Abstract
Summary: The KEGG PATHWAY database provides a widely used service for metabolic and nonmetabolic pathways. It contains manually drawn pathway maps with information about the genes, reactions and relations contained therein. To store these pathways, KEGG uses KGML, a proprietary XML-format. Parsers and translators are needed to process the pathway maps for usage in other applications and algorithms.
We have developed KEGGtranslator, an easy-to-use stand-alone application that can visualize and convert KGML formatted XML-files into multiple output formats. Unlike other translators, KEGGtranslator supports a plethora of output formats, is able to augment the information in translated documents (e.g. MIRIAM annotations) beyond the scope of the KGML document, and amends missing components to fragmentary reactions within the pathway to allow simulations on those.
Availability: KEGGtranslator is freely available as a Java™ Web Start application and for download at http://www.cogsys.cs.uni-tuebingen.de/software/KEGGtranslator/. KGML files can be downloaded from within the application.
Contact: clemens.wrzodek@uni-tuebingen.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Many academic researchers, who want to use pathway-based information, utilize the KEGG PATHWAY database (Kanehisa and Goto, 2000). The database, established in 1995, contains manually created maps for various pathways. These maps are visualized on the web and can be downloaded free of charge (for academics) as XML-files in the KEGG Markup Language (KGML). The elements in a pathway XML-file (such as reactions or genes) are usually identified by a KEGG identifier only. Thus, KEGG PATHWAY is strongly related to other KEGG databases that resolve and further describe the identifiers. However, the content of these KGML-formatted XML-files itself is limited. Gene names are often encoded as barely readable abbreviations and elements are only annotated by a single KEGG identifier. By improving the annotation and translating the KGML-files to other file formats, researchers could use the KEGG database for many applications: individual pathway pictures could be created; pathway simulation and modeling applications could be executed; graph-operations on the pathways or stoichiometric analyses could be performed (e.g. Heinrich and Schuster, 2006, chapter 3); or the KEGG pathway database could be used for gene set enrichment analyses. For these purposes, only a few converters are available: KEGGconverter (Moutselos et al., 2009) or KEGG2SBML (Funahashi et al., 2004) offer command-line or web-based conversion of KGML-files to SBML-files. KEGGgraph (Zhang and Wiemann, 2009) is able to convert KGML-files to R-based graph structures. None of these tools has a graphical user interface, is capable to validate and autocomplete KEGG reactions, adds standard identifiers (such as MIRIAM URNs) to pathway elements, or is able to write KGML files in multiple ouput formats. Along with this work, the command-line toolbox SuBliMinaL (N.Swainston et al., submitted for publication) overcomes some of these limitations.
We here present KEGGtranslator, which reads and completes the content of an XML-file by retrieving online-annotation of all genes and reactions using the KEGG API. KGML-files can be converted to many output formats. Minor deficiencies are corrected (e.g. the name of a gene), new information is added (e.g. multiple MIRIAM identifiers for each gene and reaction (Novère et al., 2005), or SBO terms describing the function) and some crucial deficiencies (like missing reactants) are addressed.
2 TRANSLATION OF KGML-FILES
In the first step of a translation, KEGGtranslator reads a given XML-file and puts all contained elements into an internal data structure. To get further information and annotation, the KEGG database is queried via the KEGG API for each element in the document (pathway, entries, reactions, relations, substrates, products, etc.). This completes the sparse XML-document with comprehensive information. For example, multiple synonyms and identifiers of many external databases (Ensembl, EntrezGene, UniProt, ChEBI, Gene Ontology, DrugBank, PDBeChem and many more) are being assigned to genes and other elements. After this initial step, various preprocessing operations are performed on the pathway. The user may choose to let KEGGtranslator correct various deficiencies automatically: Remove white nodes—KEGG uses colors in the visualization of a pathway to annotate organism-specific orthologous genes. Nodes in green represent biological entities that occur in the current organism. Nodes in white represent biological entities, corresponding to genes that occur in this pathway in other species, but not in the current one. Translating all those nodes into new models, without caring for the node color, would lead to a model, that contains invalid genes in the pathway. Remove orphans—isolated nodes without any reactions or relations are usually unnecessary for further simulations. Autocomplete reactions—another major deficiency are incomplete reactions. The XML-files only contain those components of a reaction, that are needed for the graphical representation of the pathway. Reactants that are not necessary for the visualization are usually skipped in the KGML format. Thus, the given chemical equation is sometimes incomplete (see Fig. 1). KEGGtranslator is able to lookup each reaction and amend the missing components to reactions. This leads to more complete and functionally correct pathway models, which is very important, e.g. for stoichiometric simulations. After these preprocessing steps, KEGGtranslator branches between two different conversion modes for the actual translation: a functional translation (SBML) and a graphical translation (e.g. GraphML, GML). Depending on the chosen output format, KEGGtranslator determines how to continue with the conversion.
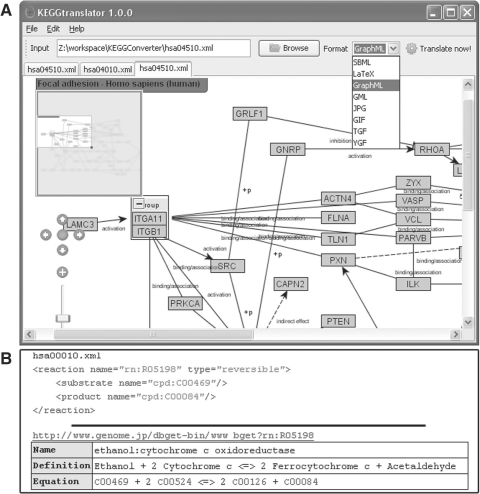
Fig. 1.
(A) Screenshot of a translated GraphML pathway in KEGGtranslator. (B) The need for autocompleting reactions: the upper half shows the KGML-file with only one substrate and product. On the lower half, the complete reaction equation is shown. As one can see, one substrate and product is missing in the XML-document.
The functional translation is performed by converting the KGML document to a JSBML data structure (Dräger et al., 2011). The focus lies on generating valid and specification-conform SBML (Level 2 Version 4) code that eases, e.g. a dynamic simulation of the pathway. Multiple MIRIAM URNs and an SBO term, which describes best the function of the element, is assigned to each entry of the pathway (pathway references, genes, compounds, enzymes, reactions, reaction-modifiers, etc.). Additionally, notes are assigned to each element with human-readable names and synonyms, a description of the element, and links to pictures and further information. The user may also choose to add graphical information by putting CellDesigner annotations to the model. But the focus in functional translation lies on the reactions in KGML documents, whereas graphical representations concentrate on relations between pathway elements. Besides the already mentioned completion of reactions, each enzymatic modifier is correctly assigned to the reaction and the reversibility of the reaction is annotated. As a final step, the SBML2LaTeX (Dräger et al., 2009) tool has been integrated into KEGGtranslator, which allows users to automatically generate a LaTeX or PDF-report, to document the SBML-code of the translated pathway. Furthermore, the user may add kinetics to the pathway by using the SBMLsqueezer (Dräger et al., 2008) tool after the translation.
In graphical translations, results can be saved as GraphML, GML or YGF and finally as images of type JPG, GIF or TGF. In this mode, the KGML data structure is being converted to a yFiles (Wiese et al., 2001) data structure. The focus here lies on the visualization of the pathway. Relations are being translated by inserting arrows with the appropriate style, which is given in the KGML document. For example, dashed arrows without heads represent bindings or associations and a dotted arrow with a simple, filled head illustrates an indirect effect. Please see the KGML specification for a complete list. As in the functional translation, GraphML allows to define custom annotation elements. KEGGtranslator makes use of those, by putting several identifiers (e.g. EntrezGene or Ensembl) and descriptions to the single nodes. From the KGML document, the shape of the node is translated as well as the colors and labels. Links to descriptive HTML pages are being setup and hierarchical group nodes are being created for defined compounds. All these features lead to a graphical representation of the pathway that provides as much information about the elements as possible.
3 DISCUSSION
KEGGtranslator is a stand-alone application with a graphical user interface that runs on every operating system for which a Java™ virtual machine is available. There are other tools for converting KGML to SBML and for converting KGML to graph structures in R. But, to our knowledge, no other KEGG converter is able to translate KGML formatted files to such a variety of output formats with important functionalities like the autocompletion of reactions or the annotation of each element in the translated file, using various identifiers. Furthermore, KEGGtranslator is simple, easy-to-use and comes with a powerful command-line and graphical user interface. The variety of output formats, combined with the translation options and comprehensive, standard-conform annotation of the pathway elements allow a quick and easy usage of files from the KEGG pathway database in a wide range of other applications.
ACKNOWLEDGEMENT
We gratefully acknowledge very fruitful discussions with Jochen Supper, Akira Funahashi and Toshiaki Katayama.
Funding: The Federal Ministry of Education and Research (BMBF, Germany) funded this work in the projects Spher4Sys (grant number 0315384C) and NGFNplus (grant number 01GS08134).
Conflict of Interest: none declared.
REFERENCES
- Dräger A., et al. JSBML: a flexible and entirely Java-based library for working with SBML. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr361. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dräger A., et al. SBMLsqueezer: a CellDesigner plug-in to generate kinetic rate equations for biochemical networks. BMC Syst. Biol. 2008;2:39. doi: 10.1186/1752-0509-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dräger A., et al. SBML2LaTeX: conversion of SBML files into human-readable reports. Bioinformatics. 2009;25:1455–1456. doi: 10.1093/bioinformatics/btp170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi A., et al. 8th Annual International Conference on Research in Computational Molecular Biology. 2004. Converting KEGG pathway database to SBML. [Google Scholar]
- Heinrich R., Schuster S. The Regulation of Cellular Systems. 2. Berlin: Springer; 2006. [Google Scholar]
- Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutselos K., et al. KEGGconverter: a tool for the in-silico modelling of metabolic networks of the KEGG pathways database. BMC Bioinf. 2009;10:324. doi: 10.1186/1471-2105-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novère N.L., et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat. Biotechnol. 2005;23:1509–1515. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- Wiese R., et al. yFiles: visualization and automatic layout of graphs. Proceedings of the 9th International Symposium on Graph Drawing (GD 2001). 2001:453–454. [Google Scholar]
- Zhang J.D., Wiemann S. KEGGgraph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics. 2009;25:1470–1471. doi: 10.1093/bioinformatics/btp167. [DOI] [PMC free article] [PubMed] [Google Scholar]