data to provide insight into the processes underlying the normal or pathological function of the heart. Furthermore, models derived from large populations of patients can provide a range of reference values against which individual patients' data can be compared.
A number of biomedical initiatives use computational modeling to integrate multi-scale anatomical, functional and clinical data from diverse sources. These include the Physiome Project (Hunter and Borg, 2003), the International Consortium for Brain Mapping (Mazziotta et al., 2001), Informatics for Integrating Biology and the Bedside (i2b2) (Murphy et al., 2006) and the Cardiac Gene Expression database (Bober et al., 2002) among others. These projects depend on large population databases for the development and validation of physiological models.
The Cardiac Atlas Project (CAP) is an international collaboration to establish a large-scale standardized database of cardiac imaging examinations and derived functional analyses. The aim is to develop a computational, structural and functional atlas of the normal and pathological heart. These atlases can be defined as a set of maps which relate scientific information to spatial coordinates at a series of scales, from genotype to phenotype (Thompson et al., 2000; Toga et al., 1996). The research objectives of CAP are to:
Establish a database of cardiac imaging examinations consisting of de-identified image files together with associated clinical data.
Develop open source software for the analysis of cardiac morphology including (i) visualization of images in 3D over time, and (ii) interactive construction of mathematical cardiac models from the images.
Develop standardized protocols for the contribution, curation, archival, classification and sharing of cardiac image data and derived analyses, including labeling of images and models in the CAP database with ontological terms.
Here, we describe the design of the computational and informatics infrastructure together with procedures for contribution of data to the CAP, de-identification, standardization and sharing of data and software tools, and policies for the protection of the rights of participants, contributors and users of the database. The article is organized as follows: Section 2 outlines the requirements, design and implementation of CAP components, accompanied by an overview of the issues underlying regulatory requirements and stake-holder rights. Section 3 gives details of the major contributing studies, the results of the validation procedures performed, current database and client functionality, XML schema and policies regarding access to the database.
2 METHODS
2.1 Stake-holders
The CAP is an international collaboration funded by the National Institutes of Health, USA. The host institutions are the University of Auckland (New Zealand) and the University of California Los Angeles (USA) with databases maintained and mirrored in both institutions. Where possible, infrastructure for data sharing has been adapted and re-used from the UCLA Center for Computational Biology and Laboratory of Neurological Imaging (LONI), which has considerable experience in the area of computational brain atlases (Mazziotta et al., 2001). Stake-holders in CAP include the following parties:
Participants: people who have participated in a research study contributing data to CAP, or have otherwise provided informed consent to contribute data to CAP.
Contributors: research study investigators who originally acquired the data and shared it by contributing it to CAP. Decisions regarding data use are typically made by the contributing study steering committee.
Users: third party researchers who access CAP data in order to undertake research into cardiovascular function and disease.
Cardiac imaging examinations and associated clinical data are contributed from a number of sources, including research studies, clinical trials and clinical centers.
2.2 Imaging and clinical data
Cardiovascular imaging provides an abundant source of detailed, quantitative data on heart structure and function. Common investigations include ultrasound, computed tomography, radionuclide imaging and MRI. Many research studies have employed MRI because it is noninvasive, well tolerated and safe (no ionizing radiation), has the ability to modulate contrast, and can provide high-quality functional information in any plane and direction (Fig. 1).
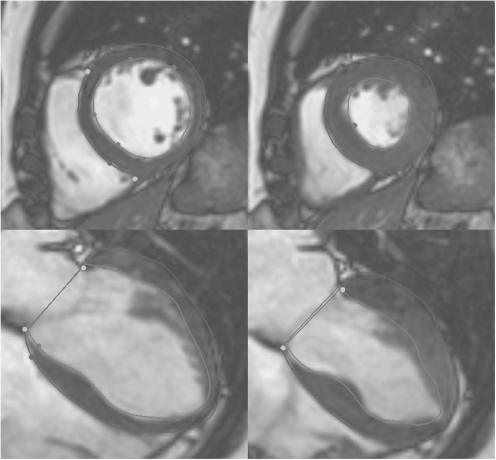
Fig. 1.
Cine MRI short- (top) and long-axis (bottom) images, at end-diastole (end of ventricular filling, left), and end-systole (end of ejection, right). Contours show inner (green) and outer (blue) boundaries of the left ventricle, and the position of the mitral valve (red).
The tomographic nature of MRI data lends itself to 3D atlas building techniques and to date, all CAP imaging data has come from MRI. These studies typically consist of 6–12 cine acquisitions in the short axis orientation, with 20–50 frames through the cardiac cycle and 1–2 mm pixel resolution. Imaging protocols include gradient recalled echo (GRE) (Boxerman et al., 1998) and steady state free precession (SSFP) (Thiele et al., 2001) techniques. Studies have also contributed core laboratory analyses of the image data, in the form of annotations and contouring (Fig. 1) of the left ventricular boundaries at end-diastole (end of filling) and end-systole (end of ejection), and de-identified text data containing the clinical status and demographics of the participants.
2.3 Regulatory compliance
Since CAP is an international, multi-institutional collaborative project, it must comply with a variety of legislative and local Institutional Review Board (IRB) requirements. Local Ethics Committee and IRB approvals were obtained for CAP at the two host institutions. In addition, CAP policy requires that all data must be obtained and contributed with the approval of a local IRB or Ethics Committee, and informed consent for data sharing must be obtained from each participant.
The Health Insurance Portability and Accountability Act (HIPAA) Privacy and Security Rules (45 CFR Parts 160, 162 and 164, available at http://www.hhs.gov/ocr/privacy/hipaa/administrative/privacyrule/index.html) regulate the use and disclosure of research participant's protected health information (PHI) in the USA. PHI are any data that could be used to identify an individual, e.g. names, dates (except for year), social security or medical record numbers, locations or other unique identifiers. To protect the identity of participants, PHI must be replaced or removed before data can be shared, a process known as de-identification. Use of de-identified data is considered by many IRBs not to constitute human subjects research.
2.4 Data de-identification
In CAP, all data must be de-identified by the contributors before upload into the database. The de-identification process removes PHI from both text and image data, and replaces subject identifiers such as the name of the individual, or the original study code, with an unrelated CAP code. Medical images contributed to CAP are stored as DICOM Objects (DICOM-PS3.5, 2009). These objects contain data attributes including information on the scanner, the imaging protocol and the scanned object. The current DICOM standard (v2009) contains more than 2800 public DICOM attributes. In addition, many DICOM and PACS manufacturers include proprietary information within private DICOM attributes.
We adapted the LONI Debabeler (Neu et al., 2005), a HIPAA compliant software tool, for the de-identification of DICOM images. We created a CAP-specific Debabeler with rules to encrypt or replace DICOM attributes that could potentially contain PHI, while retaining essential information on the image acquisition.
The output of the CAP Debabeler includes the key linking CAP codes to the original identifiers, which is then kept by the contributor. CAP personnel, and third party CAP users, do not have access to this key. The Debabeler rules are available from the CAP project site at SourceForge.net (http://sourceforge.net/projects/cardiacatlas/).
2.5 Database design
Software was designed to enable storage and retrieval of medical image and text data, ontological annotations and volumetric shape models (Fig. 2). Browsing, searching, image preview and download functionality are included.
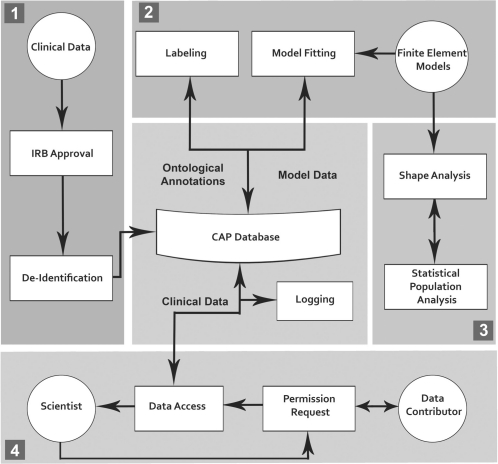
Fig. 2.
CAP workflow. Step 1: data ACQUISITION; Step 2: data processing; Step 3: data analysis and Step 4: public data access.
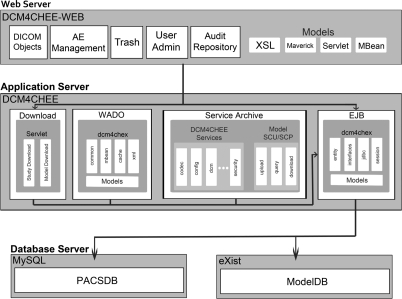
The database was developed using a three-tier architecture (web-, application- and database server) including monitoring and secure authentication with access privileges based on user need. Due to its mature architecture and code base, active development, maintenance and support, DICOM compliance, compatibility with other Java APIs and other international research projects such as the Cancer Biomedical Informatics Grid (Cimino et al., 2009) and the Cardiovascular Research Grid (CVRG; http://www.cvrgrid.org), we based the database on the open source image archive Dcm4chee, and extended its functionality. Dcm4chee is a clinical data manager system based on a J2EE (Alur et al., 2003) and JMX (Fleury and Lindfors, 2002) software architecture and is deployed within the JBoss Application Server. It provides a number of useful clinical interfaces, including:
Ability to store, query, and retrieve any type of DICOM object;
WADO (Web Access to DICOM Objects) and RID (IHE Retrieve Information for Display) interfaces to allow access from the web;
a robust user interface which runs entirely in a web browser; and
Audit Record Repository—IHE ATNA audit logging (Gregg et al., 2006).
The Dcm4chee application logic, database schema and web-application were extended to provide access to MRI specific attributes as defined in the DICOM Standard MR Image Module (DICOM-PS3.3, 2009). These included vendor and model of the scanner used to acquire the images. The database fields are populated at image import using extended methods from the Dcm4chee Enterprise Java Beans (EJBs).
The web-application extension allows all added attributes to be used as search options. A researcher might be interested in specific studies, cine series or individual images that satisfy specific search parameters. For this purpose, a search filter was added to generate result listings grouped by Patient, Study, Series or Image. To allow searching for arbitrary DICOM attributes, an XPath query (details of XPath are described at http://www.w3.org/TR/xpath20/) was implemented. An XML tree (Bray et al., 2006) representing the DICOM structure of the imported images is generated, stripped of binary and large values, and stored in the database.
The download functionality of the Dcm4chee web-application was extended to allow the download of complete DICOM studies, series and volumetric models including referenced data. This is achieved by implementing a servlet that collects the requested data from the server and provides it as a compressed archive to the user (Smart et al., 2005).
2.6 Parametric modeling of cardiac function
Atlas-based methods are well established for the statistical classification and quantification of shape and wall motion characteristics of the heart (Young and Frangi, 2009). These methods enable standardized analysis of statistical variations present within and among patient groups, and enable classification of individual phenotypes within known population distributions. In almost all cases contributed to the CAP, contours were contributed in association with the images and clinical information. These contours can be used as the input to a standardized model-based analysis to establish shape and motion with respect to a standard coordinate system, similar to the Talairach coordinate system used in the brain (Mega 2005; Tang, Hojatkashani et al. 2010). Since shape and motion are mathematically mapped, statistical tools such as principal component analysis can be used to quantify the significant modes of variation present within a population. In CAP, the parametric shape descriptors lend themselves to finite element modeling, which can then enable biophysical simulation of physiological processes, including nonlinear mechanical properties and large deformations of the heart, and solve the biophysical conservation laws linking stress, strain and energy expenditure.
By customizing mathematical models of the anatomy and function of the heart to individual cases, it is possible to construct parameter variation models describing the distribution of regional cardiac shape and function across patient subgroups. Homologous landmarks (i.e. the points that are aligned to match corresponding features in the shape) can be used to characterize shape and shape variations with the aid of a principal component analysis, or similar technique. Since mathematical models, represented by the model parameters, are a complete and efficient characterization of cardiac shape and motion, a statistical analysis of the variation inherent in the parametric shape and motion models can be formed (Young and Frangi, 2009).
2.7 CAP client design
An open-source client side software tool was developed for the visualization and analysis of the cardiac MRI images available in the database. The software allows visualization of the data in 3D + time and interactive fitting of a finite-element volumetric model to any given dataset.
The CAP client has been designed with the following objectives in mind.
Performance: to allow the user to view and manipulate large image datasets in 3D and fit models to them in an interactive manner. In order to meet the real-time 3D graphics and numerical computation requirements, the C++ programming language was chosen for its superior time efficiency and the availability of high-performance numerical libraries. The CAP client uses hardware-accelerated OpenGL API for graphics rendering and the GMM++ linear algebra library (available at http://download.gna.org/getfem/html/homepage/gmm/index.html) for model fitting.
Ease of use: the CAP client is expected to be used by nontechnical users for educational and research purposes, therefore ease of use was an important design goal. All features of the software are accessible through an intuitive graphical user interface.
Extensibility and maintainability: in order to encourage external developers to extend the CAP client to suit their needs, the source code is structured to accommodate such extensions. Various object-oriented techniques were adopted to increase the extensibility of the software. For example, the abstract factory design pattern and the adaptor design pattern (Gamma et al., 1995) were used to ease the possible replacement of the linear algebra library.
Portability: the CAP client was designed to be portable across different platforms and currently runs on Microsoft Windows, Mac OS X and Linux. This portability is achieved using cross-platform libraries such as wxWidgets (Smart et al., 2005), boost (available at http://www.boost.org) and GMM++, as well as build and testing tools such as CMake and Google Test.
The CAP client was built on the open source Cmgui library (available at http://www.cmiss.org/cmgui), an advanced visualization software library developed at the Auckland Bioengineering Institute for visualization and manipulation of general finite element models. This visualization library was employed because it has a large suite of tools available for parametric shape modeling of the heart, and is readily customizable for CAP purposes.
2.8 Semantic data model
The CAP database contains more than 1.5 million MRI images from symptomatic and asymptomatic patients. To facilitate automated fitting of volumetric models and searching for specific characteristics, CAP is labeling images using a controlled vocabulary. SNOMED-CT (Ryan et al., 2007) and the Foundational Model of Anatomy (FMA) (Rosse and Mejino, 2003) are established ontologies providing clinical and anatomical concepts. RadLex (Kundu et al., 2009) unifies and supplements these standards and provides a single source of radiology terms. From these resources, CAP has selected cardiac and MRI terms for classification of images and annotation of anatomical landmarks. Where appropriate, we have provided feedback to the resources to improve the terminology for cardiac labeling.
2.9 Policy and rights
To ensure that all data provided to CAP are managed according to well-defined principles, and in accordance with the regulatory and ethical requirements associated with de-identified human image and clinical data, policies and procedures related to data access, control and sharing have been developed. These policies apply to (i) participants from whom the data was obtained, (ii) contributors who originally collected and have contributed the data, (iii) the CAP investigators and, (iv) third-party users who wish to access CAP data. CAP contributors have made substantial monetary, intellectual and time investments for the collection of the data in a well-controlled manner (viz. original study design, recruitment, quality control, analysis, etc.), which represents a valuable scientific resource. The conditions under which data are originally acquired may vary substantially among contributors, for example ranging from public good government funded studies to privately funded trials with a commercially sensitive outcome. The terms and conditions under which data can be shared consequently vary substantially among contributors. CAP has therefore adopted a ‘bundle of rights’ approach (see www.cardiacatlas.org for further discussion) reflecting the goal of providing access as openly and widely as possible, consistent with contributor and participant consent. Data can be contributed as public domain provided the informed consent is compatible with open unrestricted access. It should be noted that an explicit objective of CAP is to provide a flexible mechanism by which data that would otherwise be inaccessible (for example, data generated by privately funded clinical trials or ongoing longitudinal studies) can now be accessed by researchers for a variety of diverse investigations. CAP has successfully achieved this aim by developing policies that not only protect the original data contributors (sponsors and investigators of privately funded or ongoing studies), but also allow third party investigators an avenue of access that would not be possible via other means. By negotiating and working through these policies with the original investigators, CAP has paved the way for future third-party access to the data.
3 RESULTS
3.1 Database
Two main studies comprise the current CAP database. The Multi Ethnic Study of Atherosclerosis (MESA; Bild et al., 2002) has contributed 2864 asymptomatic volunteers to date. MESA is investigating subclinical cardiovascular disease and the progression to clinically overt disease in a diverse, population-based sample of asymptomatic men and women aged 45–84 years. Participants with no history of cardiovascular disease were recruited from six field centers across the United States. Approximately 38% of the study's participants were white, 28% African-American, 22% Hispanic and 12% Asian, predominantly of Chinese descent.
The Defibrillators to Reduce Risk by Magnetic Resonance Imaging Evaluation (DETERMINE) trial (Kadish et al., 2009) has contributed 470 datasets from patients with myocardial infarction to date. New studies are being contributed on an ongoing basis. DETERMINE is a multicenter, randomized, clinical trial in patients with coronary artery disease (CAD) and mild-to-moderate LV dysfunction. The trial investigated whether patients with an infarct size greater than or equal to 10% of left ventricular mass, randomized to receive an implantable defibrillator plus appropriate medical therapy will have improved survival compared with patients randomized to medical therapy alone.
In MESA, MRI data were acquired on Siemens and GE 1.5T MRI scanners only. The images included cine (using the GRE pulse sequence) in short-axis planes covering from the base of the heart to the apex and in three long-axis planes. Images were analyzed using MASS 4.0 (Medis, The Netherlands) by the MESA MRI core laboratory at Johns Hopkins University School of Medicine, and ventricular contours contributed to CAP. In the DETERMINE trial, MRI data were acquired on any of Siemens, GE and Philips 1.5T or 3.0T MRI scanners. The imaging protocol included cine images (using the SSFP pulse sequence) acquired in short-axis planes from the base of the heart to the apex and in three long-axis planes, as well as delayed enhancement viability (Kim et al., 1999) used for detection and quantification of myocardial infarction, acquired in the same planes as the cine images. Images were analyzed using QMASS MR 7.2 (Medis, The Netherlands) by the DETERMINE MRI core laboratory at Northwestern University Feinberg School of Medicine, and contours contributed to CAP.
Both studies also contributed limited clinical information including: age (years), gender (M/F), height (cm), weight (kg), systolic and diastolic blood pressure (mmHg), hypertension (Y/N), heart rate (bpm), race/ethnicity and classifications for hypertension, diabetes, smoking (Y/N), alcohol (Y/N), angina (Y/N), ECG and NYHA classification.
DETERMINE included an IRB approved specific section in the participant information and consent forms to contribute de-identified data to CAP. Participants could choose to give or withhold this consent independent of their participation in DETERMINE. MESA included an IRB approved informed consent process compatible with data sharing and further IRB approval was obtained for the contribution of de-identified data to CAP.
CAP has been endorsed by the Society for Cardiovascular Magnetic Resonance (www.scmr.org). Clinical cases with appropriate informed consent can also be de-identified and made publicly available in the database.
3.2 CAP client
Given a set of cardiac MRI images from the CAP database, the CAP client software, (see Fig. 3) can be used for:
Visualization of MRI images, the mathematical model constructed from the images, and animation of the motion through time in the cardiac cycle.
Customization of a finite element model of the left ventricle to the MRI images using guide point modeling (Young, 2000). This process requires minimal human intervention, and results in a mathematical model of the heart shape and motion in 3D and time. The Client software also provides a means for users to interactively and graphically modify model parameters derived from other sources, such as semi-automatic ventricular analysis methods.
Customized model parameters, along with images and contour information, are stored in an XML file format as described in Section 3.4.
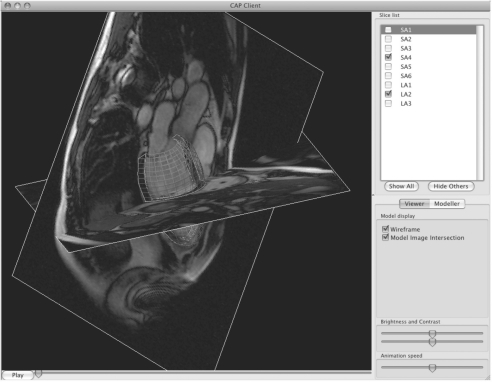
Fig. 3.
Screenshot of the CAP Client running on Mac OS X. One short axis and one long axis MRI image are visible, togther with the inner and outer surfaces of the LV model (green and red lines, respectively).
3.3 Data analysis
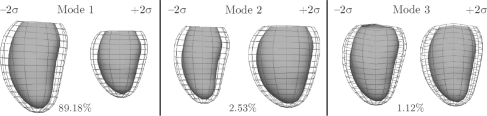
To demonstrate the utility of the database for the statistical characterization of heart shape and motion, the major modes of variation within a subset of the DETERMINE cohort were calculated using a principal component analysis. The first three modes associated with the greatest variation were found to correspond with size, sphericity and mitral valve geometry (Fig. 4). These results are encouraging since each of these modes are known measures of adverse geometric remodeling following myocardial infarction. Projection of an individual's shape and motion onto these modes (e.g. sphericity) provides a standardized method for quantifying the amount of each mode present.
Fig. 4.
First three modes of shape variation in principal component analysis of a subset of the DETERMINE cohort (n=200). Mode 1: size; Mode 2: sphericity; Mode 3: mitral geometry.
3.4 Semantic data model
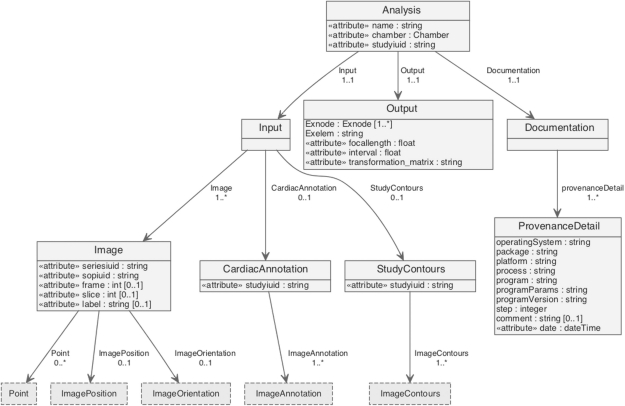
To store volumetric models (Fig. 5) and supplementary data for images such as contours and cardiac annotations using ontological concepts, we have designed a standardized data structure using XML. Storing data in an XML format allows for simple conversions using the Extensible Stylesheet Language Transformations (XSLT; described at http://www.w3.org/TR/xslt) of (i) geometrical data into other languages such as FieldML (Christie et al., 2009), and (ii) cardiac annotations into knowledge representation languages, e.g. OWL (Web Ontology Language), http://www.w3.org/TR/owl-features/, or RDF (Resource Description Framework), http://www.w3.org/standards/techs/rdf. The XML files are stored using the XML database eXist (Meier, 2003), which provides core database features, such as indexing and transaction recovery, enabling fast search and retrieval of model-related data. Import and export of XML data has been implemented by extending the Dcm4chee architecture (see Fig. 6). The extended architecture provides a vehicle to store and retrieve image, model and derived data.
Fig. 5.
In order to store volumetric models generated with the CAP client application, an XML schema has been designed representing the elements associated with volumetric shape model creation and curation. This includes input parameters, such as images, contours and markers, calculated output parameters, mesh files representing the model geometry, and provenance information.
Fig. 6.
Three-tier architecture of the CAP model implementation based on Dcm4chee. Blue boxes represent basic Dcm4chee classes, and yellow boxes represent CAP specific model extensions.
3.5 Policy and rights
Standard operating procedures were developed to manage the logistics of data sharing, including data requests and data transfer, and to maintain the rights of the stake-holders.
Participants must give informed consent compatible with data sharing to contribute their de-identified image and text data for cardiovascular research now and in the future. All data must be de-identified in a manner compatible with the HIPAA privacy rule, using the CAP de-identification process, and must occur at the Contributor's site before upload to the CAP data servers. CAP must not receive or retain the original identifiers. CAP investigators, and Users, must agree not to attempt to identify participants.
The key linking CAP codes with original identifiers must be retained by the Contributor, so that investigators of the Contributing Study can link results from CAP back to the original study if desired. Participants can request withdrawal of their data from the database at any time by requesting removal either via the CAP or directly to the Contributor. In this case the Contributor must notify CAP of which CAP data must be deleted.
Access to the data is unrestricted and open for those cases with informed consent compatible with unrestricted access. In many cases, however, the participant consent requires that access is approved by the contributor. The Contributor or Contributing Study Steering Committee then controls all data access through data distribution agreements, on a request basis. Users are required to submit a brief Research Proposal, outlining the rationale and goals of the project, timeline and data storage, to the CAP Steering Committee which includes CAP investigators from both of the partner institutions, The University of Auckland and the University of California Los Angeles, the NIH Program Officer for CAP, and investigator-representatives from each of the contributing studies. The purpose of the review is primarily to protect the rights of participants and contributors. If the proposal is within the remit of CAP, CAP will liaise with each of the Contributors whose data has been requested. Each Contributor (or nominee) must then review the proposal and, if approved, provide a Data Distribution (DDA) agreement. The User must then sign and abide by the DDAs for each Contributor. Separate DDA's are required for each Contributor because terms and conditions governing data use may differ depending on the circumstances under which the data were acquired. The DDA defines all the terms and conditions governing the use of the data, including publication policy, acknowledgements, data handling, and intellectual property.
3.6 CAP software license
All software is freely available via the CAP website, to researchers and educators in the nonprofit sector, such as educational institutions, research institutes and government laboratories. Instructions for accessing CAP source code are available at http://www.cardiacatlas.org/web/guest/tools. Developer access to CAP source projects is made available through a sourceforge Mecurial repository. CAP database and heart modeling tools, comprising database management, uploading and downloading of images, web browser interface, conversion of data formats, visualization and parametric modeling of shape and motion, are being made available using the Mozilla Public License Version 1.1. Dependent software such as Dcm4chee and Cmgui are compatible with this license. Commercialization of enhanced or customized versions of the software, or incorporation of the software or pieces of it into other software packages, is permitted subject to third party intellectual property claims. Researchers are permitted to modify the source code and are strongly encouraged to share modifications with other researchers as well as with CAP.
3.7 Use cases
CAP data may be used for a variety of purposes. Some use cases are described below:
Image analysis: a subset of the data is being provided for a left ventricular segmentation challenge at the Medical Image Computing and Computer Assisted Intervention (MICCAI) 2011 conference. This challenge enables researchers to test their automated segmentation algorithms on the same large cardiac MRI dataset, thereby facilitating comparative discussion as well as collaboration among peers on combining the results to find a better ground truth (http://cilab2.upf.edu/stacom_cesc11/index.php).
Clinical evaluation: CAP data will be used to create a statistical atlas for clinical purposes. This would be used to determine whether a particular patient fits within the normal range or how many standard deviations the patient is from normal values. Pathological processes such as LV remodeling will be examined by comparison to pathology-matched statistically predicted parameters, based on an individual's known clinical characteristics and the CAP population subgroup he or she matches most closely.
Clinical trials: CAP data will be used to test hypotheses comparing cohorts among studies in the database, or to perform metadata queries on several studies, utilizing mapping transforms to reduce bias due to study protocol.
Education: CAP data could also play an important role in biomedical education programs. The collection of a well-curated and diverse population of cardiac data is an excellent platform from which to understand normal structure and function as well as to examine the statistical differences related to age, gender, height, weight and pathology. The value of the data is enhanced by the downloadable CAP Client software for visualization in 3D and over time. Students could use the software to fit mathematical models to data, and then use the models to better understand the effects of pathology on standard clinical measurements such as ejection fraction, volumes and wall thickening parameters.
4 FUTURE WORK
In accordance with the goals of standardized classification and sharing of data and resources, CAP is developing and building upon currently available ontological schema to describe cardiac image data and derived annotations and models, with plans to federate these cardiovascular modeling software and data via the CVRG (www.cvrgrid.org). A SPARQL (http://www.w3.org/TR/rdf-sparql-query/) interface may be built when there is a substantial amount of annotated data for this purpose, which requires semantic annotations in our XML format, conversion to RDF or OWL, and implementation of a semantic storage.
The parametric modeling tools and associated ontological schema that are being developed by CAP will be expanded to facilitate data fusion between different imaging protocols and modalities as well as other data sources. Tools necessary for the statistical analysis of CAP data are also being developed and will be used for the generation of parametric distribution models.
5 CONCLUSIONS
The CAP currently hosts approximately 3000 cardiac MRI studies, derived functional analyses and associated participant data that represents a substantial and valuable resource. Tools for the de-identification of data were developed, validated and successfully deployed by the contributing studies. The necessary IRB and Ethics Committee approvals were obtained and policies were developed to protect the rights of subject participants, contributors and users of the database. Applications to use the data can now be submitted to the CAP website. Upon completion of a Data Distribution Agreement, users can browse and query the database as well as view the images, and download the data. The CAP database is compliant to the DICOM standard and provides sophisticated image attribute search options. The CAP Client software, downloadable at the Project's website, allows the user to import images from the database and customize a finite element model to the image data. Volumetric shape models are stored in XML and are available to the research community via the CAP database. CAP procedures and tools are designed to facilitate a workflow from the acquisition of CMR images toward a statistical analysis of volumetric models.
Funding: National Heart, Lung and Blood Institute, USA (R01HL087773). Support from the Center for Computational Biology (LONI) was provided by National Institutes of Health/National Center for Research Resources (U54 RR021813, P41 RR013642). MESA was supported by the National Heart, Lung and Blood Institute (N01-HC-95159 through N01-HC-95169). A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. DETERMINE was supported by St Jude Medical, Inc; and the National Heart, Lung and Blood Institute (R01HL91069). A list of participating DETERMINE investigators can be found at http://www.clinicaltrials.gov. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung and Blood Institute or the National Institutes of Health.
Conflict of Interest: none declared
REFERENCES
- Alur D., et al. Core J2EE Patterns. Upper Saddle River: Prentice Hall; 2003. [Google Scholar]
- Bild D.E., et al. Multi-ethnic study of atherosclerosis: objectives and design. Am. J. Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- Bober M., et al. CaGE: Cardiac Gene Expression Knowledgebase. Bioinformatics. 2002;18:1013–1014. doi: 10.1093/bioinformatics/18.7.1013. [DOI] [PubMed] [Google Scholar]
- Boxerman J.L., et al. Advanced MR imaging techniques for evaluation of the heart and great vessels. Radiographics. 1998;18:543–564. doi: 10.1148/radiographics.18.3.9599382. [DOI] [PubMed] [Google Scholar]
- Bray T., et al. Extensible Markup Language (XML) 1.1. W3C Recommendation; 2006. [Google Scholar]
- Christie G.R., et al. FieldML: concepts and implementation. Philos. Transact. A Math. Phys. Eng. Sci. 2009;367:1869–1884. doi: 10.1098/rsta.2009.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino J.J., et al. The caBIG terminology review process. J. Biomed. Inform. 2009;42:571–580. doi: 10.1016/j.jbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicom-PS3.3. Part 3: Information Object Definitions. National Electrical Manufacturers Association; 2009. Digital Imaging and Communications in Medicine. [Google Scholar]
- Dicom-PS3.5. Part 5: Data Structures and Encoding. National Electrical Manufacturers Association; 2009. Digital Imaging and Communications in Medicine. [Google Scholar]
- Fleury M., Lindfors J. JMX: Managing J2EE with Java Management Extensions. Indianapolis: Sams (Pearson Education); 2002. [Google Scholar]
- Gamma E., et al. Design Patterns: Elements of Reusable Object-Oriented Software. Upper Saddle River, NJ: Addison-Wesley Professional (Pearson Education); 1995. [Google Scholar]
- Gregg B., et al. Creating an IHE ATNA-based audit repository. J. Digit. Imaging. 2006;19:307–315. doi: 10.1007/s10278-006-0927-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P.J., Borg T.K. Integration from proteins to organs: the Physiome Project. Nat. Rev. Mol. Cell. Biol. 2003;4:237–243. doi: 10.1038/nrm1054. [DOI] [PubMed] [Google Scholar]
- Kadish A.H., et al. Rationale and design for the defibrillators to reduce risk by magnetic resonance imaging evaluation (DETERMINE) trial. J. Cardiovasc. Electrophysiol. 2009;20:982–987. doi: 10.1111/j.1540-8167.2009.01503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R.J., et al. Relationship of MRI delayed contrast enhancement to irreversible injury, infarct age, and contractile function. Circulation. 1999;100:1992–2002. doi: 10.1161/01.cir.100.19.1992. [DOI] [PubMed] [Google Scholar]
- Kohane I.S. The twin questions of personalized medicine: who are you and whom do you most resemble? Genome Med. 2009;1:4. doi: 10.1186/gm4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S., et al. The IR Radlex Project: an interventional radiology lexicon–a collaborative project of the Radiological Society of North America and the Society of Interventional Radiology. J. Vasc. Interv. Radiol. 2009;20:433–435. doi: 10.1016/j.jvir.2008.10.022. [DOI] [PubMed] [Google Scholar]
- Mazziotta J., et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mega M., et al. Automated brain tissue assessment in the elderly and demented population: construction and validation of a sub-volume probabilistic brain atlas. Neuroimage. 2005;26:1009–1018. doi: 10.1016/j.neuroimage.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Meier W. Lecture Notes In Computer Science: Web, Web-Services, And Database Systems. Berlin: Springer; 2003. Exist: An Open Source Native XML Database. Node 2002 Web- And Database-Related Workshops. [Google Scholar]
- Murphy S.N., et al. Proceedings of AMIA Annual Symposium. Bethesda MD: American Medical Informatics Association; 2006. Integration of clinical and genetic data in the i2b2 architecture; p. 1040. [PMC free article] [PubMed] [Google Scholar]
- Neu S.C., et al. The Loni Debabeler: a mediator for neuroimaging software. Neuroimage. 2005;24:1170–1179. doi: 10.1016/j.neuroimage.2004.10.035. [DOI] [PubMed] [Google Scholar]
- Rosse C., Mejino J.L., Jr. A reference ontology for biomedical informatics: the foundational model of anatomy. J. Biomed. Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Ryan A., et al. Toward the interoperability of HL7 v3 and SNOMED CT: a case study modeling mobile clinical treatment. Stud. Health. Technol. Inform. 2007;129:626–630. [PubMed] [Google Scholar]
- Smart J., et al. Cross-Platform GUI Programming with wxWidgets. Upper Saddle River, NJ: Prentice Hall (Pearson Education); 2005. [Google Scholar]
- Tang Y., et al. The construction of a Chinese MRI brain atlas: a morphometric comparison study between Chinese and Caucasian cohorts. Neuroimage. 2010;51:33–41. doi: 10.1016/j.neuroimage.2010.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele H., et al. Functional cardiac MR imaging with steady-state free precession (SSFP) significantly improves endocardial border delineation without contrast agents. J. Magn. Reson. Imaging. 2001;14:362–367. doi: 10.1002/jmri.1195. [DOI] [PubMed] [Google Scholar]
- Thompson P.M., et al. Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain. Hum. Brain Mapp. 2000;9:81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga A.W., et al. Proceedings of AMIA Annual Fall Symposium. Bethesda MD: American Medical Informatics Association; 1996. Informatics and computational neuroanatomy; pp. 299–303. [PMC free article] [PubMed] [Google Scholar]
- Young A.A., Frangi A.F. Computational cardiac atlases: from patient to population and back. Exp. Physiol. 2009;94:578–596. doi: 10.1113/expphysiol.2008.044081. [DOI] [PubMed] [Google Scholar]
- Young A.A., et al. Left ventricular mass and volume: fast calculation with guide-point modeling on MR images. Radiology. 2000;216:597–602. doi: 10.1148/radiology.216.2.r00au14597. [DOI] [PubMed] [Google Scholar]