Abstract
During cell division, chromosome segregation is orchestrated by the interaction of spindle microtubules with the centromere. Accurate attachment of spindle microtubules to kinetochore requires the chromosomal passenger of Aurora B kinase complex with borealin, INCENP and survivin (SUR). The current working model argues that SUR is responsible for docking Aurora B to the centromere whereas its precise role in Aurora B activation has been unclear. Here, we show that Aurora B kinase activation requires SUR priming phosphorylation at Ser20 which is catalyzed by polo-like kinase 1 (PLK1). Inhibition of PLK1 kinase activity or expression of non-phosphorylatable SUR mutant prevents Aurora B activation and correct spindle microtubule attachment. The PLK1-mediated regulation of Aurora B kinase activity was examined in real-time mitosis using fluorescence resonance energy transfer-based reporter and quantitative analysis of native Aurora B substrate phosphorylation. We reason that the PLK1-mediated priming phosphorylation is critical for orchestrating Aurora B activity in centromere which is essential for accurate chromosome segregation and faithful completion of cytokinesis.
Keywords: Aurora B, kinase sensor, phospho-CENP-A, PLK1, survivin
Introduction
The kinetochore is a super-molecular complex assembled at each centromere in eukaryotes. It provides a chromosomal attachment point for the mitotic spindle, linking the chromosome to the microtubules and functions in initiating, controlling and monitoring the movements of chromosomes during mitosis. The kinetochore of animal cells contains two functional domains; that is, the inner kinetochore, which is tightly and persistently associated with centromeric DNA sequences throughout the cell cycle and the outer kinetochore which is composed of many dynamic protein complexes that interact with microtubules only during mitosis.
During prophase Aurora B regulates global events such as changes to mitotic chromatin, whereas in prometaphase each mitotic chromosome is autonomously controlled by Aurora B activity to generate spindle checkpoint signals and correct kinetochore–microtubule attachments. Chromosomal passengers are a group of evolutionarily conserved proteins orchestrating chromosome segregation and cytokinesis (Earnshaw and Bernat, 1991; Adams et al., 2001). This protein complex containing Aurora B, survivin (SUR), INCENP and borealin, is relocated from the kinetochore to the central spindle upon anaphase onset (Earnshaw and Bernat, 1991; Li et al., 1998; Terada et al., 1998; Wheatley et al., 2001; Gassmann et al., 2004). Perturbation of their function results in defects in metaphase chromosome alignment, chromosome segregation and cytokinesis (Ruchaud et al., 2007). The current working model argues that SUR is responsible for docking Aurora B to the centromere whereas its precise role in Aurora B activation has been unclear (Vader et al., 2006). It has been recently proposed that Aurora B substrates can inhibit kinase activation, and this is relieved by phosphorylation of these substrates by the centromeric kinase polo-like kinase 1 (PLK1) (Rosasco-Nitcher et al., 2008). However, it was unclear whether and how PLK1 and Aurora B cooperate in centromere during mitosis.
Here, we identified a novel regulatory mechanism underlying Aurora B kinase priming by PLK1 phosphorylation. Our biochemical characterization demonstrates that SUR Ser20 is a substrate of PLK1 in mitosis, which is critical for orchestrating Aurora B activity at the centromere and accurate chromosome segregation. Our study shed light on the mechanism of Aurora B regulation in mitosis.
Results and discussion
SUR physically binds to PLK1 kinase domain
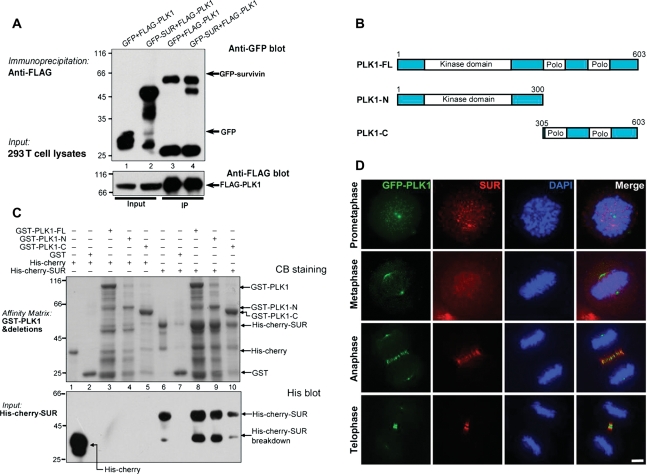
To delineate the molecular mechanism underlying PLK1 regulation in kinetochores, we immobilized bacterially recombinant proteins onto a nitrocellulose membrane to conduct a quick search for its interacting proteins using a high content Far Western assay (Fu et al., 2007; Yang et al., 2008). The PLK1-binding activity was then detected by an anti-PLK1 monoclonal antibody, in which an interaction between PLK1 and SUR was noted. To examine if PLK1 forms a cognate complex with SUR in cells, we carried out an immunoprecipitation using mitotic lysates from 293 T cells transiently transfected to express FLAG-PLK1 and green fluorescent protein (GFP)–SUR (Figure 1A). Western blot analysis demonstrated that FLAG–PLK1 interacts with GFP–SUR but not GFP tag (Figure 1A). To map the domain of PLK1 which physically binds to SUR, glutathione S-transferase (GST)-tagged recombinant PLK1 and its deletion mutants, illustrated in Figure 1B, were used as affinity matrix to isolate histidine-tagged SUR from bacteria (Figure 1C). Western blot analysis validated that the PLK1 kinase domain physically interacts with SUR. Immunoprecipitation of mitotic cell lysates with an anti-PLK1 mouse antibody pulled down endogenous SUR (Supplementary Figure S1, lane 4). SUR was not precipitated by non-specific mouse IgG (Supplementary Figure S1, lane 3), demonstrating the specific interaction of endogenous SUR with PLK1. Furthermore, immunofluorescent staining showed that PLK1 and SUR are co-localized to the centromere of metaphase cells and central spindle in anaphase and telophase cells (Figure 1D).
Figure 1.
SUR interacts and co-distributes with PLK1 in mitotic cells. (A) Aliquots of 293 T cells were transiently transfected to express SUR with wild type PLK1. Thirty-six hours after transfection, these cells were lysed and clarified cell lysates were precipitated with FLAG agarose. The beads were washed and boiled in SDS–PAGE buffer prior to electrophoresis. Western blotting analyses confirmed the co-precipitation of PLK1 with SUR. (B) Schematic illustration of human PLK1 functional domain. (C) In vitro pull-down assay with deletion mutations revealed that SUR binds to full length PLK1, kinase domain (PLK1-N) and polo-binding domain (PBD, PLK1-C). The gel was stained with CB. Western blotting analysis with an anti-histidine antibody confirmed the interaction between PLK1 with SUR. Note that the kinase domain binds stronger than PBD. (D) SUR co-distributes with PLK1 in mitotic cells. Synchronized HeLa cells transfected to expressed GFP–PLK1 (GFP) were pre-extracted, fixed and stained with SUR mouse antibody (red) and DAPI (blue). Bar, 5 µm.
SUR Ser20 is a substrate of PLK1 in mitosis
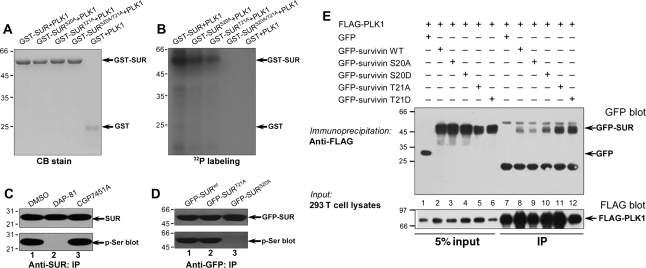
The interaction between PLK1 and SUR prompted us to examine if SUR is a substrate of PLK1. Our computational analysis suggests that Ser20 and Thr21 are potential substrates of PLK1 (Xue et al., 2008), which are conserved among eukaryotic SUR proteins. To test whether Ser20 and Thr21 are substrates of PLK1, we performed in vitro phosphorylation on recombinant GST–SUR fusion proteins, including both wild type and non-phosphorylatable SUR mutants in which Ser20 and Thr21 were both replaced by alanines (SURS20A/T21A). GST fusion proteins, wild type and mutant SUR, migrated to about the predicted 59 kDa as shown in Figure 2A. Incubation of the fusion proteins with [γ-32P]-ATP and His–PLK1 resulted in the incorporation of 32P into wild type, SURS20A and SURT21A but not SURS20A/T21A mutant and GST tag (Figure 2B). This PLK1-mediated phosphorylation is specific, as incubation of SUR with [γ-32P]-ATP in the absence of PLK1 resulted in no detectable incorporation of radioactivity into the wild type protein (data not shown). Thus, we conclude that both Ser20 and Thr21 on SUR are substrates of PLK1 in vitro.
Figure 2.
SUR is a substrate of PLK1 in dividing cells. Ser20 and Thr21 of SUR are substrates of PLK1. Bacterially recombinant GST–SUR fusion proteins were phosphorylated in vitro using [γ-32P]-ATP and active PLK1 as described under ‘Materials and methods’. Samples were separated by SDS–PAGE. (A) CB-stained gel of samples of wild type GST–SUR plus PLK1 (GST–SUR + PLK1), single and double mutant S20A/T21A GST–SUR plus PLK1 (GST–SURS20A/T21A + PLK1). (B) The same gel was dried and subsequently incubated with X-ray film. Note that there was dramatic incorporation of 32P into wild type, single site mutants (SURS20A and SURT21A) but little into the double mutant SURS20A/T21A or GST. (C) SUR is a cognate substrate of PLK1 in vivo. Extracts of nocodazole-synchronized mitotic HeLa cells were precipitated using an anti-SUR antibody. The precipitates were then fractionated by SDS–PAGE followed by immunoblotting of SUR (top panel) and serine phosphorylation (lower panel). Note that the SUR phosphorylation is a function of PLK1 but not CDK1. (D) Ser20 of SUR is phosphorylated in mitosis. GFP–SURwt, GFP–SURS20A and SURT21A were immunoprecipitated and isolated proteins were analyzed by western blotting. The membrane was first probed for GFP–SUR (upper panel) followed by detection of phosphoserine (lower panel). Lane 1, GFP–SURwt i.p.; lane 2, GFP–SURT21A i.p.; lane 3, GFP–SURS20A i.p. Note that GFP–SURS20A was not detected by phosphoserine antibody. (E) Association of PLK1 with SUR is independent of phosphorylation. Aliquots of HeLa cells were transiently transfected to express various GFP-tagged SUR plasmids with FLAG–PLK1. Thirty-six hours after transfection, these cells were lysed and clarified cell lysates were precipitated with FLAG-M2 agarose as described in ‘Material and methods’ section. Note that GFP–SURS20A weakened its association with PLK1.
If SUR is a substrate of PLK1 in vivo, suppression of PLK1 kinase activity using chemical inhibitors, such as DAP-81 (Peters et al., 2006), would alter endogenous SUR phosphorylation. Indeed, suppression of PLK1 using DAP-81 reduced serine phosphorylation of SUR immunopurified from mitotic HeLa cells (Figure 2C, lane 2). Interestingly, CDK1 inhibitor CGP7451A did not interfere with Ser phosphorylation on SUR (Figure 2C, lane 3), suggesting that SUR is a cognate substrate of PLK1 in mitosis.
To validate if Ser20 and Thr21 are PLK1 substrates in vivo, we conducted an anti-GFP immunoprecipitation of nocodazole-arrested mitotic cell lysates from HeLa cells transiently transfected to express GFP–SUR, GFP–SURS20A and GFP–SURT21A proteins. Both GFP–SUR and GFP–SURT21A proteins but not GFP–SURS20A mutant protein are phosphorylated in vivo judging by anti-phosphoserine analysis (Figure 2D). Interestingly, anti-phosphothreonine blotting indicated Thr21 is not phosphorylated at the condition that Ser20 is phosphorylated (data not shown). Thus, we conclude that SUR Ser20 is an endogenous substrate of PLK1 in prometaphase cells.
To examine if phosphorylation of Ser20 and Thr21 regulates the SUR–PLK1 association in cells, we carried out an immunoprecipitation using mitotic lysates from 293 T cells transiently transfected to express FLAG–PLK1 and phospho-mimicking mutants of SUR (Figure 2E). Western blot analysis demonstrated that FLAG–PLK1 interacts with GFP–SUR regardless of phosphorylation at Ser20 or Thr21 (Figure 2E). However, GFP–SURS20A binds PLK1 weakly compared with that of wild type or SURS20D.
Phosphorylation of Ser20 is essential for accurate chromosome segregation
To directly examine the role of PLK1-mediated phosphorylation in SUR localization to the kinetochore, we generated non-phosphorylatable and phosphomimetic SUR mutations and transiently transfected HeLa cells with GFP-tagged SUR plasmids together with mCherry-H2B. Real-time analysis of transfected HeLa cell preparations showed that the exogenously expressed GFP–SUR protein exhibits a typical inner centromere localization from prometaphase to metaphase (Figure 3A; from 00:00 to 00:33), and relocates to central spindle in anaphase cells (Figure 3A; 00:33–00:36), which is consistent with what was observed in endogenous SUR (Figure 1D). SUR is then concentrated to midbody of telophase cells until cytokinesis (Figure 3A).
Figure 3.
Phosphorylation of Ser20 of SUR is essential for accurate chromosome segregation. (A–D) Live-cell imaging of chromosome movements in HeLa cells expressing wild type and mutant SUR proteins. Aliquots of HeLa cells were transiently transfected to express various GFP-tagged SUR plasmids with mCherry-H2B. Thirty-six hours after transfection, these cells were briefly synchronized with monastrol and released for real-time imaging. Note that expression of GFP–SURS20A resulted in chromatin bridge phenotype (arrows in B and D). Bar, 5 µm. (E) Statistic analysis of mitotic phenotypes seen in live-cell imaging (mean ± SE; >20 cells of each category from three different preparations).
Our examination of GFP–SURS20A-transfected cells revealed that SURS20A localizes to kinetochore of prometaphase cells and becomes gradually dissociated from kinetochore of metaphase cells. Interestingly, expression of SURS20A caused premature anaphase as a typical metaphase alignment was not achieved prior to anaphase onset (Figure 3B). In addition, chromatin bridges are readily apparent in GFP–SURS20A-expressing cells (Figure 3D, arrows and E). On other hand, expression of SURS20D did not exhibit any apparent defects in chromosome alignment and segregation (Figure 3C). Thus, we conclude that PLK1-mediated phosphorylation of SUR at Ser20 is critical for accurate chromosome segregation.
Phosphorylation of Ser20 is essential for priming Aurora B activity in centromere
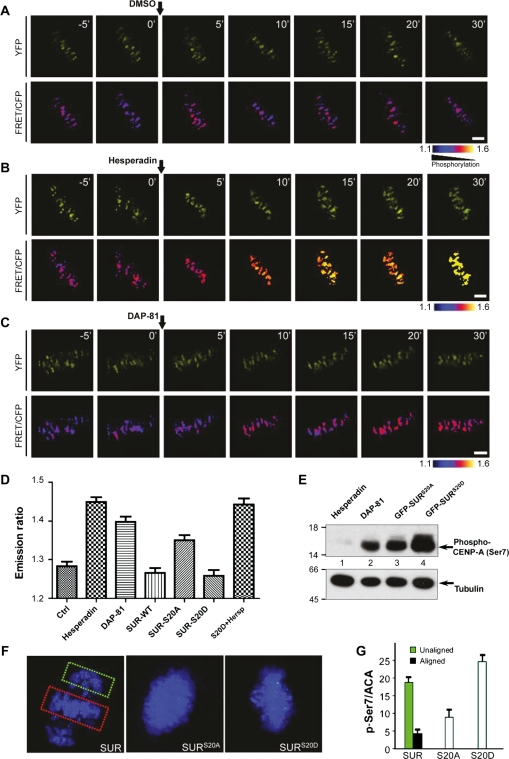
It has been recently proposed that Aurora B substrates can inhibit kinase activation, and this is relieved by phosphorylation of these substrates by the centromeric kinase PLK1 (Rosasco-Nitcher et al., 2008). To examine if phosphorylation of Ser20 of SUR retains Aurora B kinase activity, we sought to monitor Aurora B activity in centromere using fluorescence resonance energy transfer (FRET)-based sensors that report quantitative changes in substrate phosphorylation in living cells. We adapted a sensor design (Violin et al., 2003), in which changes in intra-molecular FRET between cyan and yellow fluorescent proteins (CFP–YFP) depend on changes in phosphorylation of an Aurora B substrate peptide that is conserved among members of the kinesin-13 family (Supplementary Figure S3). To mimic localizations of endogenous Aurora B substrates (Ruchaud et al., 2007), sensors were targeted to centromeres (CENP-B fusion; Figure 4A). To validate the sensor response to changes in Aurora B activity in living cells, we first imaged mitotic cells before and after kinase inhibition (experimental scheme in Supplementary Figure S4). For the sensor the YFP:CFP emission ratio increased after inhibitor hesperadin treatment (Figure 4B), consistent with dephosphorylation for this sensor design and previous reports (Liu et al., 2009). The maximal increase in emission ratio after chemical inhibition is similar to the increase during anaphase for each sensor (data not shown). The sensor is specific for Aurora B as it does not respond to other mitotic kinase inhibitors such as CGP7451A (data not shown), indicating that the measured FRET change is a faithful reporter of Aurora B kinase activity in centromere.
Figure 4.
Phosphorylation of Ser20 is essential for priming Aurora B kinase. (A) HeLa cells expressing the centromere-targeted Aurora B kinase sensor were imaged at metaphase. Upper panels, YFP images; lower panels, color-coded images of the emission ratio; timestamps (minutes) relative to drug addition. Bar, 5 µm. (B) Aurora B kinase sensor is responsive to hesperadin. Bar, 5 µm. (C) PLK1 inhibitor modulates Aurora B kinase activity in mitosis. Bar, 5 µm. (D) Statistic analyses of the YFP:CFP emission ratio indicate that SURS20D stimulates Aurora B activity (mean ± SE; >150 kinetochores of each categories from five different cells). (E) Western blotting of phospho-Ser7 of CENP-A indicated that expression of SURS20D stimulates Aurora B activity. (F) Phosphorylation of a kinetochore Aurora B substrate is stimulated by SURS20D. Left, insets show stronger p-Ser7 staining on unaligned versus aligned kinetochore. Right, overexpression of SURS20D produces stronger p-Ser7 staining on the kinetochores. (G) Statistic analyses of p-Ser7 immunofluorescence intensity at the kinetochores indicate that SURS20D promotes Aurora B activity (mean ± SE; >10 cells of each categories from three different preparations).
Examination of sensor emission ratio revealed that phosphorylation differences between individual centromeres depend on the centromere position as chromosomes congress to the equator during mitotic progression. Addition of hesperadin promotes dephosphorylation of sensor which is evident by minimized difference seen among individual centromeres as chromosomes approach metaphase alignment at the equator (Figure 4B).
If PLK1-mediated phosphorylation of SUR is a priming factor for Aurora B activation, inhibition of PLK1 would reduce Aurora B activity. As shown in Figure 4C, addition of PLK1 kinase inhibitor DAP-81 resulted in dephosphorylation of the sensor in centromere, supporting the notion that PLK1 kinase is critical for priming Aurora B activity in centromere. To examine if PLK1 phosphorylation of Ser20 is a priming factor for Aurora B, we sought to measure centromeric Aurora B activity in cells expressing SURS20D and SURS20A. Exogenous expression of mCherry–SUR exhibits a typical inner centromere similar to those of endogenous and GFP–SUR (data not shown). Similarly, mCherry–SURS20A and mCherry–SURS20D also exhibit typical centromere distribution (data not shown). As predicted, expression of mCherry–SURS20A resulted in a reduction of centromeric Aurora B kinase activity judging by an increased sensor emission ratio (Figure 4D). In contrast, expression of mCherry–SURS20D suppressed the sensor emission ratio, demonstrating that centromeric Aurora B kinase activity is a function of PLK1-phosphorylated SUR. Importantly, this SURS20D-elicited Aurora B kinase activity is minimized by hesperadin treatment (Figure 4D).
We next examined phosphorylation of Aurora B substrates using phospho-specific antibodies. First, we analyzed CENP-A phosphorylation (p-Ser7) in cells expressing SURS20D and SURS20A. As shown in Figure 4E, the p-Ser7 is minimized by treatment with Aurora B inhibitor hesperadin. Quantitative analyses indicated that expression of SURS20D resulted in an ∼2-fold increase in phosphorylation of Ser7 in CENP-A compared with that of SURS20A-expressing cells, confirming that pSer7 of CENP-A is an accurate reporter of Aurora B kinase.
We then evaluated the Aurora B kinase activity by quantitative immunofluorescence of p-Ser7 as described (Yang et al., 2008). As shown in Figure 4F and Supplementary Figure S5, the phospho-Ser7 intensity was highest in the unaligned kinetochores (green box) and lowest in the aligned kinetochores (red box), which is consistent with a recent report (Liu et al., 2009). The p-Ser7/ACA ratio was calculated for aligned (n = 147) and unaligned kinetochore (n = 113), yielding a 4-fold difference, supporting the role of Aurora B in kinetochore attachment. The ratio of p-Ser7/ACA increased 2.8-fold in SURS20D-expressing cells compared with that of SURS20A-expressing cells, consistent with our optical measurement of Aurora B activity in live cells (Figure 4D). Together, these data demonstrate that PLK1-mediated phosphorylation of SUR is a priming factor for Aurora B activity, both endogenous (p-Ser7) and exogenous (FRET sensor), during metaphase alignment.
We have shown that PLK1 phosphorylates SUR at Ser20 and this phosphorylation is important in eliciting Aurora B activity in centromere and accurate chromosome segregation in mitosis. We propose that the phosphorylation Ser20 of SUR is a priming factor for Aurora B activity in centromere, which is critical for correcting aberrant spindle microtubule attachments, provides a cooperative signaling mechanism between PLK1 and Aurora B to orchestrate accurate chromosome movements and faithful spindle microtubule attachment. Our molecular dissection has uncovered the underlying regulatory basis for temporal regulation of Aurora B activity in the centromere during mitosis.
Materials and methods
Plasmid construction
cDNA for human SUR was cloned into a T-vector (Takara Biotechnology). To generate GFP-tagged full-length SUR and PLK1, PCR-amplified cDNA was cloned into pEGFP-C2 vector (Clontech). The bacterial expression constructs of SUR were cloned into pET-22b (Novagen) and pGEX-6P (Amersham Biosciences). FLAG-tagged PLK1 cDNA was cloned by inserting the PCR product into the pcDNA3-FLAG vector (Invitrogen). The bacterial expression constructs of PLK1 and deletion mutants were cloned into pGEX-6P.
Cell culture and synchronization
HeLa and 293T cells, from American Type Culture Collection (Rockville), were maintained as subconfluent monolayers in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (Hyclone) and 100 units/ml penicillin plus 100 µg/ml streptomycin (Invitrogen) at 37°C with 8% CO2. Cells were synchronized at G1/S with 5 mM thymidine for 12–16 h and then washed with phosphate-buffered saline (PBS) four times and cultured in thymidine-free medium for 10 h to release.
Recombinant protein expression
Human PLK1 and deletion mutants were expressed in bacteria as a GST fusion protein. The GST fusion protein in bacteria in the soluble fraction was purified by using glutathione–agarose as previously reported (Wang et al., 2004). Briefly, 500 ml of Luria Bertani medium was inoculated with bacteria BL21(DE3) pLysS transformed with GST–PLK1. The protein expression was induced with 0.2 mM IPTG at 30°C for 4 h. Bacteria were then collected by centrifugation and resuspended in PBS containing a protease inhibitor mixture (Sigma) followed by sonication. The lysis solution was clarified by centrifugation for 20 min at 10000 g. The soluble fraction was applied to a column packed with glutathione–agarose beads, followed by extensive washes with PBS.
The histidine-tagged SUR proteins were purified as previous reported (Lou et al., 2004). Bacteria were harvested by centrifugation 3 h after the induction, resuspended in PBS containing proteinase inhibitors (leupeptin, pepstatin and chymostatin; 5 μg/ml), and sonicated for four bursts of 10 sec each by using a probe-tip sonicator. The lysis solution was clarified by centrifugation for 20 min at 10000 g. The soluble fraction was applied to a column packed with nickel–agarose beads, followed by extensive washes with PBS.
In vitro pull-down assay
GST pull-down was carried out as described previously (Zhu et al., 2008). GST fusion protein-bound Sepharose beads were used as an affinity matrix to isolate proteins interacting with PLK1 by using purified histidine-tagged full-length and S20A/D, T21A/D mutants of SUR that expressed in bacteria. Briefly, the GST–PLK1 fusion protein-bound Sepharose beads were incubated with purified proteins for 4 h at 4°C. After the incubation, the beads were extensively washed with PBS and boiled in SDS–PAGE sample buffer, followed by fractionation of bound proteins on 10% SDS–PAGE gel. Proteins were then transferred onto a nitrocellulose membrane for western blotting using an appropriate antibody.
In vitro phosphorylation
GST-tagged PLK1 was expressed in Sf9 cells and purified by glutathione–agarose beads. The kinase reactions were performed in 40 μl of 1× kinase buffer (25 mM HEPES, pH 7.2, 50 mM NaCl, 2 mM EGTA, 5 mM MgSO4, 1 mM DTT, 0.01% Brij35) containing 200 ng eluted PLK1 kinase, 1 μg GST-tagged or His-tagged proteins, 5 μCi [γ-32P]-ATP and 50 μM ATP. The mixtures were incubated at 30°C for 30 min. Then, the reactions were stopped with SDS sample buffer and separated by SDS–PAGE. The gel was stained with Coomassie brilliant blue, dried, and the 32P incorporation into SUR proteins was quantified by phosphorImager (Amersham BioSciences).
Transient transfection and immunoprecipitation
Two hundred and ninety-three T cells were grown to 50% confluence in Dulbecco's modified Eagle's medium and transfected with FLAG–PLK1 and GFP–SUR using Lipofectamine 2000 (Invitrogen). Cells were collected 24–36 h after transfection, and proteins were solubilized in lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM EGTA, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin and 10 μg/ml pepstatin A). Lysates were clarified by centrifugation at 16000 g for 10 min at 4°C. FLAG-tagged fusion proteins were incubated with anti-FLAG M2-linked agarose beads (Sigma). Beads were washed five times with lysis buffer and then boiled in protein sample buffer for 2 min. After SDS–PAGE, proteins were transferred onto nitrocellulose membrane. The membrane was probed with antibodies against the FLAG epitope and GFP. Immunoreactive signals were detected with an ECL kit (Pierce).
Immunofluorescence microscopy
For immunofluorescence, HeLa cells were seeded onto sterile, acid-treated 12-mm coverslips in 24-well plates (Corning Glass Works). Double thymidine-blocked and released HeLa cells were transfected with 1 μl of Lipofectamine 2000 pre-mixed with 1 ng of various plasmids as described above. In general, 36–48 h after transfection, HeLa cells were rinsed for 1 min with PHEM buffer (100 mM PIPES, 20 mM HEPES, pH 6.9, 5 mM EGTA, 2 mM MgCl2 and 4 M glycerol) and were permeabilized for 1 min with PHEM plus 0.1% Triton X-100 as previously described (Yao et al., 1997). Extracted cells were fixed in freshly prepared 4% paraformaldehyde in PHEM and rinsed three times in PBS. Cells on the coverslips were blocked with 0.05% Tween 20 in PBS (TPBS) with 1% bovine serum albumin (Sigma). These cells were incubated with various primary antibodies in a humidified chamber for 1 h and then washed three times in TPBS. Primary antibodies were visualized using fluorescein isothiocyanate-conjugated goat anti-mouse IgG or rhodamine-conjugated goat anti-rabbit IgG. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma). Slides were examined with a Zeiss LSM510 confocal scanning fluorescence microscope, and images were collected and analyzed with Image-5 (Carl Zeiss).
Quantification of the level of kinetochore-associated protein was conducted as described (Liu et al., 2008). In brief, the average pixel intensities from at least 50 kinetochore pairs from five cells were measured, and background pixel intensities were subtracted. The pixel intensities at each kinetochore pair were then normalized against ACA pixel values to account for any variations in staining or image acquisition. The values of specific siRNA-treated cells were then plotted as a percentage of the values obtained from cells transfected with a control siRNA duplex.
Live-cell imaging
Cells were cultured on coverslips in CO2-independent medium in a sealed chamber heated to 37°C and observed under a DeltaVision deconvolution microscope (Applied Precision). Images were acquired at 5 or 10 min intervals and presented in Photoshop.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grants DK-56292 and CA132389 and NIH, National Center for Research Resources (NCRR), Grant UL1 RR025008 from the Clinical and Translational Science Award Program. This work was also supported by Chinese Natural Science Foundation Grants 30500183 and 30870990 (to X.D.), and 90508002 and 90913016 (to X.Y.); Chinese Academy of Science Grants KSCX1-YW-R-65, KSCX2-YW-H-10, KSCX2-YW-R-195; the Major State Basic Research Development Program of China (973 Program) 2007CB914503 and 2010CB912103; International Collaboration Grant 2009DFA31010 (to X.D.) and Technology Grant 2006BAI08B01-07 (to X.D.); a Georgia Cancer Coalition Breast Cancer Research Grant; Atlanta Clinical and Translational Science Award Chemical Biology Grant P20RR011104 and Anhui Province Key Project Grant, 08040102005. The facilities used were supported in part by NCRR, NIH, Grant G12RR03034.
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We would like to thank members of our groups for insightful discussion and technical assistance.
References
- Adams R.R., Carmena M., Earnshaw W.C. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Bernat R.L. Chromosomal passengers: toward an integrated view of mitosis. Chromosoma. 1991;100:139–146. doi: 10.1007/BF00337241. [DOI] [PubMed] [Google Scholar]
- Fu G., Ding X., Yuan K., et al. Phosphorylation of human Sgo1 by NEK2A is essential for chromosome congression in mitosis. Cell Res. 2007;17:608–618. doi: 10.1038/cr.2007.55. [DOI] [PubMed] [Google Scholar]
- Gassmann R., Carvalho A., Henzing A.J., et al. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 2004;166:179–191. doi: 10.1083/jcb.200404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ambrosini G., Chu E.Y., et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., et al. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y., Yao J., Zereshki A., et al. NEK2A interacts with MAD1 and possibly functions as a novel integrator of the spindle checkpoint signaling. J. Biol. Chem. 2004;279:20049–20057. doi: 10.1074/jbc.M314205200. [DOI] [PubMed] [Google Scholar]
- Peters U., Cherian J., Kim J.H., et al. Probing cell-division phenotype space and polo-like kinase function using small molecules. Nat. Chem. Biol. 2006;2:618–626. doi: 10.1038/nchembio826. [DOI] [PubMed] [Google Scholar]
- Rosasco-Nitcher S.E., Lan W., Khorasanizadeh S., et al. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 2008;319:469–472. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Terada Y., Tatsuka M., Suzuki F., et al. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 1998;17:667–676. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G., Kauw J.J., Medema R.H., et al. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep. 2006;7:85–92. doi: 10.1038/sj.embor.7400562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin J.D., Zhang J., Tsien R.Y., et al. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J. Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hu X., Ding X., et al. Human Zwint-1 specifies localization of ZW10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 2004;279:54590–54598. doi: 10.1074/jbc.M407588200. [DOI] [PubMed] [Google Scholar]
- Wheatley S.P., Carvalho A., Vagnarelli P., et al. INCENP is required for proper targeting of survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 2001;11:886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- Xue Y., Ren J., Gao X., et al. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wu F., Ward T., et al. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem. 2008;283:26726–26736. doi: 10.1074/jbc.M804207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Anderson K.L., Cleveland D.W. The microtubule-dependent motor CENP-E is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 1997;139:435–447. doi: 10.1083/jcb.139.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Wang F., Yan F., et al. Septin 7 interacts with CENP-E and is required for its kinetochore localization. J. Biol. Chem. 2008;283:18916–18925. doi: 10.1074/jbc.M710591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.