SUMMARY
Tight regulation of NF-κB signaling is essential for innate and adaptive immune responses, yet the molecular mechanisms responsible for its negative regulation are not completely understood. Here we report that NLRX1, a NOD-like receptor family member, negatively regulates Toll-like receptor-mediated NF-κB activation. NLRX1 interacts with TRAF6 or IκB kinase (IKK) in an activation signal-dependent fashion. Upon LPS stimulation, NLRX1 is rapidly ubiquitinated, disassociates from TRAF6 and then binds to the IKK complex, resulting in inhibition of IKKα/β phosphorylation and NF-κB activation. Knockdown of NLRX1 in various cell types markedly enhances IKK phosphorylation and the production of NF-κB-responsive cytokines after LPS stimulation. We further provide in vivo evidence that NLRX1 knockdown in mice markedly enhances susceptibility to LPS-induced septic shock and plasma IL-6 level. Our study identifies a previously unrecognized role for NLRX1 in the negative regulation of TLR-induced NF-κB activation by dynamically interacting with TRAF6 and the IKK complex.
Keywords: Innate Immunity, NF-κB signaling, NOD-like receptors, IKK
INTRODUCTION
The innate immune system serves as the first line of defense against invading pathogens, such as bacteria and viruses, by recognizing a limited but highly conserved set of molecular structures, so-called pathogen-associated molecular patterns (PAMPs) (Akira et al., 2006). Recognition of such PAMPs relies on several classes of pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), NOD-like receptors (NLRs) and RIG-I-like receptors (RLRs) (Akira et al., 2006; Meylan et al., 2006; Rehwinkel and Reis e Sousa, 2010; Schroder and Tschopp, 2010; Takeuchi and Akira, 2010; Ting et al., 2010). Although different receptors recognize specific ligands, the ligand-receptor binding generally leads to activation of common downstream pathways like NF-κB, MAPK, and type I interferon to induce cytokine and chemokine gene expression, which facilitate pathogen clearance. Tight regulation of these key signaling pathways is essential for both innate and adaptive immunity; otherwise, aberrant immune responses may occur, leading to severe or even fatal bacterial sepsis, autoimmune and chronic inflammatory diseases (Karin et al., 2006; Liew et al., 2005).
TLR activation usually results in the recruitment of adaptor molecules, such as MyD88 and TRIF, which acts on a series of downstream signaling molecules such as TRAF6, which possesses E3 ubiquitin ligase activity. Activated TRAF6 synthesizes lysine 63 (K63)-linked polyubiquitin chains on itself and other proteins, which then serves as a scaffold to recruit TAK1 and the IκB kinase (IKK) complex through binding to the TAB2 and NEMO, respectively (Skaug et al., 2009). In contrast, activation of RIG-I and MDA-5 by double-stranded RNAs or certain viruses results in recruitment of the MAVS protein (mitochondrial antiviral signaling; also called VISA, IPS-1 and Cardif) (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). These signaling events converge on IKK complex to trigger activation of NF-κB signaling pathway and elicit inflammatory responses(Chen, 2005; Hacker and Karin, 2006) (Akira et al., 2006) (Hayden and Ghosh, 2008). The IKK complex consists of two catalytic subunits, IKKα and IKKβ, and one regulatory subunit, NEMO (also known as IKKγ). Activation of the IKK complex involves phosphorylation of serine residues on the activation loop of IKKα/β, and binding of NEMO to poly-ubiquitin chains formed by upstream signals (Skaug et al., 2009). However, the molecular mechanisms involved in the negative regulation of IKK activation are not well understood.
NLRX1, a member of the NLR family of proteins, contains an N-terminal X domain, a nucleotide binding oligomerization domain (NOD) and a C-terminal leucine-rich repeat domain (LRR). NLRX1 was shown to be a negative regulator of RIG-I-mediated antiviral response, by binding to MAVS on mitochondria and blocking its interaction with RIG-I (Moore et al., 2008). NLRX1 also modulates TNF-α and Shigella infection-induced reactive oxygen species (ROS) release (Tattoli et al., 2008), but its involvement in TLR-mediated NF-κB activation remains unknown. Here we demonstrate that NLRX1 is a negative regulator of TLR-mediated NF-κB signaling. In unstimulated cells, NLRX1 interacts with TRAF6, but after LPS stimulation, it undergoes K63-linked polyubiquitination and binds directly to the IKK complex and inhibits the phosphorylation of IKK, thus inhibiting NF-κB activation and pro-inflammatory cytokine release. We further provide in vivo evidence showing that specific knockdown of NLRX1 in mice enhances LPS-induced pro-inflammatory cytokine induction and renders mice more sensitive to LPS-induced septic shock. Thus, NLRX1 may serve as a useful target for manipulating immune responses against infectious or inflammation-associated diseases, including cancer.
RESULTS
NLRX1 negatively regulates TLR-induced NF-κB activation
To identify new molecules that might negatively regulate the NF-κB signaling pathway, we used a NF-κB-luciferase (NF-κB-luc) assay to screen a panel of candidates, including NLR family members, for their abilities to regulate NF-κB activity. TLR4, the NF-κB-luc reporter, an internal control Renilla luciferase reporter (pRL-TK-luc), and the candidate genes were co-transfected into HEK293T (293T) cells, which were then treated with the TLR4 ligand LPS for 18 h to activate NF-κB-luc activity. One of the proteins identified that inhibited NF-κB activation was NLRX1, also known as NOD9 (Figure 1A). We further confirmed NLRX1 inhibited NF-κB activation by LPS at both early (6 h) and late time points (24 h) (Figure S1A). Similar inhibitory effects were observed with 293T/TLR7 cells treated with CL-097 (a TLR7 ligand) when NLRX1 was co-expressed (Figure 1A). NLRX1 was recently shown to inhibit RIG-I-mediated type I interferon pathway by interacting with MAVS and sequestering it from interaction with RIG-I (Moore et al., 2008). Consistent with previous findings, we showed that NLRX1 inhibited RIG-I or MAVS-induced IFN-β-luc activity (Figure S1B). Since most TLRs use MyD88 as the adaptor protein for downstream signal transduction (Takeuchi and Akira, 2010), we next tested whether NLRX1 could inhibit MyD88-mediated NF-κB activation, which is independent of the MAVS-mediated pathway. As shown in Figure 1B, MyD88 strongly activated NF-κB-luc activity in 293T cells, but such activation was potently inhibited by NLRX1, suggesting that it inhibits MyD88-mediated NF-κB activation. To determine whether NLRX1 also inhibits NF-κB activation induced by downstream signaling molecules engaged in TLR signaling, we co-transfected 293T cells with TRAF6, TAK1/TAB1, IKKα, IKKβ or NF-κB p65 subunit to activate the NF-κB-luc reporter gene. We found that the activation of NF-κB by TRAF6, TAK1, IKKα, and IKKβ was markedly inhibited by NLRX1, but not by the NLR family members NOD1 or NOD2 (Figure 1B). By contrast, NLRX1 did not inhibit p65-mediated NF-κB activation, indicating that it may inhibit the NF-κB pathway immediately upstream of p65, most likely by interfering with the IKK complex.
Figure 1. NLRX1 inhibits TLR-induced NF-κB activation.
(A) 293T cells were transfected with NF-κB-luciferase (luc) reporter plasmid, TLR4 or TLR7 plasmids, together with an pcDNA3.1 empty vector or NLRX1 construct, and analyzed for NF-κB-dependent luciferase activity (fold induction) after treatment with LPS (TLR4 ligand) or CL-097 (TLR7 ligand) for 18 h. (B) 293T cells were transfected with NF-κB-luc, MyD88, TRAF6, TAK1+TAB1, IKKα, IKKβ, or p65, along with NLRX1, NOD1 or NOD2 and analyzed for NF-κB-dependent luciferase activity at 36 h post-transfection. (C) WT and MAVS−/− MEFs were transfected with an empty vector or murine NLRX1 expression vector, followed by LPS or poly (I:C) treatment. Cell supernatants were used to measure IL-6 release by ELISA. (D) MEFs and human THP-1 cells were tranfected with the NF-κB-luc reporter plasmid, along with a empty vector, murine NLRX1 or human NLRX1, and then analyzed for NF-κB-dependent luciferase activity after LPS treatment. (E) 293T cells were tranfected with IKKβ or p65 plasmids, together with an empty vector or NLRX1 plasmid. Nuclear extracts were used to detect endogenous NF-κB DNA binding activity by a gel-mobility shift assay. OCT-1/DNA complexes served as a loading control. (F) RAW264.7 cells were transfected with an empty vector or murine NLRX1 plasmid. The localization of p65 was determined by immunostaining after 15 min of LPS treatment. Data from (A–D) are plotted as means ± s.d. * P < 0.05, ** P<0.01, *** P<0.001, versus controls. All experiments were performed at least three times.
To test the possibility that MAVS is required for NLRX1-mediated inhibition of NF-κB signaling under physiological conditions, we determined IL-6 production in wild type (WT) and MAVS-deficient mouse embryonic fibroblasts (MEFs) after LPS or poly(I:C) (a TLR3 ligand) treatment (Sun et al., 2006). As shown in Figure 1C, IL-6 production was increased after LPS or poly(I:C) treatment, but such increases were potently inhibited by NLRX1 in both WT and MAVS-deficient MEFs, strongly suggesting that NLRX1 inhibits IL-6 production through a MAVS-independent pathway.
Since the mouse NLRX1 protein shares 75% amino acid similarity with human NLRX1, we examined whether the inhibitory function of NLRX1 is conserved between humans and mice. Murine NLRX1 expression markedly inhibited NF-κB-luc activation by different stimuli, suggesting a conserved biological function by which NLRX1 regulates the NF-κB pathway (Figure S1C). To test whether the findings in 293T cells can be extended to other cells, we performed NF-κB-luc reporter assays in MEFs and THP-1 cells (human monocyte cell line), which express endogenous TLR4. LPS treatment induced strong NF-κB-luc activity in both cell lines, and such activity was potently inhibited when NLRX1 was co-transfected (Figure 1D). We further examined endogenous NF-κB DNA binding ability by a gel mobility shift assay. IKKβ expression activated endogenous NF-κB to bind to 32P-labeled DNA probe containing NF-κB binding sites, but this activity was completely inhibited by NLRX1. By contrast, p65-mediated NF-κB DNA-binding activity was not affected by NLRX1, which is consistent with the luciferase reporter assay (Figure 1E). Since NF-κB activation is also associated with p65 translocation from the cytoplasm into the nucleus of cells after LPS stimulation, we examined p65 translocation in cells with or without NLRX1 expression. In cells transfected with empty vector, p65 rapidly translocated from the cytoplasm to the nucleus after LPS treatment. By contrast, p65 remained in the cytoplasm of the cells expressing NLRX1 after LPS treatment (Figure 1F). Taken together, these results suggest that NLRX1 inhibits TLR-induced NF-κB activation in various cell types.
NLRX1 dynamically interacts with TRAF6 and the IKK complex in response to LPS
To determine the molecular mechanisms by which NLRX1 negatively regulates NF-κB signaling, we tested whether NLRX1 directly interacts with the TRAF family adaptor proteins, because different NF-κB activation signals use distinct TRAF proteins to activate NF-κB signaling. Immunoprecipitation experiments showed that NLRX1 interacted with TRAF6 and TRAF3, but not with TRAF2 or TRAF5 (Figure 2A).
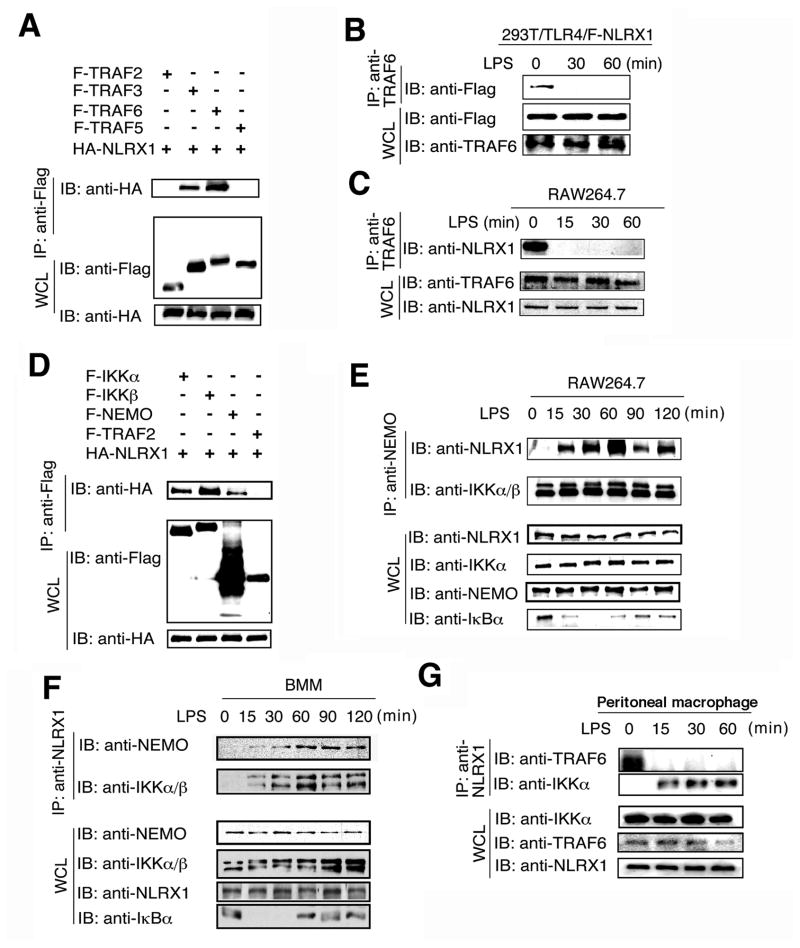
Figure 2. NLRX1 dynamically interacts with TRAF6 and IKK complex upon LPS treatment.
(A) 293T cells were transfected with F-TRAF2, F-TRAF3, F-TRAF5, F-TRAF6 and HA-NLRX1. Flag-tagged proteins was immunoprecipitated with anti-Flag followed by anti-HA immunoblotting. IP, IB and WCL denote immunoprecipitation, immunoblotting and whole cell lysate, respectively. (B–C), Cell extracts of 293T/TLR4 cells tranfected with Flag-NLRX1 (B) and RAW264.7 cells (C) were immunoprecipitated with anti-TRAF6 respectively, and then analyzed together with WCL by IB with indicated antibodies. (D) 293T cells were transfected with F-IKKα, F-IKKβ, F-NEMO, F-TRAF2 and HA-NLRX1. Flag-tagged proteins was immunoprecipitated with anti-Flag and blotted with anti-HA. (E–G) RAW264.7 cells (E), Bone marrow-derived macrophages (BMMs) (F) or peritoneal macrophages (G) were treated with LPS and the cell lysates were collected at the indicated time points and used for immunoprecipitation with anti-NEMO or anti-NLRX1, followed by immunoblot with the indicated antibodies. Results are representative of three independent experiments.
To further determine the endogenous protein interactions under physiological conditions, we developed an antibody against NLRX1 and confirmed its specificity in 293T cells, RAW264.7 cells and primary macrophages (Figure S2A–S2D). 293T/TLR4 cells transfected with Flag-tagged NLRX1 (F-NLRX1) were treated with LPS and the cell lysates were harvested at different time points. Anti-TRAF6 immunoprecipitation followed by anti-Flag immunoblotting revealed that NLRX1 interacted with TRAF6 in unstimulated cells, but dissociated from TRAF6 after LPS stimulation (Figure 2B). Similar results were obtained from RAW264.7 cells after LPS treatment (Figure 2C). To determine whether NLRX1 interacts with endogenous TRAF3, we performed similar experiments with anti-TRAF3, and found no endogenous interaction between NLRX1 and TRAF3 before or after LPS or poly(I:C) treatment, although the interaction between TRAF6 and NLRX1 was readily detected before LPS or poly(I:C) treatment (Figure S3A, S3B). These results suggest that NLRX1 interacts with TRAF6, but not TRAF3, in unstimulated cells. After activation, NLRX1 may switch from binding with TRAF6 to another target protein in the NF-κB signaling pathway to inhibit its activation. Since NLRX1 inhibits NF-κB activation induced by LPS, MyD88, TRAF6, TAK1 and IKK, but not by p65, we reasoned that it might directly interact with the IKK complex. To this possibility, we co-transfected 293T cells with HA-tagged NLRX1 together with Flag-tagged IKKα, IKKβ, and NEMO and tested their interaction. Co-immunoprecipitation with anti-Flag and Western blotting with anti-HA revealed that NLRX1 interacted with IKKα, IKKβ and NEMO, but not TRAF2 (Figure 2D). When IKK complex was immunoprecipitated from RAW264.7 cell extracts using anti-NEMO, the IKK-associated NLRX1 was barely detectable before LPS treatment, but the interaction was increased and then peaked at 60 min after LPS treatment. Importantly, anti-NEMO immunoprecipitation also pulled down IKKα and IKKβ (Figure 2E), suggesting that NLRX1 interacts with the IKK complex only after LPS stimulation. Similar results were obtained when THP1 cells were treated with LPS (Figure S3C). We also observed LPS-induced interaction between IKK and NLRX1 in bone marrow-derived macrophages, primary MEFs and primary mouse peritoneal macrophages when anti-NLRX1 was used for co-immunoprecipitation (Figure 2F, 2G, S3D). These results suggest that NLRX1 inhibits NF-κB signaling by its disassociation from TRAF6 and interaction with TLR-activated IKK complex.
NLRX1 is ubiquitinated through a K63 linkage after LPS treatment
Since NEMO binds to the K63-linked polyubiquitin chains of TRAF6 after LPS treatment, we reasoned that NLRX1 might undergo polyubiquitination, which might affect the dynamic interaction among NLRX1, TRAF6 and IKK. To test this possibility, we transfected 293T cells with plasmids expressing F-NLRX1 and HA-ubiquitin. Immunoprecipitation with anti-Flag and Western blot with anti-HA revealed that NLRX1 was strongly ubiquitinated (Figure 3A). Co-expression of NLRX1 with HA-ubiquitin (K48 only) or HA-ubiquitin (K63 only) mutant revealed that NLRX1 was preferentially ubiquitinated through K63-linked polyubiquitin chains (Figure 3A). Furthermore, we found that NLRX1 was rapidly ubiquitinated through the K63 linkage by 10–15 min after LPS treatment in 293/TLR4 and MEF cells, but was then reduced. By contrast, the K48-linked polyubiquitination of NLRX1 was not affected by LPS treatment (Figures 3B and 3C), raising the possibility that the polyubiquitin chains of NLRX1 is involved in the recruitment of NEMO and its IKK complex.
Figure 3. NLRX1 ubiquitination and its involvement in the interaction between NEMO and NLRX1.
(A) 293T cells were transfected with HA- ubiquitin, HA-ubiquitin (K48 only), HA-ubiquitin (K63 only) and F-NLRX1. F-tagged NLRX1 was immunoprecipitated with anti-Flag beads, and blotted with anti-HA. (B) 293T/TLR4 cells transfected with Flag-NLRX1 were collected at the indicated time points after LPS treatment. Flag- NLRX1 was immunoprecipitated with anti-Flag beads, and then analyzed by immunoblot with indicated antibodies. (C) MEF lysates were collected at the indicated time points after LPS treatment. Endogenous NLRX1 was immunoprecipitated with anti-NLRX1, and then analyzed by immunoblot with anti-K63-linked polyubiquitin antibody. (D) Schematic diagram showing deletion constructs of NLRX1 containing different domains. (E) 293T cells were co-tranfected with Flag-NEMO and the indicated Myc-NRLX1 deletion constructs. Flag-tagged NEMO was immunoprecipitated with anti-Flag beads, and blotted with anti-Myc. (F) 293T cells were co-transfected with HA-NLRX1 and the indicated Flag-NEMO deletion constructs. Flag-tagged proteins were immunoprecipitated with anti-Flag beads, and blotted with anti-HA. Results are representative of three independent experiments.
To test this possibility, we first determine the domains responsible for the interaction between NLRX1 and NEMO. We generated several deletion mutants of NLRX1 containing NOD and LRR domains (NLRX1-NOD-LRR), LRR domain only (NLRX1-LRR), the X and NOD domains (NLRX1-X-NOD), the NOD domain only (NLRX1-NOD) (Figure 3D). Immunoprecipitation and Western blot analyses revealed that NLRX1-FL, NLRX1-NOD-LRR, NLRX1-LRR and NLRX1-X-NOD, but not NLRX1-NOD, could interact with NEMO (Figure 3E), suggesting that the X and LRR domains are important for interaction with NEMO. Interestingly, the mutants capable of interacting with NEMO exhibited smear bands on the top of the gel, suggesting that NEMO also binds to polyubiquitinated NLRX1.
NEMO contains an ubiquitin-binding domain (UBD), which facilitates its binding to polyubiquitinatin chains of adaptor proteins and formation of protein complexes upon activation (Ea et al., 2006). To test if the UBD domain is required for NEMO binding to NLRX1, we performed experiments with NEMO deletion mutants. Co-immunoprecipitation experiments showed that smeared bands of NRLX1 protein were observed in the immunoprecipitation with anti-NEMO in the cells transfected with NEMO FL, NEMO 44–419 and NEMO 86–419, which contain UBD domain. By contrast, NLRX1 pulled down with anti-NEMO in the cells transfected with NEMO mutants without UBD domain (i.e., NEMO 1–196 and NEMO 1–302) failed to show the smear bands (Figure 3F). These results suggest that the K63-linked polyubiquitination of NLRX1 is involved in the binding of NEMO to NLRX1, and thus facilitating the initial recruitment of the NEMO/IKK complex to form a stable complex after LPS stimulation.
LRR domain of NLRX1 binds to the kinase domain of activated, but not inactive, IKKβ
To determine how NLRX1 interacts with IKKα and IKKβ, we transfected 293T cells with IKKα or IKKβ with various NLRX1 deletion mutants, and then performed co-immunoprecipitation experiments. Like NLRX1-FL, NLRX1-NOD-LRR, NLRX1-X-NOD, NLRX1-NOD interacted with IKKα and IKKβ, while NLRX1-LRR failed to interact with IKKα or IKKβ (Figures 4A, 4B), suggesting that the NOD domain of NLRX1 is required for the binding of NLRX1 to IKKα and IKKβ. However, functional assays with NF-κB-luc reporter showed that NLRX1 constructs containing LRR domain (i.e., NLRX1-NOD-LRR and NLRX1-LRR) had strong inhibitory activity, while NLRX1 mutants lacking LRR domain (i.e., NLRX1-X-NOD and NLRX1-NOD) exhibited weak or little inhibitory effect on NF-κB-luc activity (Figure 4C). These results suggest that the NOD domain of NLRX1 is involved in its interaction with IKK, but not for its inhibitory effect on IKKβ-induced NF-κB-luc activity. By contrast, the LRR domain of NLRX1 could not binds to IKKα/IKKβ, but is critically required for its inhibitory effect on IKKβ-induced NF-κB-luc activity. Thus, a key question is how the NLRX1 LRR domain inhibits IKKβ-induced NF-κB-luc activity, since it could not bind to IKKβ.
Figure 4. Mapping the interaction domains of NLRX1 and IKK subunits.
(A–B) 293T cells were tranfected with HA-IKKα (A) or HA-IKKβ (B) and the indicated F-NLRX1 deletion constructs. HA-tagged IKKα or HA-IKKβ was immunoprecipitated with anti-HA beads, and blotted with anti-Flag. (C) 293T cells were transfected with NF-κB-luc, IKKβ, along with F-NLRX1 and its deletion constructs and analyzed for NF-κB-dependent luciferase activity. Data are plotted as means ± s.d. (D) Experiment performed as in (B) except IKKβ WT was replaced by IKKβ EE mutant form. (E) Identification of the kinase domain of IKKβ interacting with NLRX1. Upper panel, schematic diagram showing protein domain structures of the IKKβ deletions. KD, kinase domain; LZ, leucine zipper; HLH, helix-loop-helix. Lower panel, 293T cells were co-tranfected with HA-NLRX1 with Flag-IKKβ and its deletion constructs indicated. Flag-tagged proteins were immunoprecipitated with anti-Flag beads, and then blotted with anti-HA. (F) Experiment performed as in (B) except IKKβ WT form was replaced by IKKβ KD mutant. Results are representative of three independent experiments.
One clue from the early results showing that NLRX1 binds to IKK only after LPS stimulation (Figures 2E–2G) raises the possibility that the NLRX1 LRR domain might bind to activated IKKα/IKKβ, rather than the inactive form of IKK. To test this possibility, we transfected 293T cells with NLRX1 mutants and IKKβ EE mutant, a constitutive active form containing serine to glutamic acid substitution on activation loop mimicking phosphorylation. Indeed, we found that NLRX1-LRR could strongly bind to IKKβ EE (Figure 4D), suggesting that the NLRX1 LRR domain can bind to the activated IKKβ. To further determine the mechanisms by which the NLRX1 LRR domain binds the activated IKKβ (IKKβ EE), we reasoned that the activated IKKβ may undergoes conformational changes and allow the LRR-interacting domain to be exposed, as previously proposed (Hayden and Ghosh, 2008). To test this possibility, we generated several IKKβ deletion mutants containing a kinase domain (KD), the leucine-zipper domain (LZ) or helix-loop-helix domain (HLH). Coimmunoprecipitation and Western blot analyses revealed that NLRX1 interacted with the kinase domain of IKKβ (IKKβ–KD), but not with IKKβ-HLH or IKKβ-LZ domain (Figure 4E). Importantly, we found that the LRR domain, but not other domains, of NLRX1 exhibited the strongest interaction with the kinase domain of IKKβ (IKKβ-KD) (Figure 4F), suggesting that the LRR domain directly binds to the kinase domain of IKKβ when it becomes accessible due to conformation changes after activation/phosphorylation.
NLRX1 inhibits IKKα/β phosphorylation and NF-κB activation
To determine the mechanisms by which NLRX1 inhibits NF-κB activation, we next cotransfected 293T cells with IKKα or IKKβ together with an empty vector or a NLRX1 expression vector. As shown in Figure 5A, expression of IKKα or IKKβ protein strongly induced IKK autophosphorylation, as detected by the phospho-specific IKK antibody. However, expression of NLRX1 markedly inhibited the phosphorylation of IKKα and IKKβ. By contrast, the phosphorylation of p38 was not affected by NLRX1 expression (Figure 5A), indicating that NLRX1 specifically inhibits IKK phosphorylation, but not p38 phosphorylation. Furthermore, NLRX1 inhibits LPS-induced IKK phosphorylation in 293T/TLR4 cells in a dose-dependent manner (Figure 5B).
Figure 5. NLRX1 inhibits IKK phosphorylation.
(A) 293T cells transfected with HA-IKKα, HA-IKKβ and HA-p38 with or without F-NLRX1 were used to analyze the phosphorylation of IKKα/β and p38. (B) 293T/TLR4 cells transfected with increasing amount of F-NLRX1 plasmid were used to analyze the phosphorylation of IKKα/β after LPS treatment. (C) RAW264.7 cells were treated with LPS, and cell lystaes were collected at the indicated time points for immunoprecipitation with anti-NLRX1 or anti-NEMO, followed by immunoblot with indicated antibodies or kinase assay (KA). (D) 293T cells were transfected with HA-IKKβ, together with an empty vector, F-NLRX1 or its deletion constructs and the phosphorylation of IKK was determined. Results are representative of three independent experiments.
Since NLRX1 or its LRR domain preferentially binds to activated form of IKKβ and inhibits IKKβ-induced NF-κB-luc activity (Figures 4C–4F), we next sought to determine the phosphorylation and kinase activity of IKKα/β in association with either NEMO or NLRX1 in LPS-stimulated RAW264.7 cells. We first immunoprecipitated NEMO-associated IKK complex with anti-NEMO and NLRX1-associated IKK complex with anti-NLRX1, respectively, and then determined their IKK phosphorylation and kinase activities for IκBα phosphorylation. We found that that NEMO-associated IKK complex displayed IKK phosphorylation and kinase activity for IκBα phosphorylation, as expected. By contrast, anti-NLRX1 immunoprecipitates contain IKK (IKKα/IKKβ and/NEMO) complex after LPS treatment, but did not exhibit any IKK phosphorylation or kinase activity for IκBα phosphorylation (Figure 5C). Consistent with the results in Figure 4C, we found that the LRR domain-containing NLRX1 constructs (i.e. NLRX1-FL, NLRX1-NOD-LRR and NLRX1-LRR) strongly inhibited IKKβ phosphorylation, while NLRX1-X-NOD and -NOD exhibited weak or no inhibitory activity (Figure 5D). Taken together, these results suggest that once NLRX1 (specifically its LRR domain) binds to the exposed kinase domain of activated/phosphorylated IKK, NLRX1-associated IKK complexes lose the phosphorylation probably through unknown phosphatases as well as their kinase activity for IκBα phosphorylation under physiological conditions.
Knockdown of NLRX1 enhances IKK phosphorylation and NF-κB-responsive genes
Since NLRX1 inhibits IKK phosphorylation and NF-κB activation, we reasoned that knockdown of NLRX1 would increase IKK phosphorylation and NF-κB-responsive gene expression. To test this prediction, we knocked down NLRX1 expression by NLRX1-specific siRNA in RAW264.7 cells and stimulated the cells with LPS (Figure S4A), and found that specific knockdown of NRLX1 markedly enhanced IKK phosphorylation, especially at 60 min after LPS stimulation, compared with RAW264.7 cells transfected with control siRNA. Kinase activity assays also showed increased and prolonged IKK kinase activity for GST-IκBα phosphorylation in NLRX1 knockdown cells compared to control siRNA-treated cells (Figure 6A). By contrast, MAPK (p38, JNK and ERK) signaling activation was largely unchanged in the cells transfected with NLRX1-specific or control siRNAs (Figure 6A). Similarly, LPS treatment induced IRF3 activation, but there was no difference in IRF3 phosphorylation between NLRX1 siRNA and control siRNA-treated cells (Figure 6A). To quantify these results, we did density scanning of the protein bands, and found that only IKK phosphorylation and kinase activity exhibited significant differences between NLRX1 knockdown and control cells. There were no appreciable differences in JNK, p38, EKR and IRF3 phosphorylation between NLRX1 knockdown and control cells (Fig. 6B). Conversely, LPS-induced IKK phosphorylation was markedly inhibited when NLRX1 was overexpressed in RAW264.7 cells (Figure S4B). Together, these results suggested that specific knockdown of NLRX1 enhances LPS-induced IKK phosphorylation and NF-κB activation.
Figure 6. Knockdown of NLRX1 enhances IKK phosphorylation and NF-κB- responsive cytokine gene expression.
(A) RAW264.7 cells were transfected with control siRNA or NLRX1-specific siRNA, and then treated with LPS for the indicated time points. LPS-induced IKK, IRF3 and MAPK (p38, JNK and ERK) activation were measured by IB with phospho-specific antibodies and IKK activity was measured by KA. (B) Quantitative comparison of signaling activation between NLRX1 knockdown and control cells by density scanning of the blots in (A). (C) RAW264.7 cells were transfected with control siRNA or NLRX1-specific siRNA, and then treated with LPS. The cytokine Tnf-α and Il-6 gene expression in RAW264.7 cells induced by LPS at different time points were determined by real-time PCR. (D) Production of cytokine TNF-α and IL-6 in culture medium of RAW264.7 cells transfected NLRX1-specific and control siRNAs after LPS treatment. Data in (C–D) are plotted as means ± s.d. * P < 0.05, ** P<0.01, versus controls. All experiments were performed at least three times with similar results.
To determine whether increased NF-κB activation in the NLRX1 knockdown cells could increase NF-κB-responsive cytokine genes, we performed knockdown experiments in RAW264.7 cells and found that Tnf-α and Il-6 gene expression were significantly higher in NLRX1 knockdown cells than those in control siRNA-transfected cells after LPS treatment (Figure 6C). Consistent with these results, the cytokine level of TNF-α and IL-6 was increased in NLRX1 knockdown cells compared to control cells (Figure 6D). Furthermore, NF-κB-responsive Ccl2 and Cxcl10 gene expression was also enhanced in NLRX1 knockdown cells compared to control cells (Figure S4C). Similar results were observed with THP-1 cells transfected with NLRX1-specific and control siRNAs (Figures S4D, S4E). Taken together, these results suggest that specific knockdown of NLRX1 increases IKK phosphorylation and NF-κB-responsive cytokine gene expression in response to LPS treatment.
NLRX1 negatively regulates NF-κB signaling and cytokine response in vivo
To substantiate the physiological function of NLRX1 in vivo, we generated NLRX1 knockdown (NLRX1-KD) transgenic mice constitutively expressing shRNA that specifically knocked down mouse NLRX1 (Figure S5A). By comparison with the ubiquitous expression of NLRX1 in the tissues of wild-type (WT) mice, mRNA transcript levels were markedly reduced in the thymus, spleen and bone marrow of NLRX1-KD mice (Figure 7A). Moreover, in MEFs generated from NLRX1-KD mice, we found a marked reduction of NLRX1 protein expression compared to WT MEFs (Figure 7B). It has been reported that knockdown of human NLRX1 in 293T cells enhances antiviral immunity and inhibits virus replication (Moore et al., 2008). To determine whether knockdown of mouse NLRX1 expression also contributes to antiviral immune responses, we infected MEFs with vesticular stomatitis virus (VSV)-enhanced green fluorescence protein (eGFP). VSV-eGFP infection was strongly inhibited in NLRX1-KD MEFs compared with WT cells, as evaluated by GFP signal in the cells (Figure 7B). Similar results were obtained in Nlrx1−/− MEFs (Figure S5B). NLRX1-KD mice also produced more interferon-β (IFN-β) in plasma, compared with WT mice after intravenous injection of poly (I:C) (Figure 7C). To further determine if NLRX1 knockdown affects mouse survival in response to viral infection, we injected NLRX1-KD and WT mice with VSV-eGFP via the tail vein and then monitored their survival. NLRX1-KD mice showed slightly but not significantly increased mouse survival compared to WT mice (Figure S5C). Consistent with this observation, we did not detect appreciable differences in serum viral titers and cytokine (IFN-β and IL-6) levels (Figures S5D–S5F). Together, these results suggest that NLRX1 knockdown has some effects on type I interferon and antiviral immunity in vitro, but not in vivo, thus raising the possibility that NLRX1 may have a predominant regulatory role in TLR-induced NF-κB signaling.
Figure 7. NLRX1 negatively regulates NF-κB signaling and cytokine responsein vivo.
(A) Knockdown of murine NLRX1 expression in different tissues of NLRX1-KD mice as measured by quantitative real-time PCR. (B) NLRX1 protein expression was determined by immunoblot in WT and NLRX1-KD MEFs. WT and NLRX1-KD MEFs were infected by VSV-eGFP, and was visualized by fluorescent microscopy. (C) WT and NLRX1-KD mice (n=10 per group) were injected intravenously with poly(I:C) (200 μg per mouse) and then sera were collected at indicated times for IFN-β. measurement. (D) LPS-induced IKK and MAPK activation in peritoneal macrophages from WT and NLRX1-KD mice. The quantified results are shown after band density scanning. (E) IL-6 production in WT and NLRX1-KD MEFs after treatment with LPS, Pam3CSK4 or infected with VSV-eGFP. (F) IL-6 and TNF-α production in peritoneal macrophages from NLRX1-KD or WT mice after treatment with LPS. (G) Survival of NLRX1-KD and WT mice (n=9 per group) after peritoneal injection with LPS (30 mg/kg). (H) Plasma IL-6 levels from WT and NLRX1-KD mice (n=9 per group) 3 h after peritoneal injection with LPS (30 mg/kg). Data in (A, C, E and F) are plotted as means ± s.d. * P<0.05, ** P<0.01, versus controls. All experiments were performed at least three times with similar results,
To test this possibility, we isolated peritoneal macrophages from WT and NLRX1-KD mice and examined IKK phosphorylation and MAPK signaling activation after LPS treatment. Consistent with the results obtained with RAW264.7 cells, IKK phosphorylation was much higher and prolonged in NLRX1-KD macrophages compared to WT cells after LPS treatment, while p38 and JNK phosphorylation was largely unchanged (Figure 7D). To define the physiological function of NLRX1 in response to other TLR ligands, we found that IL-6 production was enhanced in NLRX1-KD MEFs after stimulation with LPS, Pam3CSK4 (a TLR2 ligand) and infection with VSV-eGFP, compared to WT MEFs (Figure 7E). Similarly, we found that IL-6 expression was increased in Nlrx1−/− MEFs compared to WT MEFs treated with LPS, Pam3CSK4, poly (I:C) or CL-097 (Figures S6A, S6B). Furthermore, IL-6 and TNF-α production in NLRX1-KD macrophages was significantly higher than those in WT cells after treatment with LPS, Pam3CSK4, poly (I:C), CL-097 and CpG (a TLR9 ligand) (Figures 7F, S6C, S6D). These results suggest that NLRX1-KD and Nlrx1−/− cells markedly enhance NF-κB signaling and cytokine (IL-6 and TNF-α) production after TLR stimulation.
To determine the specificity of NLRX1 in inhibiting NF-κB activation, we compared NF-κB activation in NLRX1-KD and WT cells induced by TNFR, T cell receptor (TCR) or B cell receptor (BCR) stimulation. We did not observe an appreciable difference in IL-6 production between NLRX1-KD and WT cells after TNF-α treatment (Figure S6E). This result is consistent with immunoprecipitation experiments showing that NLRX1 interacted neither with TRAF2 nor with IKK in cells treated with TNF-α (Figures 2A and S6F). Furthermore, we failed to find differences in TCR- or BCR-induced NF-κB signaling between NLRX1-KD and WT cells (Figures S6G, S6H), suggesting that NLRX1 is not involved in TCR- or BCR-induced NF-κB activation.
We next sought to determine the function of NLRX1 in LPS-induced septic shock. NLRX1-KD and WT mice were injected with a high dose of Escherichia coli LPS (30 mg/kg) intraperitoneally and monitored for their survival. As shown in Figure 7G, more than 80% of the NLRX1-KD transgenic mice died within 18 h, whereas all WT mice remained alive. All NLRX1-KD transgenic mice died within 30 h compared to about 50% of the WT mice, which continued to survive after 36 h (Figure 7G). Enhanced LPS toxicity in NLRX1-KD mice correlated with markedly increased plasma IL-6 level (Figure 7H). However, we did not observe significant difference in plasma TNF-α level (data not shown). These results provide in vivo evidence that NLRX1 negatively regulates NF-κB activation, NF-κB-responsive cytokine IL-6 and LPS-induced septic shock.
DISCUSSION
NLRX1 is a member of the NOD-like family of proteins, which are characterized by a conserved NOD and LRR regions, and are involved in the activation of diverse signaling pathways (Akira et al., 2006; Inohara et al., 2005; Meylan et al., 2006; Ting and Davis, 2005). Among these proteins, NOD1, NOD2 and NALP3 have been extensively studied and shown to activate inflammasome pathway once they encounter relevant PAMPs (Akira et al., 2006; Inohara et al., 2005; Kobayashi et al., 2005; Meylan et al., 2006; Shaw et al., 2008; Ting and Davis, 2005). However, the biological function of other NLR proteins remains poorly understood (Ting et al., 2010). Previous report shows that NLRX1 functions as a negative regulator of cellular antiviral immunity by interfering with viral infection-induced RIG-I-MAVS complex formation, or modulates Shigella infection-induced reactive oxygen species (ROS) generation (Moore et al., 2008; Tattoli et al., 2008). Our studies, however, indicate that NLRX1 functions as a negative regulator that inhibits TLR-induced NF-κB activation. The findings presented here identify a previously unrecognized role for NLRX1 in the negative regulation of NF-κB signaling by targeting TRAF6 and IKK complex. Furthermore, we provide compelling evidence that NLRX1 negatively regulates NF-κB signaling, cytokine production and LPS-induced septic shock in vivo.
In response to TLR stimulation, most TLRs recruit MyD88 and activate the downstream molecules such as TRAF6, which acts as an E3 ubiquitin ligase to catalyze K63-linked polyubiquitin chain on itself. The polyubiquitin chains of TRAF6 function as a scaffold to recruit the TAK1 and IKK complexes through binding to the regulatory subunits, TAB2 and NEMO, respectively (Skaug et al., 2009). TLR-induced NF-κB signaling pathway has been well characterized, but it is still not completely understood that how the activated signals are dampened to prevent pathological consequences, such as septic shock and inflammatory disorders. The increased list of negative regulators of TLR signaling (Komuro et al., 2008; Liew et al., 2005) includes diverse proteins that regulate the TLR signaling pathway at different stages. The deubiquitinating enzymes A20 and CYLD inhibit NF-κB signaling by targeting TRAF6 upstream of IKK (Kovalenko et al., 2003; Liew et al., 2005; Trompouki et al., 2003; Wertz et al., 2004), while a recent report shows that CUEDC2 inhibits IKK activity by recruiting a phosphatase PP1 and keeps IKK in an inactivated status (Li et al., 2008). We recently reported that NLRC5 inhibits TLR-induced NF-κB activation by constitutive interaction with IKKα/β, but not NEMO (Cui et al., 2010). However, our results here show that NLRX1 interacts with TRAF6 in unstimulated cells, but disassociates from TRAF6 upon LPS stimulation. By contrast, there was no interaction between NLRX1 and TRAF2 or TRAF3 before or after stimulation. In addition, NLRX1 did not show any inhibitory effect on TNFR-, TCR- or BCR-induced NF-κB signaling, which is independent of TRAF6. These results further suggest that NLRX1 specifically inhibits TLR-induced TRAF6-dependent NF-κB signaling through targeting TRAF6. Although the molecular mechanisms responsible for its disassociation from TRAF6 are not known, we found that NLRX1 is rapidly ubiquitinated at 10–15 min after LPS stimulation. Interestingly, like TRAF6, NLRX1 also undergoes K63-linked, but not K48-linked, polyubiquitination after LPS stimulation. The interaction between NLRX1 and the IKK complex is induced after LPS stimulation. Thus, the molecular mechanisms by which NLRX1 inhibits NF-κB signaling are distinct from NLRC5. It appears that several negative regulators specifically target TRAF6 and/or IKK to control NF-κB activation through distinct mechanisms.
Our results show that the K63-linked polyubiquitination of NLRX1 may be involved in the dynamic interaction of NLRX1 with TRAF6 or with the IKK complex. NLRX1 polyubiquitination may alter its interaction with TRAF6, and serve as a scaffold to recruit the IKK complexes through binding to the regulatory subunit NEMO. This notion is further supported by our results showing that NEMO mutants lacking UBD domain fail to pull down the polyubiquitinated NLRX1 (the smear bands). Furthermore, NLRX1 interacts only with constitutively active IKKβ. IKK activation has been proposed to depends on phosphorylation of IKKβ on its activation loop, which in turn might induce the complex conformation change and exposure of kinase domain (Hayden and Ghosh, 2008). Indeed, we show that the LRR domain in NLRX1 interacts strongly with the kinase domain of IKKβ when IKKβ becomes accessible due to conformational changes after IKK activation or phosphorylation. These results suggest that the interactions of NLRX1 with TRAF6 or with the IKK complex are complicated and are signaling-dependent. Based on these findings, we propose a working model: NLRX1 associates and disassociates with TRAF6, depending on the activation status of cells. Upon stimulation with LPS, both TRAF6 and NLRX1 are rapidly ubiquitinated through the K63 linkage and they dissociate from each other. The K63 linked polyubiquitination of TRAF6 leads to the recruitment of the TAK1 complex and IKK complex to polyubiquitin chains, activating IKK for its phosphorylation, which becomes the target of NLRX1. The polyubiquitin chains of NLRX1 may serve as a scaffold to recruit NEMO, and then NLRX1 binds to activated IKK and forms a relatively stable NLRX1-IKK complex through the interaction between the NLRX1 LRR domain and the IKKβ kinase domain. Meanwhile, the phosphorylation of IKK is removed by unknown phosphatases, resulting in formation of a NLRX1-IKK complex without IKK phosphorylation and kinase activity. Thus, the interactions of NLRX1 with TRAF6 or with the IKK complex are a dynamic process and rely on TLR-induced activating signal. To determine whether TRAF6 is responsible for NLRX1 ubiquitination, we performed experiments with WT and Traf6−/− MEFs and found that NLRX1 is equally ubiquitinated in both types of cells after LPS treatment (data not shown), suggesting that TRAF6 may not be the E3 ligase responsible for NLRX1 ubiquitination. Thus, further studies are needed to identify such E3 ubiquitin ligases for the ubiquitinatination of NLRX1.
Although NLRX1 has been reported as a mitochondria-associated protein, it is evident that some NLRX1 protein is present in the cytoplasm to function as a negative regulator of NF-κB signaling. Similar proteins such as Stat3 and Bcl-2 have been reported to have different biological functions in the cytoplasm and mitochondria (Gough et al., 2009; Hockenbery et al., 1993; Jacobson et al., 1993; Wegrzyn et al., 2009). To define the biological function of NLRX1 in vivo, we generated NLRX1-KD mice with shRNA targeting NLRX1 expression. Our study using NLRX1-KD mice, as well as cells derived from both NLRX1-KD and NLRX1 knockout mice, further provide evidence that NLRX1 is a critical negative regulator of TLR-induced NF-κB signaling pathway. Macrophages and MEFs from NLRX1-KD mice enhance IKK phosphorylation and produce more proinflammatory cytokines in response to TLR stimulation. More importantly, NLRX1-KD mice are more sensitive to LPS-induced septic shock, with association of increased level of plasma IL-6. Although we observed enhanced antiviral immunity against VSV-eGFP infection in NLRX1-KD and Nlrx1−/− MEFs and increased IFN-β production in NLRX1-KD mice after poly(I:C) injection, we did not observe appreciable differences in mouse survival, viral titers and serum cytokine levels after VSV-eGFP infection. These results suggest that NLRX1 may play a predominant role in TLR-induced NF-κB signaling, compared with type I IFN pathway.
Collectively, the findings presented here identify a previously unrecognized role for NRLX1 in the negative regulation of NF-κB signaling and provide insights into molecular mechanisms by which NLRX1 dynamically interacts with TRAF6 and IKK in a signal-dependent fashion. Thus, NLRX1 may serve as a therapeutic target for the treatment of infectious and autoimmune diseases, as well as cancer.
EXPERIMENTAL PROCEDURES
Transfection and reporter assay
293T cells, MEFs and THP-1 cells were transfected with plasmids encoding NF-κB luciferase or IFN-β luciferase, pRL-TK Renilla luciferase and different expression or control vectors. Lipofectamine 2000, lipofectamine LTX with PLUS reagent (Invitrogen, Carlsbad, CA) and Amaxa nucleofector kit V (Lonza Amaxa Inc. Gaithersburg, MD) were used for tranfection of 293T cells, MEFs and THP-1 cells, respectively. The luciferase activity was determined by a dual luciferase assay kit (Promega, Madison, WI) with a Luminoskan Ascent luminometer (Thermo Scientific, Waltham, MA).
Immunoprecipitation and immunoblot analyses
For immunoprecipitation experiments, whole cell extracts were prepared after transfection or stimulation and incubated with indicated antibodies together with Protein A/G beads (Pierce, Rockford, IL) for overnight. For anti-Flag or anti-HA immunoprecipitation, anti-Flag or anti-HA agarose gels (Sigma, St. Louis, MO) were used. Beads were then washed 4 times with lysis buffer, and immunoprecipitates were eluted with SDS loading buffer (Cell Signaling Technology Inc., Danvers, MA) and resolved in SDS-PAGE gels. The proteins were transferred to PVDF membrane (Bio-Rad) and further incubated with the indicated antibodies. LumiGlo Chemiluminescent Substrate System from KPL (Gaithersburg, MD) was used for protein detection.
Generation of NLRX1-KD transgenic mice
Plasmid DNA for NLRX1-specifc shRNA1 from Open biosystems (Huntsville, AL) was linearized by restriction enzymes Pvu I and Xba I digestion and injected into fertilized mouse eggs of C57BL/6 strain through a Transgenic Core. Positive founders and offspring were identified by genotyping PCR of the tail DNA using the primers 5′-ACGTCGAGGTGCCCGAAGGA (forward) and 5′-AAGCAGCGTATCCACATAGCGT (reverse). Three transgenic lines were maintained by crossing founders to C57BL/6. All the mice were maintained in a pathogen-free animal facility. All animal studies performed were approved by the BCM Institutional Animal Care and Use Committee.
Statistical analysis
Unless indicated otherwise, all data were plotted as means ± SD. Significant differences between groups were determined by two-tailed Student’s t test or two-way ANOVA. In the mouse endotoxic shock study, Kaplan-Meier survival curves were generated and analyzed for statistical significance with Graphpad Prism 4.0 software.
Supplementary Material
NLRX1 negatively regulates Toll-like receptor-mediated NF-κB activation.
NLRX1 disassociates from TRAF6 and then binds to the IKK complex after TLR stimulation.
NLRX1Knockdown enhances NF-κB activation and cytokine production after LPS stimulation.
NLRX1 knockdown in mice enhances susceptibility to septic shock and plasma IL-6 level.
Acknowledgments
We thank Drs. Jenny Ting for Nlrx1−/− MEF cells, Michael Karin, Bing Su, Shao-Cong Sun, and Matsumoto Kunihiro for IKKα, IKKβ, NEMO, p38, p100, and TAB1 constructs, Hongbin Shu for TRAF constructs. We would also like to thank and Dr. Adebusola Aboseda Alagbala for generating bone marrow-derived macrophages and critical reading of the manuscript. This work was supported in part by grants from National Cancer Institute, National Institutes of Health and Cancer Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006:re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hockenbery DM, Oltvai ZN, Yin XM, Milliman CL, Korsmeyer SJ. Bcl-2 functions in an antioxidant pathway to prevent apoptosis. Cell. 1993;75:241–251. doi: 10.1016/0092-8674(93)80066-n. [DOI] [PubMed] [Google Scholar]
- Hornung V, Schlender J, Guenthner-Biller M, Rothenfusser S, Endres S, Conzelmann KK, Hartmann G. Replication-dependent potent IFN-alpha induction in human plasmacytoid dendritic cells by a single-stranded RNA virus. J Immunol. 2004;173:5935–5943. doi: 10.4049/jimmunol.173.10.5935. [DOI] [PubMed] [Google Scholar]
- Inohara Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Burne JF, King MP, Miyashita T, Reed JC, Raff MC. Bcl-2 blocks apoptosis in cells lacking mitochondrial DNA. Nature. 1993;361:365–369. doi: 10.1038/361365a0. [DOI] [PubMed] [Google Scholar]
- Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Li HY, Liu H, Wang CH, Zhang JY, Man JH, Gao YF, Zhang PJ, Li WH, Zhao J, Pan X, et al. Deactivation of the kinase IKK by CUEDC2 through recruitment of the phosphatase PP1. Nat Immunol. 2008;9:533–541. doi: 10.1038/ni.1600. [DOI] [PubMed] [Google Scholar]
- Liew FY, Xu D, Brint EK, O’Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–286. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shaw MH, Reimer T, Kim YG, Nunez G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr Opin Immunol. 2008;20:377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tattoli I, Carneiro LA, Jehanno M, Magalhaes JG, Shu Y, Philpott DJ, Arnoult D, Girardin SE. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JP, Davis BK. CATERPILLER: a novel gene family important in immunity, cell death, and diseases. Annu Rev Immunol. 2005;23:387–414. doi: 10.1146/annurev.immunol.23.021704.115616. [DOI] [PubMed] [Google Scholar]
- Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–290. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.