Abstract
A substantial proportion of the genome encodes membrane proteins that are delivered to the endoplasmic reticulum by dedicated targeting pathways1. Membrane proteins that fail targeting must be rapidly degraded to avoid aggregation and disruption of cytosolic protein homeostasis2,3. The mechanisms of mislocalized protein (MLP) degradation are unknown. Here, we reconstitute MLP degradation in vitro to identify factors involved in this pathway. We find that nascent membrane proteins tethered to ribosomes are not substrates for ubiquitination unless they are released into the cytosol. Their inappropriate release results in capture by the Bag6 complex, a recently identified ribosome-associating chaperone4. Bag6 complex capture depends on unprocessed or non-inserted hydrophobic domains that distinguish MLPs from potential cytosolic proteins. A subset of these Bag6 clients is transferred to TRC40 for membrane insertion, while the remainder are rapidly ubiquitinated. Depletion of the Bag6 complex impairs efficient ubiquitination selectively of MLPs. Thus, by its presence on ribosomes synthesizing nascent membrane proteins, the Bag6 complex links targeting and ubiquitination pathways. We propose that such coupling permits fast-tracking of MLPs for degradation without futile engagement of cytosolic folding machinery.
Protein targeting and translocation into the ER are not perfectly efficient5,6, thereby necessitating pathways to degrade MLPs released inappropriately into the cytosol. For example, mammalian prion protein (PrP), a widely expressed GPI-anchored cell surface glycoprotein, displays ~5–15% translocation failure in vitro, in cells, and in animals2,3,5–10. This non-translocated population of PrP is degraded efficiently by a proteasome-dependent pathway, limiting cytosolic PrP levels at steady state2,3,9,10. Prompt degradation is essential since mislocalized PrP can aggregate, make inappropriate interactions, and cause cell death and neurodegeneration2,11–14. The pathways for efficient disposal of MLPs are not known.
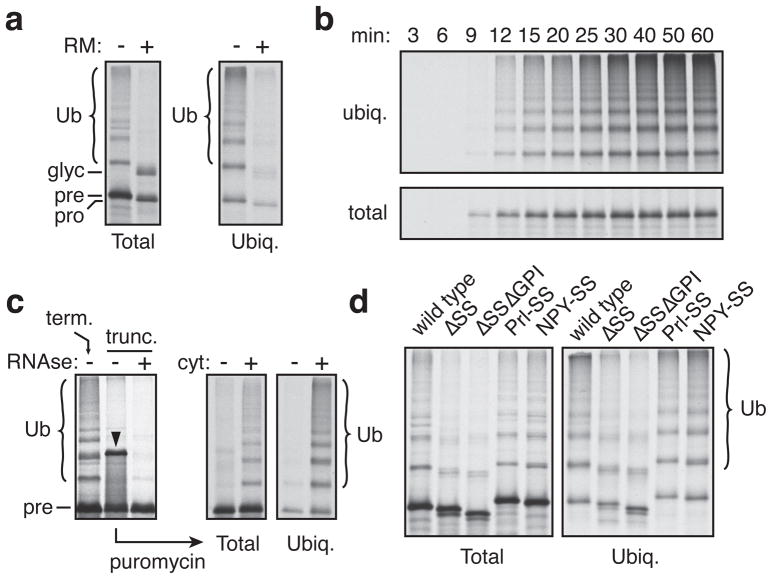
To study this problem, we reconstituted ubiquitination of mislocalized PrP in vitro. Radiolabeled PrP synthesized in rabbit reticulocyte lysate (RRL) supplemented with ER-derived rough microsomes (RMs) was predominantly translocated, processed and glycosylated (Fig. 1a). However, various conditions that reduced translocation, including omitting RMs, inactivating SRP-dependent targeting, or blocking translocation through the Sec61 translocon, all resulted in increased PrP ubiquitination in a lysine-dependent manner (Fig. 1a, Sup. Fig. S1–S3). Other mislocalized secretory and membrane proteins were also similarly ubiquitinated in the cytosol (Sup. Fig. S4). Ubiquitination of mislocalized PrP closely parallels PrP synthesis (Fig. 1b), suggesting that ubiquitination is rapid. Yet, ubiquitination occurred strictly post-translationally, since full length PrP tethered as a nascent peptidyl-tRNA to the ribosome was not ubiquitinated until released into the cytosol with puromycin (Fig. 1c, Sup. Fig. S5). An unrelated membrane protein behaved similarly (Sup. Fig. S6).
Fig. 1. Non-translocated PrP is rapidly ubiquitinated.
(a) PrP translations in reticulocyte lysate, without or with rough microsomes (RMs), were analyzed directly (left) or after isolation of ubiquitinated products (right). (b) Time course of PrP synthesis and ubiquitination in vitro. (c) PrP containing (‘term.’) or lacking (‘trunc.’) a termination codon was translated in vitro. Truncated PrP was released with puromycin without or with cytosol and ubiquitination analyzed. Arrowhead indicates tRNA-containing PrP, which can be digested by RNAse. (d) Wild type PrP or constructs lacking the signal sequence (ΔSS) or both the signal sequence and GPI anchor (ΔSSΔGPI) were analyzed for ubiquitination. Prl-SS and NYP-SS contain signals from Prolactin and Neuropeptide Y, respectively.
Efficient ubiquitination of PrP was strongly dependent on unprocessed hydrophobic signals at the N- and C-terminus (Fig. 1d). Conversely, GFP became a ubiquitination substrate when hydrophobic targeting signals were added (Sup. Fig. S4). Ubiquitination was therefore not solely a consequence of protein misfolding, since PrP lacking the N-terminal targeting signal (ΔSS) and C-terminal GPI-anchoring signal (ΔGPI) is still misfolded due to its lack of glycosylation and disulfide bond formation. This suggested a specialized pathway for hydrophobic domain-containing MLPs that works more rapidly than traditional quality control pathways that engage only after repeated failed folding attempts15,16.
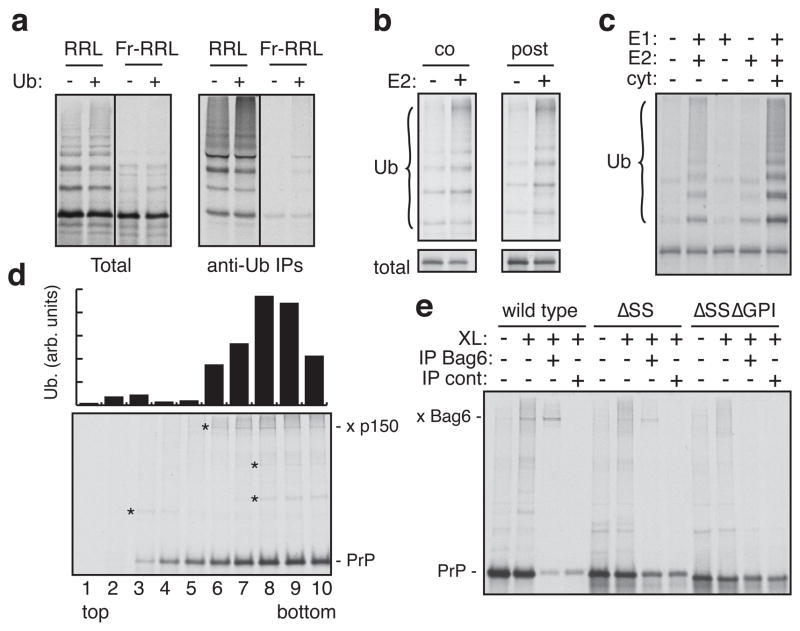
To identify factors involved in the MLP degradation pathway, we combined biochemical fractionation and functional reconstitution. We produced a translation-competent fractionated RRL (Fr-RRL; Sup. Fig. S7) with reduced ubiquitination selectively of non-translocated PrP (Fig. 2a) and other MLPs (Sup. Fig. S8), but not ubiquitination in general (Sup. Fig. S7). The missing factor in Fr-RRL (other than ubiquitin, which we included in all assays) proved to be the E2 enzyme UbcH5 (Fig. 2b, Sup. Fig. S8, S9). Because UbcH5 restored ubiquitination equally well when added after PrP translation (Fig. 2b), we surmised that at least some population of PrP remains in a ubiquitination-competent state. Indeed, PrP and other MLPs affinity purified from Fr-RRL under native conditions could be ubiquitinated on the beads simply by adding purified E1, UbcH5, ubiquitin, and ATP (Fig. 2c, Sup. Fig. S10).
Fig. 2. Bag6 interacts with MLPs via hydrophobic domains.
(a) PrP translated in RRL or Fr-RRL, without or with 10 uM ubiquitin, was analyzed directly (left) or after anti-ubiquitin immunoprecipitation (right). (b) PrP translated in Fr-RRL is ubiquitinated when UbcH5a (E2; 250 nM) is included co-translationally or added post-translationally. Total synthesis (bottom) and ubiquitinated products (top) are shown. (c) PrP was immunoaffinity purified under native conditions, and incubated with the indicated components (‘cyt’ is cytosol; E1 was at 100 nM; E2 was UbcH5a at 250 nM). All reactions contained His-ubiquitin and ATP. Purified ubiquitinated products are shown. (d) PrP translated in Fr-RRL was separated into 10 fractions on a 5–25% sucrose gradient and subjected to chemical crosslinking (bottom gel) or ubiquitination assays (top graph). Asterisks indicate crosslinks. (e) Crosslinking reactions of in vitro synthesized PrP or deletion constructs were analyzed directly or after immunoprecipitation.
To identify factors that maintain ubiquitination-competence of MLPs, the Fr-RRL translation products were separated by size on a sucrose gradient, and each fraction subjected to parallel ubiquitination and chemical crosslinking analyses (Fig. 2d; Sup. Fig. S11). The fractions retaining maximal ubiquitination-competence of two different substrates correlated well with a ~150 kD crosslinking partner (Fig. 2d, Sup. Fig. S11). This interaction was direct (Sup. Fig. S12) and strongly dependent on unprocessed N- and C-terminal signals on PrP (Fig. 2e, Sup. Fig. S13), correlating with requirements for ubiquitination (Fig. 1d). Based on molecular weight, dependence on hydrophobic domains for interaction, and migration position on the sucrose gradient, we surmised the ~150 kD crosslink might be Bag6 (also called Bat3 or Scythe), a hypothesis subsequently verified by immunoprecipitation (Fig. 2e, Sup. Fig. S13, S14). Bag6 was recently identified as part of a three-protein ribosome-interacting chaperone complex (composed of Bag6, TRC35, and Ubl4A)4 involved in tail-anchored (TA) membrane protein insertion into the ER4,17. A combination of crosslinking, affinity purification, and immunoblotting studies verified that all three subunits of this complex are associated with MLPs (Sup. Fig. S14, S15, and data not shown). Thus, the Bag6 complex binds multiple MLPs via their hydrophobic domains and has broader specificity than binding only TA proteins.
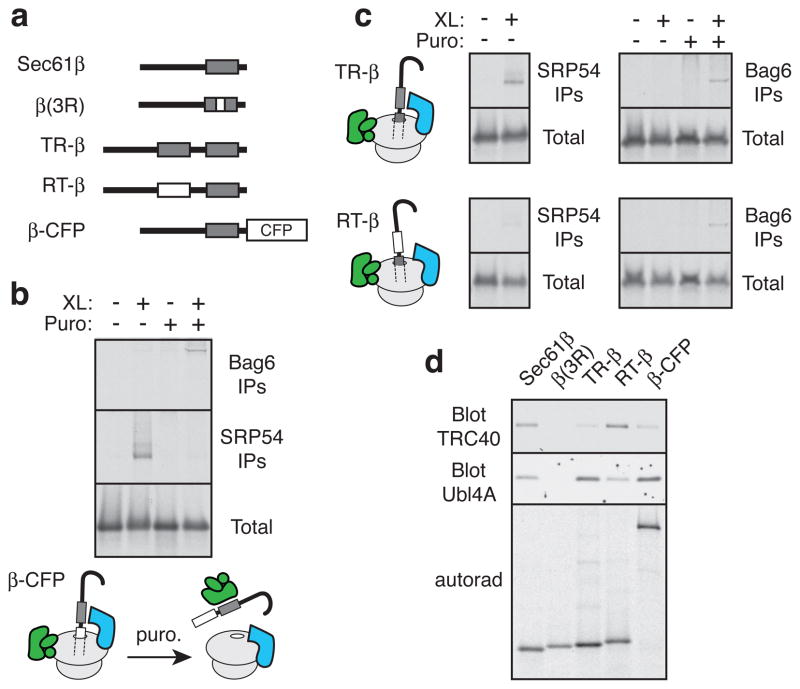
To understand when the Bag6 complex first captures MLPs, we analyzed ribosome-nascent chains (RNCs) synthesizing membrane proteins. When a transmembrane domain (TMD) emerges from the ribosomal tunnel, a direct interaction with SRP54 (the signal sequence binding subunit of SRP) could be detected by crosslinking (Fig. 3a–3c). By contrast, Bag6, even though it was found to reside on such RNCs and is abundantly present in the cytosol4, does not make direct contact with the substrate (Fig. 3b, 3c). When the TMD was still inside the ribosomal tunnel, the nascent chain was not crosslinked to either Bag6 or SRP54 (Fig. 3c), even though both complexes can be recruited to such ribosomes4,18. Upon release of each of these nascent chains from the ribosome with puromycin, Bag6 crosslinks were observed (Fig. 3b, 3c). Thus, the Bag6 complex captures substrates concomitant with or after ribosomal release of nascent chains; these same hydrophobic domains are bound by SRP as long as the TMD is exposed as a RNC19.
Fig. 3. Bag6 captures MLPs released from the ribosome.
(a) Diagram of constructs derived from Sec61β, with transmembrane domains shown as grey boxes and hydrophilic changes in white boxes. (b) RNCs of β-CFP with the TMD outside the ribosome were subjected to crosslinking before or after release with puromycin, and analyzed directly (bottom) or after immunoprecipitation with anti-Bag6 or anti-SRP54. Diagram of results; Bag6 complex is green, SRP is blue. (c) As in panel b, but using TR-β and RT-β in the top and bottom panels, respectively. (d) The indicated constructs were translated in vitro, immunoaffinity purified via the N-terminus, and immunoblotted with anti-TRC40 or anti-Ubl4A (to detect the Bag6 complex). Autoradiograph shows equal recovery of the translated substrates.
Earlier analysis of TA and non-TA membrane proteins had shown that only the former are efficiently loaded onto TRC40, the targeting factor for TA protein insertion into the ER20. Indeed, modifying a TA protein by either placing polypeptide sequences after the TMD (β-CFP; see Fig. 3a), or by adding an extra TMD (TR-β), reduced TRC40 interactions while simultaneously increasing interactions with the Bag6 complex (Fig. 3d). Similarly, comparison of the crosslinking partners of PrP and those of the TA protein Sec61β showed that both interact with the Bag6 complex, but only the latter is primarily found bound to TRC40 (Sup. Fig. S15). Given that TA protein loading onto TRC40 depends on the Bag6 complex4, these data suggest that the Bag6 complex is acting as a triage factor: it captures a relatively broad range of membrane proteins upon their ribosomal release, but transfers only a subset of them (i.e., TA proteins) to TRC40 for post-translational membrane insertion. The remainder are apparently targeted for ubiquitination by virtue of their persistent interaction with Bag6.
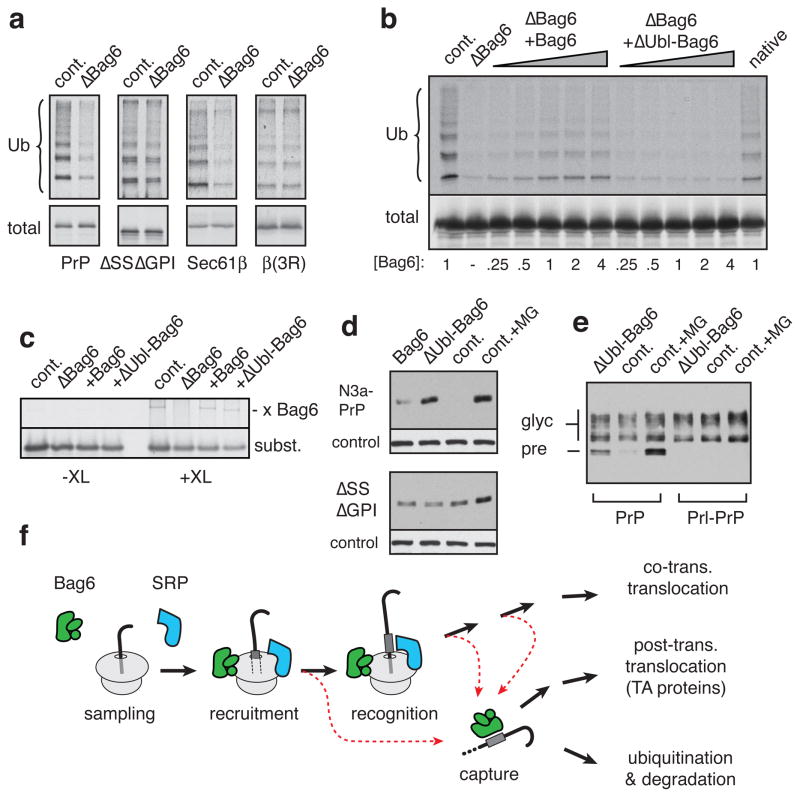
To illustrate this, we immunodepleted the Bag6 complex from RRL (Sup. Fig. S16) and showed that ubiquitination of several different MLPs is reduced (Fig. 4a; Sup. Fig. S17). By contrast, the control protein GFP was not ubiquitinated in RRL, but became a substrate when it was appended with either a ubiquitin (Ub-GFP) or any of several hydrophobic ER-targeting domains (Sup. Fig. S18). Only the latter were Bag6-dependent in their ubiquitination, consistent with their interaction with Bag6 by crosslinking analysis (Sup. Fig. S13). Conversely, ΔSSΔGPI-PrP, which does not interact with Bag6 (Fig. 2e), was ubiquitinated (albeit slowly and less efficiently) in a Bag6-independent manner (Fig. 4a). Disrupting the TMD of Sec61β with three Arginines (which disrupts Bag6 interaction4) also resulted in less ubiquitination, which was no longer Bag6-dependent (Fig. 4a). Thus, the Bag6 complex is not required for ubiquitination of misfolded proteins per se, but is especially important for efficient ubiquitination of MLPs.
Fig. 4. Maximal ubiquitination of MLPs requires Bag6.
(a) Various constructs were assayed for ubiquitination in lysates containing or lacking Bag6. The ubiquitin gels of ΔSSΔGPI and β(3R) were exposed ~3 times longer than PrP and Sec61β. (b) Bag6-depleted lysates were replenished with recombinant Bag6 (Sup. Fig. S16), ΔUbl-Bag6, or native Bag6 complex, and tested for ubiquitination of TR-β. Relative Bag6 levels are indicated. (c) TR-β interacts with recombinant Bag6 and ΔUbl-Bag6 by crosslinking. (d) The indicated PrP constructs were co-transfected with Bag6 complex, ΔUbl-Bag6 complex, or irrelevant plasmid (see Sup. Fig. S20) and detected by immunoblotting. One sample was treated with proteasome inhibitor (MG132) for 4 h. Loading control is also shown. (e) Effect of ΔUbl-Bag6 complex on wild type PrP and Prl-PrP. Unglycosylated precursor PrP is preferentially stabilized by either ΔUbl-Bag6 complex overexpression or proteasome inhibition. (f) Model: the Bag6 complex captures ribosomally released hydrophobic proteins and triages them between post-translational targeting (TA proteins) and ubiquitination.
Recombinant Bag6 (Sup. Fig. S16) added to translation extracts depleted of the Bag6 complex restored ubiquitination of a model MLP (Fig. 4b) and interacted with it in crosslinking assays (Fig. 4c). Bag6 lacking its N-terminal Ubl domain (ΔUbl-Bag6) was inactive in restoring ubiquitination (Fig. 4b) despite interacting normally with substrate (Fig. 4c). This suggested that Bag6 may recruit the ubiquitination machinery to substrates via its Ubl domain. To test this, FLAG-tagged recombinant Bag6 or ΔUbl-Bag6 was added to the Fr-RRL translation system lacking the UbcH5 E2 enzyme (Sup. Fig. S7). Bag6-substrate complexes were immunopurified via the FLAG tag and incubated with purified E1 and E2 enzymes, ubiquitin, and ATP. Substrate ubiquitination was observed with Bag6, but not ΔUbl-Bag6, verifying that the Ubl domain recruits ubiquitination machinery (Sup. Fig. S19) to the substrate. Indeed, Bag6 has been observed to interact with an E3 ubiquitin ligase via its Ubl domain21.
Fig. 4b and 4c indicate that ΔUbl-Bag6 should act as a dominant-negative and partially stabilize Bag6 substrates, providing a selective tool for in vivo analysis. Bag6 complex or ΔUbl-Bag6 complex was over-expressed (~2-fold; Sup. Fig. S20) in cultured cells and the levels of a co-expressed MLP substrate assessed. A translocation-impaired signal sequence mutant of PrP (termed N3a-PrP; ref. 5) was stabilized by ΔUbl-Bag6 complex, but hardly affected by wild type Bag6 complex (Fig. 4d). Importantly, ΔSSΔGPI-PrP, which does not interact with Bag6 (Fig. 2e), was unaffected by either Bag6 or ΔUbl-Bag6 overexpression (Fig. 4d), and showed higher steady state levels than N3a-PrP (data not shown). This suggests degradation by a different quality control pathway, consistent with its failure to be recognized as an MLP (Fig. 2e).
Wild type PrP, whose translocation is slightly inefficient in vivo2,3,6,8–10, showed preferential stabilization of a non-glycosylated species when co-overexpressed with ΔUbl-Bag6 complexes (Fig 4e; Sup. Fig. S21). This same species was stabilized by proteasome inhibition and shown in earlier studies to represent non-translocated PrP precursor2,3,9,10. Replacing the slightly inefficient PrP signal sequence with the efficient signal from Prolactin (Prl-PrP) precluded generation of non-glycosylated PrP with either proteasome inhibition or ΔUbl-Bag6 overexpression (Fig. 4e). Although the extent of stabilization seems modest, it is comparable to that seen with two hours of proteasome inhibition (Sup. Fig. S21). Partial knockdown of Bag6 with shRNA similarly stabilized a non-glycosylated species of PrP (Sup. Fig. S22). Thus, not only are MLPs generated in vivo2,3,6,8–10, but their maximally efficient degradation requires functional Bag6.
Our results reveal a pathway for MLP degradation and identify an unexpectedly close link with protein targeting (Fig. 4f). Ribosomes synthesizing nascent membrane proteins can recruit both SRP and Bag6 upon entry of the first hydrophobic segment into the ribosomal tunnel4,18. This is a potential targeting complex for the ER membrane in both the co-translational and post-translational membrane protein insertion pathways. We now find that such ribosomes are also potential degradation complexes since the first component of this degradation pathway is already poised to act in the event of failed targeting or inappropriate release from the ribosome. Bag6 therefore imposes a degradative fate for membrane proteins that is only avoided by productive targeting.
Because membrane proteins would never fold in the cytosol, their direct degradation by a specialized pathway may be important to avoid unnecessarily occupying essential cellular folding pathways, particularly under conditions of stress. MLPs are distinguished from nascent cytosolic proteins by relatively long linear hydrophobic stretches, a feature that is key to Bag6 recognition. Indeed, mutagenesis shows that even modest reductions of TMD hydrophobicity sharply curtail Bag6 interaction4. This distinguishes Bag6 from more general chaperones like Hsp70, whose substrate binding pocket seems more suited to shorter and moderately hydrophobic segments that typify nascent cytosolic proteins. This differential specificity probably explains how MLPs are triaged differently than other potential substrates of cytosolic quality control15,16,22–28. These pathways could intersect or cooperate in still undefined ways given that Bag6 and Hsp70 were observed to co-immunoprecipitate26.
In addition to this degradation role, the Bag6 complex also facilitates loading of TA proteins onto TRC40 for post-translational insertion into the ER4. As expected, TA proteins are also ubiquitinated via Bag6 in the absence of or saturation of TRC40 (Sup. Fig. S23). Thus, substrates of both the co- and post-translational targeting pathways are ubiquitinated via Bag6 when targeting fails. After ubiquitination, Bag6 could chaperone its polyubiquitinated substrates to the proteasome, a function recently proposed on the basis of co-immunoprecipitation with Bag6 of polyubiquitinated proteins26. The Bag6 complex is therefore a multi-purpose triage factor for chaperoning especially aggregation-prone hydrophobic proteins through the aqueous cytosol. This view would conceptually link its roles in TA protein targeting4,17, the MLP pathway (this study), a chaperone for newly dislocated proteins during ERAD27,28, and delivery of terminally misfolded proteins to the proteasome26.
Methods Summary
Reagents and standard methods
Plasmids, antibodies, in vitro translations, sucrose gradients, chemical crosslinking, immunoprecipitations, and immunodepletions have been described2–8,14,20,29,30. Pulldowns with Co+2 immobilized on chelating sepharose were performed on samples denatured in boiling 1% SDS, followed by 10-fold dilution into cold 0.5% Triton X-100, 25 mM Hepes, 100 mM NaCl, and 10 mM imidazole. Culture, transfection, and immunoblot analysis of N2a cells (dominant-negative inhibition experiments) and Hela cells (for shRNA experiments) were as before2,3. Full length Bag6 (or ΔUbl-Bag6, lacking residues 15-89) tagged at the C-terminus with a FLAG epitope was overexpressed by transient transfection into HEK-293T cells and purified with anti-FLAG resin under high salt (400 mM KAc).
Modified translation extracts
Fr-RRL contained native ribosomes (isolated from RRL) mixed with a DEAE-elution fraction prepared from ribosome-free RRL (Sup. Fig. S7). Fr-RRL was adjusted to the following final conditions for translation: 72 mM KAc, 2.5 mM MgAc2, 10 mM Hepes, pH 7.4, 2 mM DTT, 0.2 mg/ml liver tRNA, 1 mM ATP, 1 mM GTP, 12 mM creatine phosphate, 40 ug/ml creatine kinase, 40 uM each amino acid (except Methionine), and 1 uCi/ul 35S-Methionine.
Ubiquitination assays
For full length proteins, translations containing 10 uM His-ubiquitin were for 1 h at 32°C. In Fr-RRL, post-translational ubiquitination was initiated by adding E2 enzyme to 250 nM and incubation for 1 h. For RNCs, samples were supplemented with E1 enzyme (85 nM), E2 enzyme (usually 250 or 500 nM), cytosol (RRL or Fr-RRL), 10 uM His-ubiquitin, ATP regenerating system (1 mM ATP, 10 mM creatine phosphate, 40 ug/ml creatine kinase), and 1 mM puromycin. Reaction conditions were 100 mM KAc, 50 mM Hepes, pH 7.4, 5 mM MgCl2, and 1 mM DTT. Incubation was for 1 h at 32°C. On-bead ubiquitination of affinity purified products was the same, except without puromycin.
Methods
Plasmids and antibodies
The SP64 vector-based constructs encoding bovine preprolactin, PrP, ΔSS-PrP (lacking residues 2-22), ΔSSΔGPI-PrP (additionally lacking residues 232-254), and HA-tagged PrP (with the epitope inserted at codon 50) have been characterized,3,5,29–32. Prl-PrP and NPY-PrP encode versions in which the N-terminal signal sequence (residues 1-22) of PrP was replaced5 with that of either bovine preprolactin or human neuropeptide Y. N3a-PrP contains a mutated signal sequence (WL → DD at residues 7 and 8) that is translocation deficient5. The lysine-free version of PrP was provided by C. Ott and made by standard mutagenesis methods. Wild type Sec61β (appended at the C-terminus with an epitope recognized by the 3F4 antibody), Sec61β(3R), Sec61β-CFP and CFP-Sec61β have been described4,20. Sec61β-TR (referred to as TR-β in the text and figures) contains the TMD of human transferrin receptor (IAVIVFFLIGFMIGYLGY) at codon 50 in the cytosolic domain of Sec61β4. This positions the TMD outside the ribosomal tunnel when the Sec61β TMD is inside the tunnel4. RT-β contains an irrelevant hydrophilic sequence (YPKYPIMNPIKKKTITAI) at the same position4. GFP, SS/GPI-GFP (containing the N-terminal signal sequence of bovine preprolactin and C-terminal GPI anchoring sequence of PrP), ManII-GFP (containing the N-terminal type II signal anchor domain of Golgi α-Mannosidase II), and SiT-GFP (containing the type II signal anchor domain of sialyl transferase) have been described32–34. The plasmid encoding Vpu (a type I signal anchored membrane protein from HIV-1), and was obtained from J. Bonifacino and J. Magadan35. An expression plasmid for bovine Rhodopsin has been characterized29. For translations of full-length products, the open reading frames were PCR amplified using a forward 5′ primer annealing to or encoding an SP6 or T7 promoter, and a reverse primer in the 3′ UTR at least 100 nucleotides beyond the stop codon. For RNCs, the reverse primer annealed in the coding region and lacked a stop codon. PrP and Vpu RNCs included the entire open reading frame except the stop codon. RNCs of β-CFP encoded 46 residues beyond the TMD such that it would have fully emerged from the ribosome. Similarly, the RNCs of TR- β and RT- β encoded up to and including the TMD of Sec61β such that the TR and RT sequences had emerged from the ribosome. Bag6-FLAG and ΔUbl-Bag6-FLAG (lacking residues 15-89 of Bag6) encoding human Bag6 containing a C-terminal FLAG epitope were subcloned into a mammalian expression vector by standard methods. Expression vectors for human TRC35 and Ubl4A containing C-terminal FLAG tags were obtained from Origene. Expression vectors for shRNAs directed against human Bag6 were from Origene. The target sequences were TGACGGCTCTGCTGTGGATGTTCACATCA and CAGCTATGTCATGGTTGGAACCTTCAATC. The irrelevant sequence used as a control was GCACTACCAGAGCTAACTCAGATAGTACT. Antibodies to Bag6, TRC40, TRC35, Ubl4A, and Sec61β have been described4,36. Anti-SRP54 (BD Biosciences), anti-Ubiquitin (BioMol), and 3F4 anti-PrP monoclonal (Signet) were purchased.
In vitro translation
In vitro transcription and translation in RRL was with minor modifications of published procedures30. The most notable change was the inclusion in most experiments of 10 uM His-tagged ubiquitin (Boston Biochem) to facilitate subsequent isolation of ubiquitinated products. Preliminary experiments showed that at this concentration, endogenous ubiquitin was more than 90% competed, resulting in little or no untagged ubiquitinated products. Translation times, unless otherwise indicated, were for 1 h at 32°C. Shorter times for TA proteins (as used in our earlier studies) resulted in very little ubiquitination4,20, presumably because saturation of TRC40 is required before substrates occupy the Bag6 complex4. To generate RNCs, translation times were typically reduced to 30 min to minimize spontaneous release or hydrolysis of the tRNA. Translocation assays into RMs5, inhibition by cotransin29, and inactivation with NEM37 treatment was as before. For direct analysis or downstream immunoprecipitation, translation reactions were stopped and the proteins denatured using 1% SDS and heating to 100°C. For other applications requiring native complexes (e.g., crosslinking, affinity purification, or downstream assays), samples were placed on ice and subsequent manipulations performed at 0–4°C.
Sucrose gradient separation and crosslinking
To generate RNCs, translation reactions (typically 200 ul volume) were chilled on ice and immediately layered onto 2 ml 10–50% sucrose gradients in physiologic salt buffer (PSB: 100 mM KAc, 50 mM Hepes, pH 7.4, 2 mM MgAc2). Centrifugation was for 1 h at 55,000 rpm at 4°C in the TLS-55 rotor (Beckman), after which 200 ul fractions were removed from the top. The peak ribosomal fractions (6 and 7) were pooled, and used as the RNCs. These were used immediately or flash frozen in liquid N2 for later use in RNC crosslinking or ubiquitination experiments. Chemical crosslinking experiments were essentially as described4,20. Chilled translation reactions were layered onto 2 ml 5–25% sucrose gradients in PSB and centrifuged for 5 h at 55,000 rpm at 4°C in the TLS-55 rotor (Beckman), after which 200 ul fractions were removed from the top. Crosslinking employed 250 uM of BMH, except in experiments to detect SRP interaction, which employed 200 uM DSS. Reactions were for 30 min at either 0°C (BMH) or 25°C (DSS), and quenched with 25 mM 2-mercaptoethanol (BMH) or 100 mM Tris (DSS). The samples were subsequently denatured and subjected to direct analysis or immunoprecipitation as described below. Photocrosslinking employed published methods38, except that we used the Fr-RRL system for translation and benzophenone-modified lysyl-tRNA (from tRNA Probes). The absence of endogenous charged tRNAs and Hemoglobin increased photocrosslinker incorporation and photolysis, respectively. Photolysis was for 15 min on ice, and the samples were analyzed directly.
Modified translation extracts
Fr-RRL was typically prepared from 25 mls RRL (from Green Hectares) that had first been treated with Hemin and micrococcal nuclease. Its characterization will be described in a future publication, but its preparation is as follows. All procedures were on ice or at 4°C. The lysate was centrifuged at 100,000 rpm for 40 min in the TLA100.4 rotor (Beckman). The supernatants were pooled, and the tubes rinsed (without disrupting the ribosomal pellet) with an equal volume of column buffer (20 mM Tris, pH 7.5, 20 mM KCl, 0.1 mM EDTA, 10% glycerol), which was added to the supernatant. The pellet was resuspended by dounce homogenization in ribosome wash buffer (RWB: 20 mM Hepes, pH 7.5, 100 mM KAc, 1.5 mM MgAc2, 0.1 mM EDTA), layered onto a 1 M sucrose cushion in RWB, and re-isolated by centrifugation at 100,000 rpm for 1 h in the TLA100.4 rotor. The final pellet was resuspended in one-tenth the original lysate volume, and defined as native ribosomes. The ribosome-free supernatant from above was applied to a 10 ml DEAE column at a flow rate of ~1 ml/min, and washed with column buffer until the red hemoglobin was removed (~50 mls). The washed column was eluted in a single step with 50 mls of column buffer containing 300 mM KCl. The eluate was adjusted slowly with solid ammonium sulfate to 75% saturation (at 4°C) with constant stirring. After 1 h of mixing, the precipitate was recovered by centrifugation at 15,000 rpm in the JA-17 rotor. The supernatant was discarded and the pellet dissolved in a minimal volume (~8 mls) of dialysis buffer (20 mM Hepes, pH 7.4, 100 mM KAc, 1.5 mM MgAc2, 10 % glycerol, 1 mM DTT). This was dialyzed against two changes of dialysis buffer overnight, recovered, adjusted to 10–12 mls (i.e., twice the original concentration), and flash-frozen in N2. To make a translation competent Fr-RRL, the native ribosomes and dialyzed DEAE-eluate were adjusted to 72 mM KAc, 2.5 mM MgAc2, 10 mM Hepes, pH 7.4, 2 mM DTT, 0.2 mg/ml liver tRNA, 1 mM ATP, 1 mM GTP, 12 mM creatine phosphate, 40 ug/ml creatine kinase, 40 uM each amino acid (except Methionine), and 1 uCi/ul 35S-Methionine. The concentration of ribosomes and lysate was the same as that of RRL. Immunodepletions of RRL were as before4.
Ubiquitination assays
The human E1 conjugating enzyme and all mammalian E2 enzymes were obtained from Boston Biochem. For full length proteins, translations containing 10 uM His-ubiquitin were for 1 h at 32°C. In Fr-RRL, post-translational ubiquitination was initiated by adding E2 enzyme to 250 nM and further incubation for 1 h. For RNCs, samples were supplemented as indicated in the figures with E1 ubiquitin activating enzyme (85 nM), E2 enzyme (usually 250 or 500 nM), cytosol (RRL or Fr-RRL, at the same concentration as in translations), 10 uM His-ubiquitin, ATP regenerating system (1 mM ATP, 10 mM creatine phosphate, 40 ug/ml creatine kinase), and 1 mM puromycin. Reaction conditions were 100 mM KAc, 50 mM Hepes, pH 7.4, 5 mM MgCl2, and 1 mM DTT. Incubation was for 1 h at 32°C. On-bead ubiquitination of affinity purified products was the same, except without puromycin. To prepare the affinity purified substrate, translation reactions in Fr-RRL were chilled on ice, diluted to 1 ml in PSB, and incubated with immobilized antibodies against the HA-epitope (for PrP-HA and Vpu-HA) or Sec61β. In Sup. Fig. S19, the translation reactions were supplemented with FLAG-tagged Bag6 or ΔUbl-Bag6 (each added to 2-fold excess above endogenous Bag6 levels), and anti-FLAG beads (Sigma) were used for the pulldown. After 1 h, the resin was washed five times in PSB, and residual buffer carefully removed before adding the ubiquitination components as above. The reaction was incubated with constant low-level shaking (in the Eppendorf Thermomixer) at 32°C for 1 h. 1% SDS was added directly to the reactions, which were analyzed directly as well as after ubiquitin pulldowns.
Cell culture studies
Culture, transfection, and immunoblot analysis of N2a cells (dominant-negative inhibition experiments) and Hela cells (for shRNA experiments) were as before2,3. Cells were seeded in 24-well dishes the day prior to transfection. For the dominant-negative experiments, the plasmids were mixed in the ratios as indicated in Sup. Fig. S20 and transfected using Lipofectamine 2000 as directed by the supplier (Invitrogen). 24 h after transfection, the cells were harvested in 1% SDS, the DNA sheared by vortexing and boiling, and the total sample analyzed by SDS-PAGE and immunoblotting. For shRNA experiments, each well received a mixture of 550 ng of the shRNA plasmid, 200 ng of the PrP expression plasmid, and 50 ng of CFP expression plasmid. Transfection was with Lipofectamine 2000. Examination of CFP fluorescence verified at least 50% transfection efficiency. The cells were cultured for ~100 h before harvesting and analysis by immunoblotting.
Bag6 purification
Full length Bag6 or ΔUbl-Bag6 tagged at the C-terminus with a FLAG epitope was overexpressed by transient transfection into HEK-293T cells. The Trans-IT reagent (Mirus) was used, and after three days of expression, the cells harvested in 50 mM Hepes, pH 7.4, 150 mM KAc, 5 mM MgAc2, and 1% Deoxy-BigCHAP. The soluble extract was incubated with immobilized anti-FLAG antibodies (Sigma) with constant mixing, and the resin was washed four times in high salt lysis buffer containing 400 mM KAc, and twice with detergent-free lysis buffer containing 230 mM KAc. Elution was carried out with 1 mg/ml competing peptide at room temperature. The final protein was checked by colloidal coomassie blue (Sup. Fig. S16), and its concentration relative to that in RRL determined by immunoblotting of serial dilutions. Blotting also confirmed lack of TRC35 and Ubl4A in Bag6 prepared by this method.
Miscellaneous biochemistry
Immunoprecipitation was as before5,36. Pulldowns with Co+2 immobilized on chelating sepharose were performed on samples denatured in boiling 1% SDS, followed by 10-fold dilution into cold 0.5% Triton X-100, 25 mM Hepes, 100 mM NaCl, and 10 mM imidazole. The complete denaturation step is essential for samples containing RRL because the Hemoglobin is a strong Co+2-binding protein in its native state. Typically, 10 ul of packed resin was used per sample, and after incubation for 1–2 h at 4°C, the resin was washed three times in the above buffer, and eluted in SDS-PAGE sample buffer containing 20 mM EDTA. SDS-PAGE was on 8.5% or 12% Tricine gels. Figures were prepared using Adobe Photoshop and Illustrator.
Supplementary Material
Acknowledgments
We are grateful to E. Whiteman and X. Li for initial experiments on parts of this project, S.W. Kang, S. Shao, and Z. Zhang for useful discussions, P. Sengupta, J. Magadan, and C. Ott for constructs, J. Taunton and J. Garrison for cotransin, S. Shao for comments on this manuscript, and Y. Ye for useful discussions and sharing results prior to publication. This work was supported by the Intramural Research Program of the National Institutes of Health (RSH) and a postdoctoral fellowship from The Wenner-Gren Foundations (TH).
Footnotes
Supplementary Information consisting of Technical Notes and 23 figures accompanies the paper.
Author contributions – TH performed most experiments, with contributions from AS (ubiquitination assays in modified lysates), MM (defining Bag6 substrate-specificity), HE (characterization of the Fr-RRL system), EG (Bag6 crosslinking analysis), and RSH (in vivo studies). RSH conceived the project, guided experiments, and wrote the paper with input from all authors.
The authors are not aware of any competing interests. Reprints and permissions information is available at www.nature.com/reprints
References
- 1.Cross BC, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- 2.Rane NS, Yonkovich JL, Hegde RS. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang SW, et al. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariappan M, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SJ, Mitra D, Salerno JR, Hegde RS. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- 6.Levine CG, Mitra D, Sharma A, Smith CL, Hegde RS. The efficiency of protein compartmentalization into the secretory pathway. Mol Biol Cell. 2005;16:279–291. doi: 10.1091/mbc.E04-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SJ, Hegde RS. Cotranslational partitioning of nascent prion protein into multiple populations at the translocation channel. Mol Biol Cell. 2002;13:3775–3786. doi: 10.1091/mbc.E02-05-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rane NS, Chakrabarti O, Feigenbaum L, Hegde RS. Signal sequence insufficiency contributes to neurodegeneration caused by transmembrane prion protein. J Cell Biol. 2010;188:515–526. doi: 10.1083/jcb.200911115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orsi A, Fioriti L, Chiesa R, Sitia R. Conditions of endoplasmic reticulum stress favor the accumulation of cytosolic prion protein. J Biol Chem. 2006;281:30431–30438. doi: 10.1074/jbc.M605320200. [DOI] [PubMed] [Google Scholar]
- 10.Drisaldi B, et al. Mutant PrP is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP undergoes retrotranslocation prior to proteasomal degradation. J Biol Chem. 2003;278:21732–21743. doi: 10.1074/jbc.M213247200. [DOI] [PubMed] [Google Scholar]
- 11.Ma J, Lindquist S. Conversion of PrP to a self-perpetuating PrPSc-like conformation in the cytosol. Science. 2002;298:1785–1788. doi: 10.1126/science.1073619. [DOI] [PubMed] [Google Scholar]
- 12.Chakrabarti O, Hegde RS. Functional depletion of mahogunin by cytosolically exposed prion protein contributes to neurodegeneration. Cell. 2009;137:1136–1147. doi: 10.1016/j.cell.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma J, Wollmann R, Lindquist S. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science. 2002;298:1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- 14.Rane NS, Kang SW, Chakrabarti O, Feigenbaum L, Hegde RS. Reduced translocation of nascent prion protein during ER stress contributes to neurodegeneration. Dev Cell. 2008;15:359–370. doi: 10.1016/j.devcel.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 16.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leznicki P, Clancy A, Schwappach B, High S. Bat3 promotes the membrane integration of tail-anchored proteins. J Cell Sci. 2010;123:2170–2178. doi: 10.1242/jcs.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berndt U, Oellerer S, Zhang Y, Johnson AE, Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc Natl Acad Sci U S A. 2009;106:1398–1403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 20.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 21.Lehner B, et al. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics. 2004;83:153–167. doi: 10.1016/s0888-7543(03)00235-0. [DOI] [PubMed] [Google Scholar]
- 22.Park SH, et al. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisele F, Wolf DH. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nillegoda NB, et al. Ubr1 and ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell. 2010;21:2102–2116. doi: 10.1091/mbc.E10-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minami R, et al. BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol. 2010;190:637–650. doi: 10.1083/jcb.200908092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst R, et al. Enzymatic blockade of the ubiquitin-proteasome pathway. PLoS Biol. 2011;8:e1000605. doi: 10.1371/journal.pbio.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, et al. A chaperone holdase maintains polypeptides in soluble states for proteasome degradation. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrison JL, Kunkel EJ, Hegde RS, Taunton J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature. 2005;436:285–289. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- 30.Sharma A, Mariappan M, Appathurai S, Hegde RS. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol Biol. 2010;619:339–363. doi: 10.1007/978-1-60327-412-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emerman AB, Zhang ZR, Chakrabarti O, Hegde RS. Compartment-restricted Biotinylation Reveals Novel Features of Prion Protein Metabolism In Vivo. Mol Biol Cell. 2010 doi: 10.1091/mbc.E10-09-0742. doi:10.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashok A, Hegde RS. Retrotranslocation of prion proteins from the endoplasmic reticulum by preventing GPI signal transamidation. Mol Biol Cell. 2008;19:3463–3476. doi: 10.1091/mbc.E08-01-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole NB, et al. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- 34.Wu MM, et al. Organelle pH studies using targeted avidin and fluorescein-biotin. Chem Biol. 2000;7:197–209. doi: 10.1016/s1074-5521(00)00088-0. [DOI] [PubMed] [Google Scholar]
- 35.Magadan JG, et al. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 2010;6:e1000869. doi: 10.1371/journal.ppat.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krieg UC, Walter P, Johnson AE. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc Natl Acad Sci U S A. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.