Abstract
Motivation: Infectious disease research is generating an increasing amount of disparate data on pathogenic systems. There is a growing need for resources that effectively integrate, analyze, deliver and visualize these data, both to improve our understanding of infectious diseases and to facilitate the development of strategies for disease control and prevention.
Results: We have developed Disease View, an online host–pathogen resource that enables infectious disease-centric access, analysis and visualization of host–pathogen interactions. In this resource, we associate infectious diseases with corresponding pathogens, provide information on pathogens, pathogen virulence genes and the genetic and chemical evidences for the human genes that are associated with the diseases. We also deliver the relationships between pathogens, genes and diseases in an interactive graph and provide the geolocation reports of associated diseases around the globe in real time. Unlike many other resources, we have applied an iterative, user-centered design process to the entire resource development, including data acquisition, analysis and visualization.
Availability and Implementation: Freely available at http://www.patricbrc.org; all major web browsers supported.
Contact: cmao@vbi.vt.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Infectious diseases are complex systems, involving multiple organisms (e.g. pathogens and hosts) interacting across different environments and time scales. Much of the data that we have related to infectious disease is multidimensional, incomplete and likely to be biased in ways we do not fully understand. In addition, these data are often not integrated nor interoperable making it difficult for researchers from different disciplines to communicate and relate their work to others. Integrating these data remains a formidable challenge, and although a handful of individual parts of the system have been modeled with some success, a global model of infectious disease does not yet exist. The lack of such a framework makes it difficult to design effective analytical algorithms to assist in the development of diagnostics and therapeutics.
Bioinformatic resources have been developed for specialized areas in infectious disease research; however, they do not fully interoperate. For example, numerous online resources exist to support host organism researchers [e.g. Immport (www.immport.org) and InnateDB (www.innatedb.ca)], offering rich datasets related to host immune response to microbial infection; however, they do not yet provide structured access to associated pathogen data. Similarly, there are many databases containing comprehensive information about pathogens [e.g. PATRIC (www.patricbrc.org), ViPR (www.viprbrc.org), IRD (www.fludb.org), EuPathDB (www.eupathdb.org) and VectorBase (www.vectorbase.org)], but these do not yet provide structured access to mammalian host data (e.g. host genes). Still more online resources offer data related to infectious disease outbreaks [e.g. CDC (www.cdc.gov), WHO (www.who.int) and HealthMap (healthmap.org)], yet they mainly focus on epidemiological information and provide little or no host or pathogen data. Some resources, such as BiologicalNetworks, PHIDIAS, PHI-base and PIG, integrate host–pathogen interactions at different levels, but lack disease or outbreak information (Driscoll et al., 2009; Kozhenkov et al., 2011; Winnenburg et al., 2008; Xiang et al., 2007). These are a few recent examples of attempts to integrate different types of host, pathogen and disease information; a comprehensive review is beyond the scope of this article. As the infectious disease community turns its attention to an integrative view of disease, infection and health, it becomes critical to have access to integrated host, pathogen and disease data. As such, an integrated information system that brings together the many facets of infectious disease is urgently needed.
Also needed are methods for designing and developing user interfaces to communicate complex integrated infectious disease data to diverse users, and provide powerful and meaningful ways to interact with these data. The growing depth and breadth of the data requires a thoughtful approach to integrating and presenting it in a useful manner, one that takes into account combinations of data, interfaces and services from multiple resources, and provides value added over the sum of its parts. To guide the process of understanding which data sources to use, how to integrate data and how to portray data to users, we employ a well-established approach to developing user interfaces (UIs). Specifically, we apply usability engineering: a cost-effective, user-centered iterative process that ensures a high level of effectiveness and efficiency in complex interactive user interfaces (Gabbard et al., 2003; Hix and Hartson, 1993).
Our ultimate goal is to use informatics frameworks to help reduce barriers in infectious disease research and development by providing a broader spectrum of infectious disease, pathogen, host and outbreak data to enable infectious disease-centric access and analysis of host–pathogen interactions. Toward this end, we have developed Disease View, a host–pathogen data and visualization resource that integrates diverse data sources, including host, pathogen, host–pathogen interactions and disease outbreak, and provides a mechanism for infectious disease-centric data analysis and visualization. We instantiate Disease View as a component of PATRIC: a National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID) bioinformatics resource center containing a wealth of resources related to bacterial pathogens (Snyder et al., 2007).
2 USABILITY ENGINEERING PROCESSES FOR PATRIC'S DISEASE VIEW
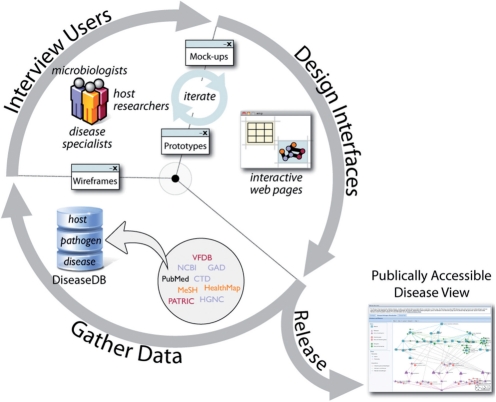
When applying usability engineering (UE) techniques, it is often the case that the specific UE methods must be tailored to meet specific development goals. For Disease View, we used a combination of domain analysis, structured interviews, iterative conceptual design and various usability evaluation methods as described below. Figure 1 depicts our UE approach, where we iteratively gather data, interview users and design interfaces. Within each activity, Figure 1 also shows a number of relevant components, such as users, databases, interfaces, and so on. Sections 2 through 4 describe in detail the role of these components. For illustrative purposes, we also provide a graphical summary of the evolution of PATRIC's Disease View user interfaces, from whiteboard sketches to refined working prototypes, in Supplementary Figure S1.
Fig. 1.
An iterative process of gathering data, interviewing representative users, and evolving user interface designs to develop and publically release PATRIC Disease View.
We began by reviewing and gathering data from existing online resources containing host, pathogen and infectious disease data, with a specific eye toward methods by which researchers and/or resources have connected these traditionally disparate information realms. We identified specific databases (such as VFDB, CTD, GAD—see Section 3) as candidate sources to leverage in response to specific queries/summaries of interest to infectious disease researchers. We then explored how these data sources could be logically (and technically) integrated to provide value-added above and beyond each singleton data resource. From this work, we generated a ‘strawman’ data model and tabular display to demonstrate the technical feasibility of a meaningful set of integrated data; where ‘meaningful’ is measured by representative users (as opposed to developers).
We then conducted structured interviews with researchers currently working with diseases related to bacterial pathogens. From these interviews, we identified high-level findings such as what type of researchers will benefit from Disease View, what are the goals of researchers when using these data, and thus what types of tasks should we aim to support via Disease View. We also identified important specific findings such as the types of online information resources typically used, how these information are used, what tools are employed to use the data and specifically how users take information related to hosts, disease and pathogens and manually integrate it to address their research questions. For these interviews, we used the initial strawman to illustrate concepts, verify which data pieces were most relevant and which were of less (or no) value. From these insights, we generated user-centered requirements, a specific set of most relevant host–pathogen data, and a specific set of useful tasks we could support using these data; all of which we used to inform an initial set of conceptual user interface design sketches. Through iterative sketching and internal expert usability evaluations (Section 1 in Supplementary Material), three distinct components began to evolve: a table representing which diseases are related to the pathogen(s) of interest, a visual representation of both host and pathogen genes related to disease and a map depicting location-based disease information (e.g. recent or historical outbreaks, etc.). We then created low-fidelity ‘static’ prototypes of Disease View UIs (manifested as PowerPoint drawings)—allowing us to conduct early expert usability evaluations without having yet done any software development; a cost-effective approach that allows us to make important conceptual changes with very little cost. At this stage of development, the expert evaluation was less focused on specific detailed UI elements, but instead on page layout, information layout and interaction with different disease view elements.
Based on expert evaluation, we evolved the wireframes into refined web-based prototypes that incorporated both static and interactive components. This approach allowed us to identify obvious usability problems and improve the UIs prior to conducting user-based evaluation (Section 1 in Supplementary Material). As a result, our user-based evaluations yield more valuable, domain-specific usability findings such as to what degree UI designs cognitively support completion of important tasks. We conducted several user-based usability evaluations using these prototypes with representative users from PATRIC's scientific working group. These users included researchers working with bacterial pathogen-related diseases such as brucellosis, tuberculosis and helicobacter infection. We worked with several different user classes including principal investigators, post-doctoral assistants, graduate students and lab technicians. During these evaluations, we observed that users had a difficult time digesting the volume of information displayed, and attempted to make (unintended) connections between disparate blocks of information; for example, trying to associate a table depicting MeSH (Medical Subject Headings) terms and disease with a map depicting outbreak information. Moreover, users wanted to allocate more screen real estate for each of the three main components since each conveyed a large amount of important information. As such, we made a design decision to break the three components into three separate screen/areas: Summary, Disease-Pathogen Visualization and Disease Map as shown at http://www.patricbrc.org/portal/portal/patric/DiseaseOverview?cType=taxon&cId=590.
3 DATA INTEGRATION
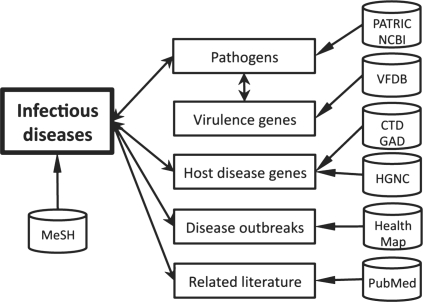
A primary challenge facing bioinformatics researchers is to combine data from different biological repositories into a single, unified view on the data (data integration). Through our usability engineering process, we incorporated into Disease View six primary data types of interest to infectious disease researchers: pathogens, virulence genes, host disease genes, disease outbreaks, related literature and relationships among infectious disease terms. The relationships among these modules and their data sources in Disease View are illustrated in Figure 2.
Fig. 2.
Modules and data sources of Disease View. Modules are represented as rectangles. Data sources are shown as cylinders.
To integrate the infectious disease, host and pathogen data obtained from various sources, we developed DiseaseDB, a relational database implemented in Oracle 11g. See the Supplementary Figure S2 for the database schema model. The schema includes data structures for several types of disease data including: disease–pathogen mappings, pathogens, virulence genes and human genes associated with infectious diseases. Disease outbreaks and literature information are dynamically fetched on the fly from HealthMap and PubMed, respectively, but not stored in the DiseaseDB. Data types and sources are described below.
3.1 Bacterial infectious disease–pathogen mapping
To incorporate and integrate infectious disease, pathogen virulence and host data, our first step was to map infectious diseases with corresponding pathogens in a computer recognizable format. We used MeSH (www.nlm.nih.gov/mesh) infectious disease terms as controlled vocabulary for infectious disease names and NCBI taxonomy names for the pathogen names. MeSH is a controlled vocabulary thesaurus used for biomedical and health-related document indexing. MeSH disease terms are subset of MeSH terms that have been widely used in disease studies. Many databases such as Genetic Association Database (GAD, geneticassociationdb.nih.gov) and Comparative Toxicogenomics Database (CTD, ctd.mdibl.org) use MeSH disease terms to extract and map disease information in biomedical literature (Davis et al., 2011; Lin et al., 2006). The latest 2011 MeSH version contains 225 distinct bacterial infectious disease terms. The hierarchal MeSH structure allows more general disease terms to be distinguished from subclasses. Based on MeSH disease term definitions and online medical reference, these bacterial infectious disease terms were manually curated and mapped to bacterial pathogens with taxonomy level assigned depending on how specific or general a disease term is in the MeSH hierarchy (Table 1; Supplementary Table S1 for MeSH bacterial infectious disease terms). This mapping enables association between pathogen-related information such as genomics, postgenomics, virulence and their corresponding infectious disease terms.
Table 1.
Bacterial infectious diseases and pathogen mapping
| Pathogen taxon level | Number of infectious disease terms |
|---|---|
| Species or subspecies | 121 |
| Genus | 54 |
| Family | 18 |
| Order | 4 |
| Superkingdom | 28 |
3.2 Pathogens and pathogen virulence genes
Pathogens, pathogen genomes and pathogen genes were retrieved directly from PATRIC. The internal ids in PATRIC were mapped and referenced in DiseaseDB. We incorporated the pathogen virulence gene information to facilitate understanding the mechanisms of infection and pathogenesis. Pathogen virulence genes were collected from two sources: curated virulence genes from Virulent Factor Database (VFDB, www.mgc.ac.cn/VFs) and the homologs of VFDB curated virulence genes from blast search within the same genera to which the curated virulence genes belong (Altschul et al., 1990; Yang et al., 2008). The blast cutoff was 95% for identity and 80% for length coverage. The curated virulence genes and virulence factor descriptions were downloaded as a flat file from the VFDB website and then parsed and stored in DiseaseDB. As of March 2011, we have integrated 2295 curated virulence genes from VFDB and 36490 taxon-scoped homologs in DiseaseDB. The homolog information is important for identifying unknown virulence genes in related genomes, since some homologs are annotated as ‘hypothetical protein’ in the original genome annotations.
3.3 Human genes associated with infectious diseases
GAD and CTD are two online resources that provide information about host gene–disease association studies. GAD associates human genes with diseases using genetic evidence derived from literature curation. CTD provides manually curated data from published literature describing cross-species chemical– and gene–disease relationships to illuminate molecular mechanisms by which environmental chemicals affect human disease. Both resources incorporate MeSH disease terms in their gene–disease mapping. The latest data from GAD and CTD were downloaded as flat files and then parsed and stored in DiseaseDB for data integration. We extracted a subset of the human genes (281 genes from GAD and 2286 genes from CTD) that are associated with bacterial infectious diseases from these sources. We also integrated these sources together to compare and contrast gene–disease associations that are identified genetically and chemically. Human genome and gene data were downloaded from HUGO (Human Genome Organization) Gene Nomenclature Committee (HGNC, www.genenames.org) and also stored in DiseaseDB.
3.4 Reports of global infectious disease outbreaks
Global infectious disease outbreak reports were dynamically retrieved from HealthMap (Freifeld et al., 2008). HealthMap is an automated electronic information system that brings together disparate data sources to provide a unified view of the current global impact of infectious diseases. Sources used in HealthMap include news aggregators, scientific reports such as ProMED-mail, World Health Organization, Eurosurveillance, Wildlife Disease Information Node, individual accounts from users and others. The infectious diseases associated with NIAID's 22 watchlist bacterial pathogens were manually mapped to the disease names defined in HealthMap (Supplementary Table S2).
The infectious disease, pathogen and host datasets within the DiseaseDB allow for multiple points of access. For example, a database query may start with particular diseases and retrieve virulence factors in linked pathogens or a query may retrieve human genes involved in human–pathogen interactions from a set of pathogenic organisms. From a user's perspective, querying data is only the first step. Data must be presented in a way to support a researcher's broader goals of understanding the interaction of components of disease phenomenon. Disease View relies heavily on visualization to meet this requirement.
4 VISUALIZING INFECTIOUS DISEASE RELATIONSHIPS
A primary goal of Disease View is to enable researchers to quickly identify potentially interesting relationships within infectious disease data. A powerful method for visualizing entity relationships is the graph, where nodes represent discrete entities and edges join nodes that are related in some way. Protein interaction networks are commonly represented as graphs, for example, with nodes representing individual proteins and edges joining pairs of proteins that interact. Graphs can be powerful analytical tools, either through topological analysis using standard graph theoretical algorithms like centrality and connectivity (Goh et al., 2007; Jeong et al., 2001) or by using our own visual pattern recognition system to extract meaning, derive new hypotheses, identify structure in the data or improve our confidence in the results of quantitative analyses.
Disease View represents infectious disease relationships as a multipartite graph (or multigraph) called the Disease-Pathogen Visualization (see Section 3 in Supplemental Material for full details of the graph construction). In brief, the Disease-Pathogen Visualization uses nodes to represent diseases, pathogens, virulence genes and disease-associated host genes. Node identity is double encoded as shape and color in order to clarify the multiple types in the graph. Relationships between nodes are represented as edges: directed edges indicate hierarchical relationships among diseases (based on MeSH) and among pathogens (based on NCBI taxonomy); undirected edges indicate associative relationships between diseases and human genes, and between pathogens and virulence genes.
We chose to represent integrated infectious disease data as a multigraph for several reasons: (i) it allows us to integrate multiple data sources within a single lossless visualization; (ii) it allows us to combine a high-level perspective on a set of complex relationships with the ability to drill down to more detailed information; (iii) it allows us to take advantage of human perceptive cognition to suggest avenues for further exploration and discovery; and (iv) it became clear as a result of our iterative design process that extracting simple graphs from Disease View data was not as useful as presenting the entire multigraph, despite the loss of applicability of standard topological analyses in the latter.
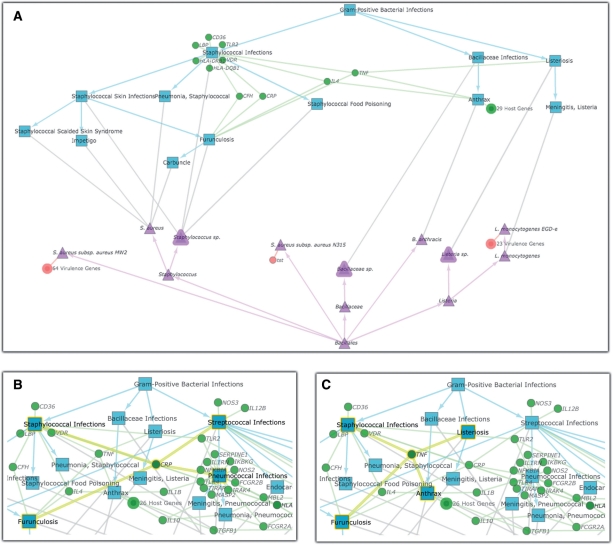
The underlying structure of Disease View data consists of two interacting hierarchies (pathogen taxonomy and MeSH disease hierarchy), each ‘decorated’ with a non-overlapping set of genes. It became clear during our iterative design process that the most useful knowledge was found in the relationships between the two hierarchies, and in the non-hierarchical relationships between members of a hierarchy (i.e. via shared genes). To emphasize these features, we developed a novel graph layout algorithm called mirrored tree. The mirrored-tree algorithm partitions the graph into three distinct subsets: the rooted disease hierarchy (based on MeSH); the rooted taxonomy hierarchy (based on NCBI); and the genes (disease-associated host genes and pathogen-associated virulence genes). The two hierarchical partitions are laid out as trees, with their roots at opposite ends of the visible space and their leaves aligned along the center. Genes (or aggregations of genes) that are unique to a pathogen or disease are arranged proximal to the associated disease or pathogen node. Genes that are shared among multiple diseases or pathogens are placed at the weighted mean distance from all associated diseases or pathogens. Consistent coloring of nodes and edges within each of the disease and pathogen hierarchies is used to emphasize these substructures (see Fig. 4A for an example of the mirrored-tree algorithm applied to the order Bacillales).
Fig. 4.
Disease-Pathogen visualizations for order Bacillales (A) and partial views for class Bacili (B and C); components of PATRIC Disease View for Use Case 2. Disease nodes are represented as blue rectangles, pathogen nodes as purple triangles and genes as green (host) and red (pathogen) circles.
By applying different layout schemes to different partitions of the Disease-Pathogen Visualization, we use our knowledge of the underlying graph structure to add clarity to the representation. By visually segregating the disease and pathogen hierarchies, the mirrored-tree layout draws special attention to the high-value disease–pathogen edges: they are the only edges that cross the center of the visible space. Furthermore, disease–pathogen edges that join leaf nodes in the two hierarchies indicate highly specific associations, whereas disease–pathogen edges that travel into the interior of the hierarchies indicate a more general (i.e. less informative) assignment. The mirrored-tree layout also emphasizes informative non-hierarchical relationships in the Disease-Pathogen Visualization. Positioning a disease or pathogen node in close proximity to its set of uniquely associated genes forms easily identifiable clusters of related nodes. The size of each cluster suggests how much is known at the genetic level about the given disease or pathogen. This rudimentary clustering also facilitates the identification of genes that may be involved in shared host-response or infection events: these genes are not part of any clusters.
In addition to layout, the Disease-Pathogen Visualization also uses aggregation to reduce visual clutter without losing information. First, if a disease or a pathogen has >10 unique associated genes, the genes are aggregated into a single Collection node, and individual genes in the Collection are accessible by clicking the node. Second, many diseases are associated with taxonomic levels of genus or higher, reflecting non-specificity in the data. For these cases, the Disease-Pathogen Visualization includes an edge from each disease to a pathogen species Collection node.
5 DISEASE VIEW IN PATRIC
We decided early on to implement Disease View as a web-based component of PATRIC. Currently, PATRIC provides rich data and analysis tools for all bacterial species including NIAID category A–C priority bacterial pathogens. It also includes comprehensive bacterial data such as genome annotation, comparative genomics, phylogeny, metabolic pathways, transcriptomics, proteomics, protein structures, protein–protein interactions and published literature. By leveraging these existing PATRIC resources, we improved the utility of Disease View as an integrative, web-based view onto infectious disease. Through our user-centered, iterative usability engineering process (Section 2), we produced a set of Disease Views accessible via PATRIC's taxon landing pages—dedicated pages for each NCBI taxonomic level, from the bacteria superkingdom to individual genus pages. These detailed designs were then polished to employ PATRIC interaction metaphors and visual style sheets. PATRIC's web developers implemented these designs and incorporated the Disease View into PATRIC's website.
From the PATRIC home page (http://www.patricbrc.org), a user can choose a PATRIC taxon of interest using the Organisms menu or via the tag cloud of ‘Most Viewed Bacteria’. This will open the taxon landing page for that taxon. Clicking the Diseases tab on the taxon landing page will open the Disease View, which is in three subsections: the Disease View Summary, the Disease-Pathogen Visualization and the Disease Map. Note that each Disease View in PATRIC presents infectious disease data for the given taxon as well as all child taxa (i.e. data from lower taxa are rolled up and represented at the parent level).
The Disease View Summary provides an overview of disease, pathogen and gene information. It features an Infectious Disease Overview, containing pathogens and their associated MeSH disease terms, links to tables of known virulence factors and links to tables of associated human genes. From this Overview, users may access: National Library of Medicine MeSH Descriptor data by clicking on any MeSH disease term; the taxon landing page for a specific PATRIC organism by clicking on its name; a Virulence Disease table by clicking on a number in the Pathogen Virulence Genes column; tables of human genes associated with each disease, including genetic and chemical evidence for the associations, by clicking on a number within one of the Human Disease Genes columns. The Disease View Summary also includes a hyperlinked list of PubMed literature specifically related to the associated diseases for the given pathogens.
The Disease-Pathogen Visualization contains an interactive graph of the relationships between pathogens, diseases, virulence genes and disease-associated host genes. Users may click on nodes and edges in the graph to view more specific information, including links to external resources. Controls along the top of the image allow users to filter the graph, adjust the view and the layout, export the graph as either an image or a Cytoscape-compatible graph file and access PATRIC FAQs. Controls along the bottom of the graph afford interactive panning and zooming.
The Disease Map provides a real-time view of recent reports of global disease outbreaks, geolocated on an interactive global map. It also includes the number of recent alerts, time periods, related diseases, locations, data sources and report categories, including New & Ongoing Outbreaks, Warnings and International Significance. This feature is made available through a collaboration between PATRIC and HealthMap.
To view each of these components for Staphylococcus, for example, visit http://www.patricbrc.org/portal/portal/patric/DiseaseOverview?cType=taxon&cId=1279.
6 USE CASES OF PATRIC DISEASE VIEW
6.1 Use Case 1: multiple perspectives on an infectious disease
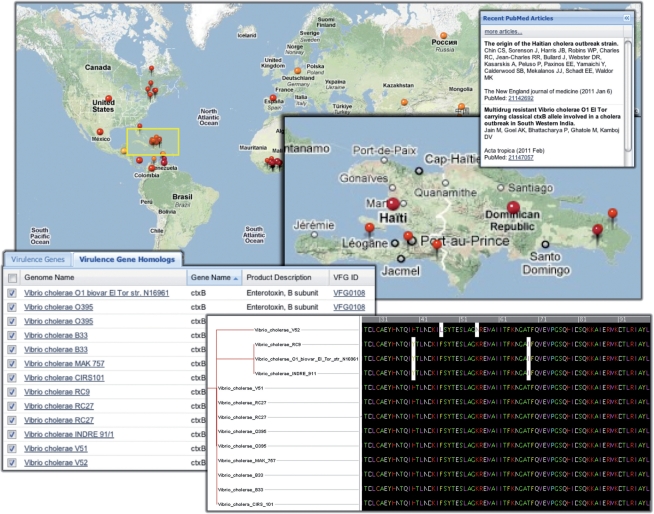
Disease View positions PATRIC as a single point of contact for a multifaceted exploration of infectious diseases because it integrates multiple types of data related to an infectious disease. One illustrative example is Vibrio cholerae, the water-borne bacterial pathogen that causes cholera. Given the recent outbreak of cholera in the aftermath of the 2010 earthquake in Haiti, we may be interested in learning more about this disease. Figure 3 shows a few of the steps in a possible exploration of this disease. From the PATRIC home page, we can click to the landing page for Vibrio, and then the Disease View (the Diseases tab). Switching to the Disease Map brings up a map of outbreak reports related to Vibrio-associated diseases (Fig. 3, map). A preponderance of dark red markers (high activity) around Haiti and the Dominican Republic reflects the still-active outbreak of cholera in this region; clicking any of these markers will reveal a continuously updated list of related news reports (not shown in the figure). Switching to the Summary, we might see that the Recent PubMed Articles (Fig. 3, upper right) contains a link to the publication (Chin et al., 2011) that determined the origin of the Haiti strain (since this feature of PATRIC Disease View is continuously updated, the article may no longer be visible but still accessible by clicking the ‘more articles’ link). According to Chin et al. genetic evidence from various key locations in the genome indicate that the Haiti isolate is most closely related to V.cholerae CIRS 101 (El Tor biotype of O1 serogroup), from an outbreak in Bangladesh in 2002 and 2008.
Fig. 3.
Disease View components showcased in Use Case 1; clockwise from top includes a map, PubMed insert, multiple sequence alignment and tabular data view.
The V.cholerae O1 serogroup includes two well-known biotypes: El Tor and classical. These biotypes are distinguished on the basis of biochemical, phenotypic and genetic differences. For example, cholera caused by classical strains is considered more severe, whereas the El Tor biotype is better able to survive in the environment. Also, the two biotypes have different alleles for the ctxB gene (Safa et al., 2008), which together with ctxA produces the cholera toxin that is responsible for the disease symptoms. In addition, classical strains of V.cholerae O1 do not contain the rtxC gene of the repeat in toxin (RTX) cluster that is responsible for activating cytotoxicity (Lin et al., 1999).
Unlike previous global cholera pandemics, which were dominated by more severe, classical strains, the current pandemic has been characterized almost entirely by El Tor strains. Recent evidence, however, indicates the rise of hybrid strains around the world that carry the classical cholera toxin gene in an El Tor background (Safa et al., 2008). This shift in the dominant cholera toxin has alarming implications. Since it is the primary contributor to the disease, we may see more severe symptoms associated with the classical toxin; compounding this problem, a genetic shift in the ctxB gene may render existing vaccines less effective, and lead to an increase in antibiotic resistance.
Which type of toxin can we expect from the Haiti isolate? PATRIC Disease View allows us to explore this question at the genetic level. We can see in the Summary table that 92 virulence genes are associated with V.cholerae strains. Clicking that number will bring up a table of all the virulence gene homologs within PATRIC (Fig. 3, table). From this table, we can select the ctxB homologs from sequenced Vibrio species (13 total at the time of writing), and generate a multiple sequence alignment (Fig. 3, alignment). The alignment results clearly show amino acid substitution events at positions 39 and 68 of the CtxB protein. In contrast to the three strains (V.cholerae O1 bioviar El Tor str. N16961, V.cholerae RC9 and V.cholerae INDRE 911) harboring the El Tor cholera toxin gene, all other strains in the list, including V.cholerae CIRS 101, carry Y39H and I68T mutations. These mutations are one way to distinguish the more severe classical toxins from the El Tor toxins (Popovic et al., 1994). Since the Haiti isolate is most closely related to V.cholerae CIRS 101, which carries the classical toxin, we may hypothesize that the Haiti isolate also produces the classical toxin. Chin et al. confirmed that the two sequenced Haiti isolates contain the Y39H and I68T mutations, which supports this hypothesis.
6.2 Use Case 2: identifying host genes common to multiple diseases
PATRIC Disease View facilitates the identification of subtle relationships in the data by aggregating host–pathogen infectious disease data at different taxonomic levels. For example, the taxonomic class Bacilli includes several well-known gram-positive pathogens, including Staphylococcus, Streptococcus, Bacillus anthracis and Listeria. Using the Disease View Summary on the Bacilli landing page, we can see the diseases associated with all of these pathogens, including Anthrax, Meningitis, Pneumonia, Scarlet Fever and more. What are the connections between these diseases? More specifically, do they share any host genes in common? Identifying host genes shared by multiple diseases from related pathogens may suggest potential areas of interest for developing therapeutics.
The Disease-Pathogen Visualization for Bacilli can reveal inter-relationships among these pathogens from the perspective of their associated diseases. The layout of the graph itself reveals multiple host genes shared among more than one Bacilli-related disease. For example, by rolling over the node that represents the CRP (C-Reactive Protein) gene, we can see it is associated with Staphyloccocal, Streptococcal and Pneumococcal Infections (Fig. 4B). CRP is a general host-response protein produced by hepatocytes and normally found at very low levels in blood serum. During acute phase protein reaction (a non-specific response to inflammatory stimuli like bacteria and viruses), the concentration of CRP rapidly increases several hundredfold, and it undergoes Ca2+-dependent binding to various compounds including specific target molecules from bacteria. Consequently, elevated CRP concentration is routinely used to diagnose infectious disease in a clinical setting; it can also be used to differentiate between bacterial and viral infections.
From the Disease-Pathogen Visualization of Disease View, we might tentatively conclude that CRP would be useful in diagnosing Streptococcal, Staphylococcal and Pneumonococcal infections, since CRP is associated with these disease classes. Published studies do support this conclusion (Lindback et al., 1989), although care must be taken to avoid over-interpretation of the Disease View relationships. For example, it may be tempting to hypothesize further that Listeria infection does not lead to elevated CRP levels, since there is no known disease–gene association in the graph; however, this is more likely due to a lack of data, since several studies support just such an association (Kleemann et al., 2009; Saleem et al., 2008).
In another example from the Bacilli, rolling over the node that represents the TNF (Tumor Necrosis Factor) gene reveals that it is associated with Staphylococcal infections, Listeriosis (Listeria) and Anthrax (Bacillus anthracis) (Fig. 4C). TNF is a cytokine involved in inflammation and the regulation of immune cells, and it has been identified as an important mediator of septic shock (Beutler et al., 1985), a serious and often fatal condition that results from bacterial infection. Because sepsis can also arise from bacterial infections in general, we would expect the TNF gene to be associated with more than just Bacilli. By navigating up the taxonomic tree to the Firmicutes, we can use the Disease-Pathogen Visualization to verify an additional relationship between TNF and Tetanus (Clostridium tetani). In addition, despite a more complicated picture, the Disease-Pathogen Visualization for Proteobacteria reveals that TNF is implicated in multiple gram-negative diseases as well (figure not shown).
7 CONCLUSIONS AND FUTURE WORK
We have developed Disease View, an online host–pathogen resource that provides easy access to integrated infectious disease, host, pathogen and disease outbreak data in a user-friendly web interface via the PATRIC resource. This resource enables the analysis of large integrated host–pathogen–disease datasets to facilitate our understanding of the disease mechanisms and to assist in the development of diagnostics and therapeutics. In comparison with many other resources mentioned in Section 1, the strengths of our approach are as follows: (i) Disease View enables infectious disease centric access of host, pathogen and outbreak information; (ii) diseases can be viewed at any level of the bacterial taxonomic tree; (iii) interactive graphs are provided online to enable visual analysis of the host–pathogen–disease network; in addition, the graphs can be downloaded as static images or exported to Cytoscape; and (iv) a well-established usability engineering process has been applied throughout this work to ensure the high level of effectiveness and quality of the software.
We present the following lessons learned while developing Disease View: (i) when dealing with a large amount of heterogeneous data, care must be taken not to overload user interfaces such that users either become disoriented or draw erroneous conclusions between pieces of unrelated data. Do not assume you know what users want; ask them. And subsequently focus the data and interfaces to serve these focused communities and/or use cases. (ii) Do not underestimate the power of visual analysis. Initially, we attempted to devolve the disease–pathogen–gene multigraph into sets of bipartite graphs for numerical analyses, but user feedback told us very clearly that the multigraph was the more useful analytical tool in this case.
In the future, we will continue to improve Disease View by providing customized searches and data analysis tools, adding host–pathogen interactions and additional data from animal models. Through our user interviews, we also see a need to develop disease-centric landing pages (e.g. a Tuberculosis page) to include information about diseases such as symptoms, diagnostics, vaccines, treatments and host response.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Chendong Zhang and Dan Sullivan for their help with Disease View database production; Ronald Kenyon and Rebecca Will for PATRIC project administrative support; and Susan Aman for assistance with HealthMap integration. We would like to thank all researchers for providing direction for, and feedback on, our evolving Disease View concepts and user interfaces including Josep Bassaganya-Riera, Lindsay Cowell, Ward Fleri, Raquel Hontecillas-Magarzo, Tom Kawula, Liwu Li, Cecilia Lindestam Arlehamn, Eric LoVullo, Brittany Mortensen, Bjoern Peters, Endang Purwantini, Alessandro Sette, Rebecca Wattam and Daniela Weiskopf.
Funding: This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200900040C, awarded to BWS Sobral.
Conflict of Interest: none declared.
REFERENCES
- Altschul S.F., et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beutler B., et al. Passive immunization against cachectin/tumor necrosis factor protects mice from lethal effect of endotoxin. Science. 1985;229:869–871. doi: 10.1126/science.3895437. [DOI] [PubMed] [Google Scholar]
- Chin C.S., et al. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P., et al. The Comparative Toxicogenomics Database: update 2011. Nucleic Acids Res. 2011;39:D1067–D1072. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll T., et al. PIG–the pathogen interaction gateway. Nucleic Acids Res. 2009;37:D647–D650. doi: 10.1093/nar/gkn799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifeld C.C., et al. HealthMap: global infectious disease monitoring through automated classification and visualization of Internet media reports. J. Am. Med. Inform. Assoc. 2008;15:150–157. doi: 10.1197/jamia.M2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbard J.L., et al. Engineering for Usability, In Proceedings of Human Systems Integration Symposium 2003. Vienna, VA: American Society of Naval Engineers; 2003. Usability engineering for complex interactive systems development. [Google Scholar]
- Goh K.I., et al. The human disease network. Proc. Natl Acad. Sci. USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hix D., Hartson H.R. Developing User Interfaces: Ensuring Usability Through Product and Process. New York, NY: John Wiley & Sons; 1993. [Google Scholar]
- Jeong H., et al. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Kleemann P., et al. Chronic prosthetic joint infection caused by Listeria monocytogenes. J. Med. Microbiol. 2009;58:138–141. doi: 10.1099/jmm.0.004234-0. [DOI] [PubMed] [Google Scholar]
- Kozhenkov S., et al. BiologicalNetworks–tools enabling the integration of multi-scale data for the host-pathogen studies. BMC Syst. Biol. 2011;5:7. doi: 10.1186/1752-0509-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B.K., et al. Tracking the epidemiology of human genes in the literature: the HuGE Published Literature database. Am. J. Epidemiol. 2006;164:1–4. doi: 10.1093/aje/kwj175. [DOI] [PubMed] [Google Scholar]
- Lin W., et al. Identification of a vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl Acad. Sci. USA. 1999;96:1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindback S., et al. The value of C-reactive protein as a marker of bacterial infection in patients with septicaemia/endocarditis and influenza. Scand. J. Infect. Dis. 1989;21:543–549. doi: 10.3109/00365548909037883. [DOI] [PubMed] [Google Scholar]
- Popovic T., et al. Detection of cholera toxin genes. In: Wachsmuth I.K., et al., editors. Vibrio Cholerae and Cholera: Molecular to Global Perspectives. Washington: American Society for Microbiology; 1994. pp. 41–52. [Google Scholar]
- Safa A., et al. Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerg. Infect. Dis. 2008;14:987–988. doi: 10.3201/eid1406.080129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem B.R., et al. Periaortic endograft infection due to Listeria monocytogenes treated with graft preservation. J. Vasc. Surg. 2008;47:635–637. doi: 10.1016/j.jvs.2007.09.029. [DOI] [PubMed] [Google Scholar]
- Snyder E.E., et al. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–D406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnenburg R., et al. PHI-base update: additions to the pathogen host interaction database. Nucleic Acids Res. 2008;36:D572–D576. doi: 10.1093/nar/gkm858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z., et al. PHIDIAS: a pathogen-host interaction data integration and analysis system. Genome Biol. 2007;8:R150. doi: 10.1186/gb-2007-8-7-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., et al. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 2008;36:D539–D542. doi: 10.1093/nar/gkm951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.