Abstract
The neuron-specific isoform of the TAF1 gene (N-TAF1) is thought to be involved in the pathogenesis of DYT3 dystonia, which leads to progressive neurodegeneration in the striatum. To determine the expression pattern of N-TAF1 transcripts, we developed a specific monoclonal antibody against the N-TAF1 protein. Here we show that in the rat brain, N-TAF1 protein appears as a nuclear protein within subsets of neurons in multiple brain regions. Of particular interest is that in the striatum, the nuclei possessing N-TAF1 protein are largely within medium spiny neurons, and they are distributed preferentially, though not exclusively, in the striosome compartment. The compartmental preference and cell type-selective distribution of N-TAF1 protein in the striatum are strikingly similar to the patterns of neuronal loss in the striatum of DYT3 patients. Our findings suggest that the distribution of N-TAF1 protein could represent a key molecular characteristic contributing to the pattern of striatal degeneration in DYT3 dystonia.
Keywords: TAF1, DYT3 dystonia, Striosome, Neurodegeneration, Transcription dysregulation syndrome
INTRODUCTION
Among the monogenic primary dystonias (Müller, 2009), DYT3 dystonia, also named X-linked dystonia-parkinsonism (XDP/DYT3, MIM314250), is the result of disrupted alternative splicing regulation. A series of linkage analyses (Haberhausen et al., 1995; Nolte et al., 2003) identified the disease locus of the DYT3 gene as Xq13.1, including TAF1 [TATA box-binding protein (TBP) associated factor 1], formerly called TAFII250. TAF1 is the largest subunit of the transcription factor IID complex (TFIID), which is composed of TBP and thirteen different TAFs. TAF1 appears to function as a major scaffold by which TBP and other TAFs interact in the assembly of TFIID. TAF1 is an essential component of the transcription machinery and is known to be a key regulator for RNA polymerase II (RNAPII)-dependent gene transcription that involves conversion of cellular signals provided by gene-specific activator proteins into the synthesis of mRNA (Wassarman and Sauer, 2001). Makino et al. (2007) recently reported that the TAF1 gene is the causative gene of DYT3 dystonia and showed that there is a specific reduction of the neuron-specific isoform of the TAF1 gene (N-TAF1) in DYT3 patients. DYT3 dystonia thus can be classified as one of the neurodegenerative disorders associated with transcriptional dysregulation.
DYT3 dystonia is clinically characterized by an adult-onset movement disorder that manifests progressive and severe dystonia followed by overt parkinsonism in the later years of life (Lee et al., 2002). The neuropathology of DYT3 (Goto et al., 2005) involves a primary and progressive degeneration of medium spiny neurons (MSNs) in the striatum, with sparing of large cholinergic neurons. There is also differential neurodegeneration in the striatal compartments, judging by immunostaining, with a predominant loss of neurons in striosomes relative to the surrounding matrix compartment in the DYT3 striatum. How such impaired transcription is related to the neurodegeneration and symptoms characteristic of DYT3 dystonia is still not well understood.
As a first step to addressing this issue, we generated a monoclonal antibody (mAb) that specifically recognizes N-TAF1 protein for the purpose of studying the spatial distribution of N-TAF1 protein in the brain. The antibody successfully stained neurons expressing N-TAF1 in rat brains, and here we show that N-TAF1 is located exclusively in the nuclei of neurons in multiple brain regions. Notably, within the striatum, N-TAF1-positive (N-TAF1+) nuclei were largely within neurons of the medium spiny type, and there was a marked tendency of these N-TAF1-positive neurons to lie in the striosomal compartment in the striatum. As striosomes may be a primary site of neurodegeneration early in the progression of neuropathology in DYT3 patients (Goto et al., 2005), these data suggest a link between N-TAF1 expression and specific patterns of neuropathology in DYT3 dystonia.
EXPERIMENTAL PROCEDUES
All procedures involving the use of animals and analysis of brain anatomy were approved by the Institutional Care and Use Committees of the University of Tokushima and Shiga University of Medicines.
Production of antibodies
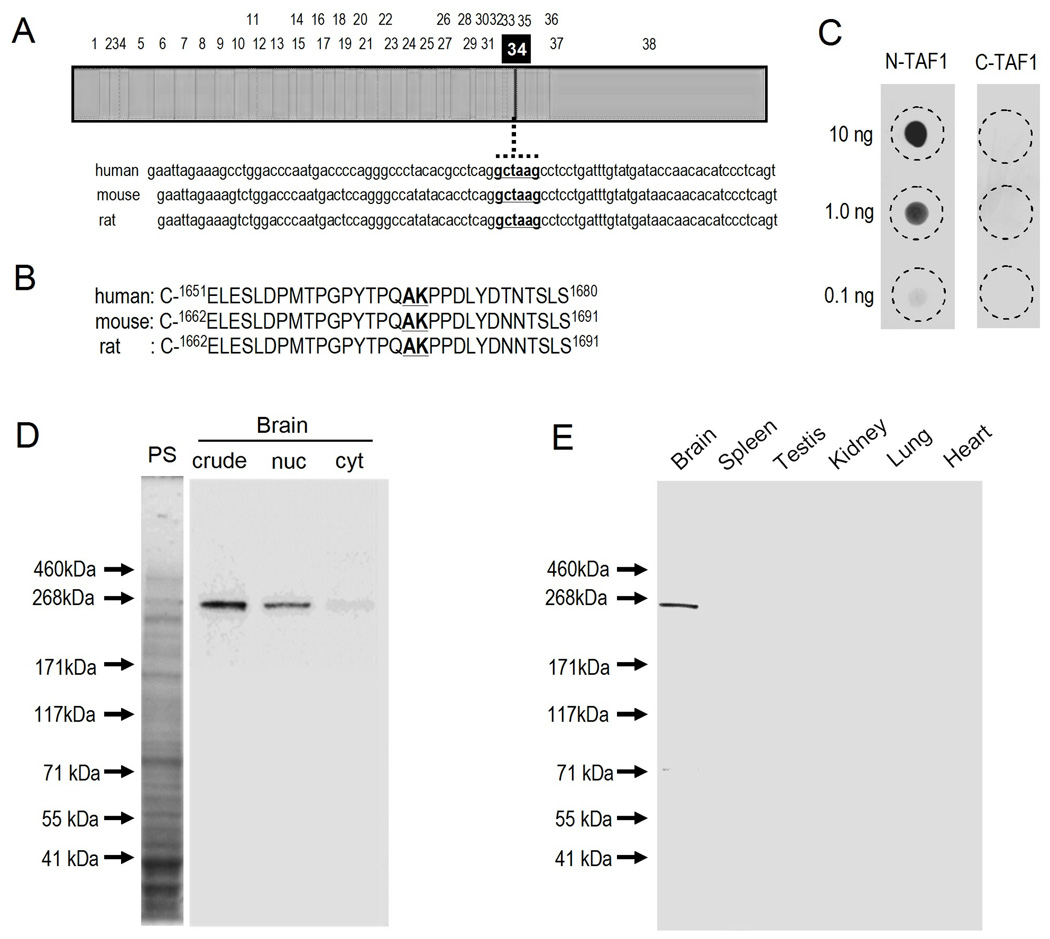
Sequence data were used for antibody production. Fig. 1A illustrates the nucleotide sequence of a full-length cDNA for N-TAF1 and that of neighboring exon 34’. Based on a comparison of the amino acid sequence of N-TAF1 (GenBank, accession number AB300418) to that of the common form of TAF1 (C-TAF1) (GenBank, accession number NM_004606), the N-TAF1-peptide for immunization was chosen and synthesized as C-1659TPGPYTPQAKPPDLY1673, for which the residues between E-1651 and S-1680 of humans are highly conservative across species (Fig. 1B). This epitope peptide corresponds to the peptide for C-TAF1 (C-1659TPGPYTPQPPDLY1671) with an insertion of 2 amino acids (Ala-Lys) between Q-1666 and P-1667. N-TAF1- and C-TAF1-synthetic peptides were chemically conjugated to the carrier protein keyhole limpet hemocyanin (KLH) and bovine serum albumin (BSA), respectively. The KLH-conjugated N-TAF1-peptide was used to produce mouse mAbs against N-TAF1 by hybridoma technology. Among 4 positive hybridoma cell lines identified by screening and purified by limiting dilution and single-cell cloning, we chose mAb-3A11F for this study.
Fig 1. Production and characterization of monoclonal antibody against N-TAF1 protein.
(A) Nucleotide sequence neighboring exon 34’ in a full-length cDNA for N-TAF1.
(B) The amino acid residues neighboring exon 34’ of human, mouse, and rat.
(C) Dot blot assay. Indicated amounts of KLH-conjugated N-TAF1-peptide (N-TAF1) or BSA-conjugated C-TAF1-peptide (C-TAF1) were spotted onto a nitrocellulose membrane and detected by dot immunobinding assay with anti-N-TAF1 antibody mAb-3A11F.
(D) Western blot assay on the brain extracts. Crude homogenates (20 µg of protein) extracted from a rat brain (crude) were loaded onto 10% SDS-PAGE and then processed for the transimmunoblot technique using mAb-3A11F. Nuclear (nuc) and cytosol (cyt) fractions (20 µg of protein) of a rat brain were also processed for the transimmunoblot. PS, protein staining.
(E) Western blot assay on the multiple organ extracts. Crude homogenates (20 µg of protein) extracted from a rat brain, spleen, testis, kidney, lung, and heart were loaded onto 10% SDS-PAGE and then processed for the transimmunoblot technique using mAb-3A11F.
Dot blot assay
Peptide samples were spotted onto nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CL). The air-dried membranes were then blocked with 5% skim milk in 0.01 M Tris-buffered saline containing 0.1% Tween 20 and were incubated overnight at room temperature with anti-N-TAF1 antibody mAb-3A11F (0.2 µg/ml). We detected the bound antibodies using the Elite ABC kit (Vector Laboratory, Burlingame, CA) with diaminobenzidine (0.6 mg/ml), H2O2 (0.001%), and (NH4)2Ni(SO4)2·6H2O (2.5 mg/ml).
Western blot assay
Adult Sprague-Dawley rats (250–300 g, 63–70 days old) were used. The animals were deeply anaesthetized with a lethal intraperitoneal dose of pentobarbital and were perfused transcardially with 0.9% saline in 0.01 M phosphate buffer, pH 7.2 (PBS). Protein lysates extracted from the brains and other organs were obtained according to the method described previously (Sako et al., 2010). To prepare subcellular fractionations, brain tissues were homogenized in 0.05 M Tris-HCl, pH 7.2 containing 0.025 M KCl, 0.005 M MgCl2, and 0.32 M sucrose. The crude brain homogenate was then centrifuged at 600 × g for 10 min to obtain the nuclear and cytosol fractions. The precipitated pellet was the nuclear fraction and sonicated in ultrapure distilled water containing Complete Mini (Roche Applied Science, Basel, Switzerland). Protein lysates (20 µg of protein) were subjected to 10% SDS-PAE and proteins were then transferred to a polyvinylidene difluoride membrane. The membranes were incubated with anti-N-TAF1 antibody mAb-3A11F (0.02 µg/ml) and Can Get Signal (Toyobo, Osaka, Japan) and were then with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG. Bound antibodies were detected by chemiluminescence staining (ECL plus kit, GE Healthcare, Buckingham, UK).
Tissue preparation for histology
Deeply pentobarbital-anaesthetized adult Sprague-Dawley rats (250–300g) were perfused transcardially with PBS followed by cold 4% paraformaldehyde in 0.1 M PBS, pH 7.2. The brains were removed, post-fixed overnight in the same fixative at 4°C, and cut on a vibratome (Leica, Germany). Sections were stored in cold PBS containing 0.01% NaN3 until use.
Immunostaining for N-TAF1
Immunohistochemical labeling was done as previously reported (Sato et al., 2008; Sako et al., 2010). Free-floating 25 µm-thick sections were incubated with mAb-3A11F (0.02 µg/ml) overnight at room temperature. Bound antibodies were detected by the Elite ABC protocol. The final peroxidase reaction was developed for 2 hr at 25°C in 0.01 M Tris-buffered saline containing DAB (0.6 mg/ml), H2O2 (0.001%), and (NH4)2Ni(SO4)2·6H2O (2.5 mg/ml). For immunofluorescence staining, sections were incubated with mAb-3A11F (0.002 µg/ml) overnight at room temperature. Bound antibodies were detected by the Histofine Simple Stain Kit (Nichirei, Tokyo, Japan) and Tyramide Signal Amplification system with Cyanine3 (Perkin Elmer, Shelton, CT). Controls included omission of primary antibodies and preabsorption of antibodies with excess of KLH-conjugated N-TAF1-peptide. The preabsorption examination was carried out with the ABC protocol as follows. Before application of the antibody to the sections, 0.02 µg/ml of anti-N-TAF1 antibody (mAb-3A11F) was incubated with 10 µg/ml of KLH-conjugated N-TAF1-peptide at 4°C overnight.
Immunofluorescence staining for the dopamine and cAMP-regulated phosphoprotein of 32 kDa (DARPP-32), choline acetyltrasferase (ChAT), parvalbumin (PV), calcium- and diacylglycerol-regulated guanine nucleotide exchange factor I (CalDAG-GEFI), µ-opiate receptor (MOR) and NeuN
Rabbit polyclonal antibodies against DARPP-32 (Cell Signaling, Danver, MA; 1:200) (Sako et al. 2010), ChAT (Chemicon International, Temecula, CA, USA; 1:200), and PV (Abcam, Cambridge, UK, 1:5,000), CalDAG-GEFI (1:20,000) (Crittenden et al. 2009) and MOR (Millipore, Billerica, MA, USA; 1:10,000), and mouse monoclonal antibody to NeuN (Millipore, 1:1,000) were used as primary antibodies. Bound antibodies were detected by goat anti-rabbit or mouse IgG conjugated with Alexa 488 (Invitrogen, Carlsbad, CA).
Digital imaging and densitometry
Digital microscopic images from immunostained sections were acquired with Meta-Morph software (Molecular Devices, Tokyo, Japan), imported into Adobe Photoshop CS4, and processed digitally. Cell and nuclear density analyses were made at a standard mid-level of the striatum in the anterior-to-posterior coordinate (0.0 to +0.2 mm relative to bregma) (Paxinos and Watson, 1986) for each rat (n = 5). To estimate the density of NeuN+, DARPP-32+ and N-TAF1+ cells in the caudoputamen, we counted these cells within a 1 mm × 1 mm field in the striatum. Among N-TAF1+ cells, the percent population of those cells colocalized with DARPP-32, ChAT, or PV was also calculated. For each animal, measurements were made in 5 striatal fields from 5 sections. Measurements of the density of N-TAF1+ nuclei in striatal striosome and matrix compartments were made on the sections doubly-stained for N-TAF1 and MOR. We counted the number of N-TAF1+ nuclei within the striosomes (n = 25) and in the matrix areas (n = 25) from 5 striatal fields of each rat (n = 5), and calculated the density of N-TAF1+ nuclei/mm2 in each compartment. For statistical analysis we used Student’s two tailed t-test and P-values less than 0.05 were considered as statistically significant.
RESULTS
Identification of N-TAF1 protein in rat brains
Dot blot assays (Fig. 1C) demonstrated that anti-N-TAF1 antibody (mAb-3A11F) reacted with the KLH-conjugated N-TAF1-peptide in a dose-dependent manner and showed no cross-reactivity to the BSA-conjugated C-TAF1-peptide. On immunoblots of rat brain extracts (Fig. 1D), a protein band with an approximate molecular mass of 250 kDa, corresponding to the predicted size of the native N-TAF1 protein, was selectively detected with mAb-3A11F, and N-TAF1 protein is enriched in nuclear fraction of brain. No immunoreactive band corresponding to N-TAF1 protein is found in the other organs such as spleen, testis, kidney, lung, and heart (Fig. 1E). These findings strengthen our previous data that N-TAF1 mRNA transcripts are expressed exclusively in the neuronal tissues (Makino et al., 2007).
Localization of N-TAF1 protein in multiple brain regions
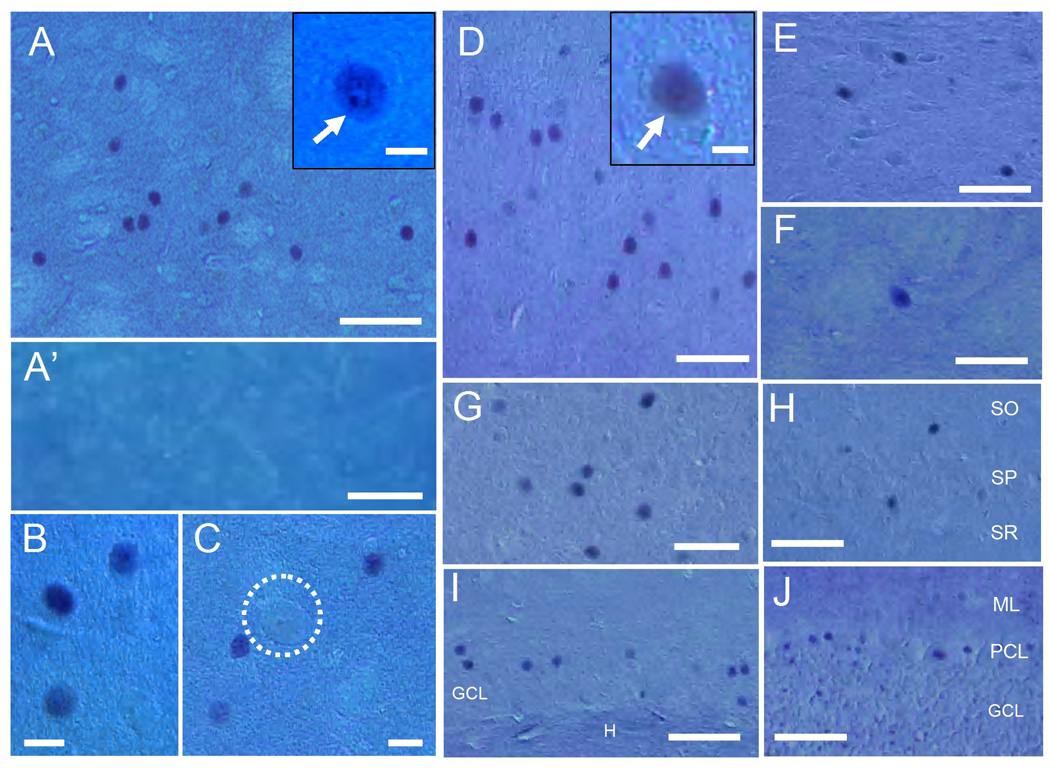
The applicability of the mAb-3A11F for histological study was determined by immunohistochemistry (Fig. 2) and immunofluorescence (Fig. 3) assays of immunostained rat brain sections. Consistent with the general concept that TAF1 is an essential protein in the transcription machinery (Wassarman and Sauer, 2001), immunohistochemical staining revealed that N-TAF1 labeling located exclusively in the nuclei of immunolabeled cells (Fig. 2). These cells were distributed in many regions of the brain, including the striatum (Fig. 2A–C), cerebral cortex (Fig. 2D), substantia nigra (Fig. 2E), globus pallidus (Fig. 2F), thalamus (Fig. 2G), hippocampus proper (Fig. 2H) and dentate gyrus (Fig. 2I), and cerebellum (Fig. 2J). Thus, N-TAF1 protein appears as a nuclear protein within subsets of neurons in multiple brain regions. We focused our study on the striatum because of the severe neurodegeneration that occurs in this region in DYT3 dystonia (Goto et al., 2005).
Fig. 2. Immunohistochemical staining of N-TAF1 under Nomarski optics in multiple brain regions.
(A–C) Striatal section stained for N-TAF1 shows that N-TAF1+ nuclei are in medium-sized cells. It is also shown that large (giant)-sized cells (C, dashed open circle) are devoid of N-TAF1 labeling. A high-power image of the neuron possessing strong N-TAF1 labeling in its nucleus and nucleoli (arrow) is shown in the inset in A. Note that no N-TAF1+ nuclei are found in a striatal section incubated with the anti-N-TAF1 antibody (mAb-3A11F) preabsorbed with excess of KLH-conjugated N-TAF1-peptide (A’).
(D–J) Distribution of N-TAF1+ nuclei in non-striatal brain regions that include the cerebral cortex (D), substantia nigra (E), globus pallidus (F), thalamus (G), hippocampus proper (H) and dentate gyrus (I), and cerebellum (J). A high-power image of cortical neuron possessing strong N-TAF1 labeling in its nucleus is shown in the inset in D.
Scale bars: A, A’ and D–J, 100 µm; B, C, 20 µm; insets in A and D, 10 µm.
SO, stratum oriens; SP, stratum pyramidale; SR, stratum radiatum, GCL, granule cell layer; H, hilus dentata; ML, molecular layer; PCL, Purkinje cell layer.
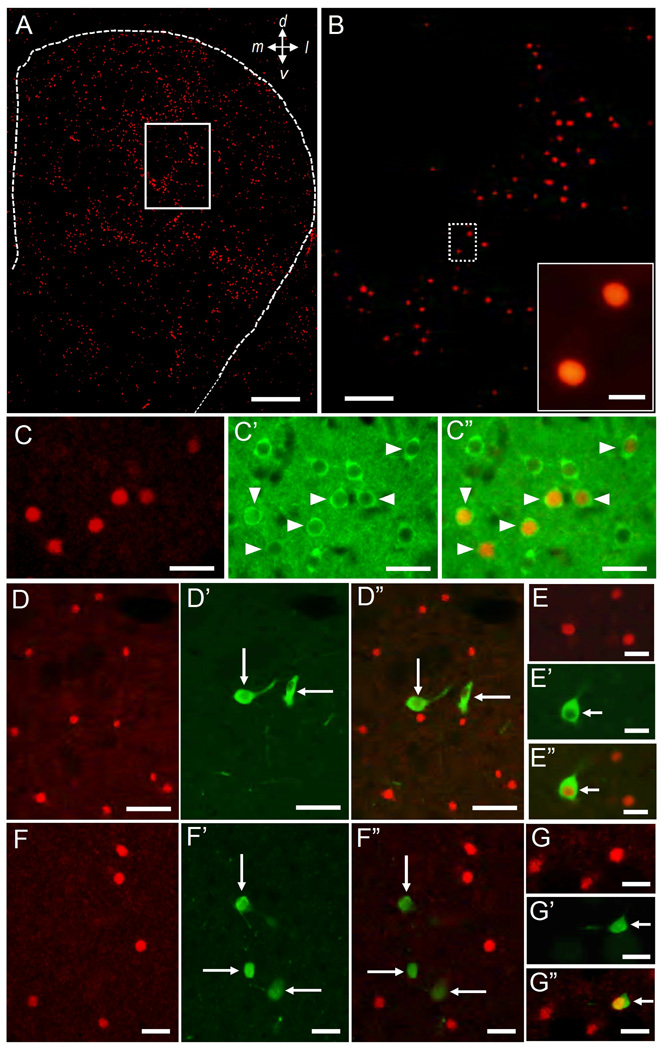
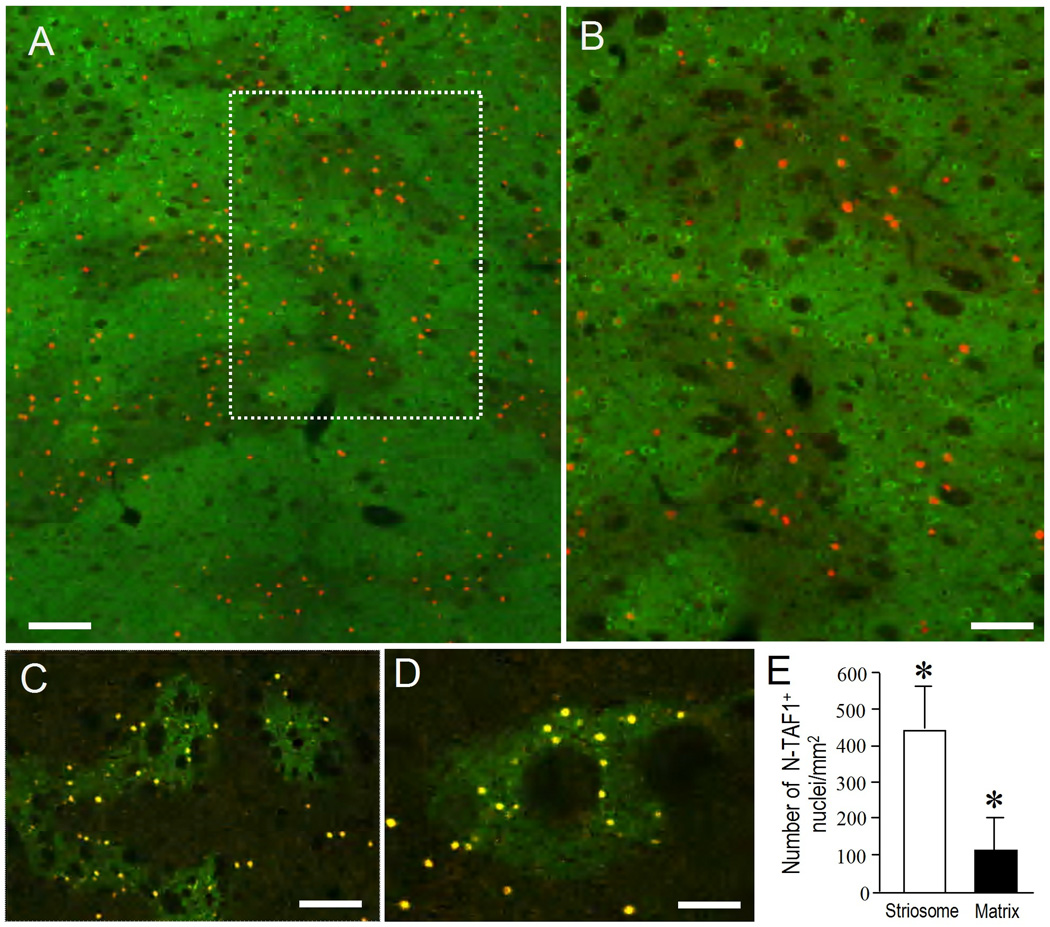
Fig. 3. Cellular localization of N-TAF1 in the striatum.
(A, B) Low-power image of Immunofluorescence staining for N-TAF1 protein. Region shown in open box in (A) is illustrated at higher magnification in (B). Region shown in dashed open box (B) is illustrated at higher magnification in the inset in B.
(C) Double immunofluorescence staining for N-TAF1 (C) and DARPP-32 (C’), and merged (C”). DARPP-32+ cells having N-TAF1+ nuclei are indicated by arrowheads in C’ and C”.
(D, E) Double immunofluorescence staining for N-TAF1 (D, E) and ChAT (D’, E’), with merged image (D”, E”). Cholinergic cells with no N-TAF1+ nuclei are shown in D’ and D” (long arrows). A cholinergic cell having N-TAF1+ nucleus is also shown in E’ and E” (short arrows).
(F, G) Double immunofluorescence staining for N-TAF1 (F, G) and PV (F’, G’), with merged image (F”, G”). PV+ cells with no N-TAF1+ nuclei are shown in F’ and F” (long arrows). A PV+ cell having N-TAF1+ nucleus is also shown in G’ and G” (short arrows).
Scale bars: A, 500 µm; B, 250 µm; inset in B, 20 µm; C, E, F and G, 50 µm; D, 100 µm.
Cellular localization of N-TAF1 in the striatum
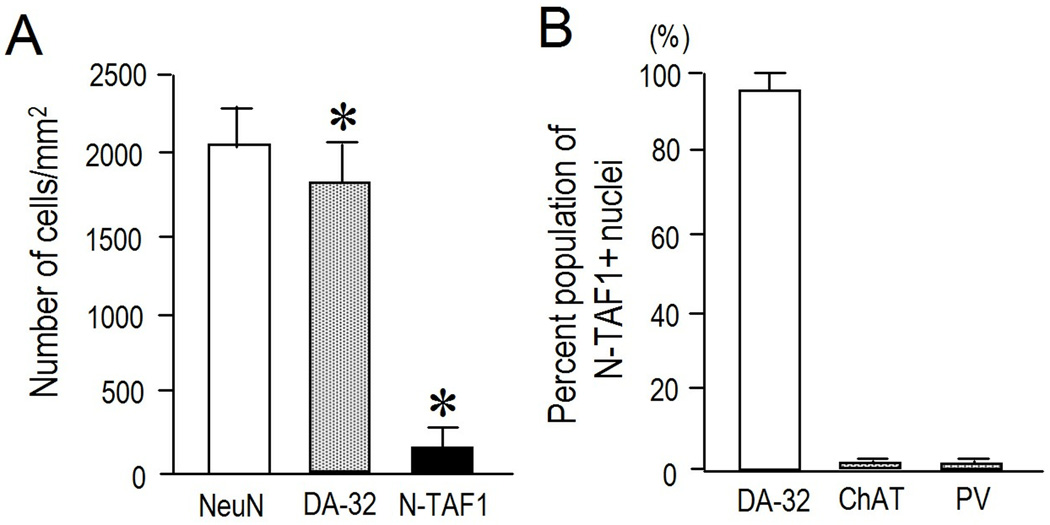
With immunohistochemical findings (Fig. 2A–C), immunofluorescence staining (Fig. 3) with the mAb-3A11F demonstrated a striking pattern of cellular expression of N-TAF1 protein in the striatum. At low-power magnifications, N-TAF1 labeling appeared as dots that showed a scattered but uneven distribution in the striatum (Fig. 3A and B). Higher power microscopic observation showed that there was a distinct subset of striatal cells with strong N-TAF1 labeling in their nuclei (Fig. 2A–C) and occasionally in their nucleoli (inset in Fig. 2A). Most N-TAF1+ cells were of medium size (Fig. 2C). Double-immunofluorescence staining (Fig. 3C) demonstrated that a large majority of the N-TAF1+ nuclei examined were within neurons positive for DARPP-32, a protein phosphatase inhibitor highly expressed in striatal MSNs (Langley et al., 1997). Moreover, double-labeling demonstrated that most of the N-TAF1+ nuclei were not found within the neurons positive for ChAT, a marker of the large cholinergic interneurons of the striatum (Pisani et al., 2007) (Fig. 3D), nor in neurons that stained for PV, a marker of medium aspiny interneurons thought to correspond to the fast-firing neurons of the striatum (Kawaguchi et al., 1995) (Fig. 3F). Only a few N-TAF1+ cells were simultaneously labeled for ChAT (Fig. 3E) or for PV (Fig. 3G). Cell density analysis (Fig. 4A) revealed that N-TAF1+ neurons comprised approximately 10% of all the striatal neurons positive for DARPP-32 (p<0.001). Among N-TAF1+ nuclei, more than 95% of nuclei were of DARPP-32+ cells, while less than 0.5% of nuclei were within cholinergic cells or PV+ cells (Fig. 4B). Together these data suggest that in the rat striatum, N-TAF1 is a nuclear protein expressed preferentially in a distinct subpopulation of striatal MSNs.
Fig. 4. Quantitative study on cellular localization of N-TAF1+ nuclei in the striatum.
(A) Density measurements of NeuN+ cells, DARPP-32+ cells (DA-32) and N-TAF1+ nuclei. Data are shown as means ± SEM (bars) values (n = 25). * indicates P < 0.001 DARPP-32 vs. N-TAF1.
(B) Percent population of N-TAF1+ nuclei co-localized with DARPP-32, ChAT, and PV. Data are shown as means ± SEM (bars) values (n = 25).
Localization of N-TAF1+ nuclei in the striosome and matrix compartments of the striatum
To determine whether the striatal neurons possessing N-TAF1+ nuclei were present in both the striosome and the matrix compartments, we next used dual antigen-immunostaining (Canales and Graybiel, 2000; Sato et al., 2008; Sako et al., 2010) for N-TAF1 and CalDAG-GEFI (Fig. 5A and B), and for N-TAF1 and MOR (Fig. 5C and D), markers for the matrix and striosome compartments, respectively. The N-TAF1+ nuclei were preferentially localized in the striosomes, with far fewer in the nearby matrix. Nuclear density analyses (Fig. 5E) confirmed this visual impression that the N-TAF1+ nuclei were significantly (p<0.01) concentrated in the striosomes relative to the matrix compartment. However, not all of the neurons in the striosomes were stained for N-TAF1; and, as shown in Fig. 5A–D, some N-TAF1+ neurons were also scattered through the extrastriosomal matrix area. Thus the distribution was one of striosome-predominance.
Fig. 5. Differential localization of N-TAF1 protein in the striatal compartments.
(A, B) Low-power image of a section doubly-stained for N-TAF1 (red) and CalDAG-GEFI (green), a marker for matrix compartment (A). Region shown in dashed open box in (A) is illustrated at higher magnification in (B).
(C, D) Lower (C) and higher (D) photomicrographs of sections double-stained for N-TAF1 and MOR, a marker for striosomes. Immunofluorescence-staining shows N-TAF1+ nuclei as yellow dots, and striosomes as green-patchy zones.
(E) Density measurements of N-TAF1+ nuclei in the striosome and matrix compartments. Data are mean ± SEM (bars) values (n = 25). * indicates P < 0.01 striosome vs. matrix. Scale bars: A and C, 200 µm; B and D, 100 µm.
Discussion
Transcriptional dysregulation mechanisms (Vermeulen et al., 1994) have been proposed as primary etiologic factors in neurodegenerative disorders, including Huntington’s disease (HD), dentatorubral pallidoluysian atrophy, and spinocerebellar ataxia 17, all of which have expansions of polyglutamine (polyQ) repeats in the proteins encoded by the affected genes. These expansions differentially interfere with RNAPII-dependent gene transcription, resulting in an increased neuronal susceptibility to certain genotoxic insults (Freman and Tjian 2002). With genomic sequencing analysis followed by expression analysis of DYT3 brain tissues, we previously found a disease-specific SVA retrotransposon insertion in an intron of the TAF1 gene, leading to the loss of the N-TAF1 transcript (Makino et al., 2007). Given that N-TAF is critical for the regulation of RNAPII-dependent gene transcription, and that there is reduced neuron-specific expression of the TAF1 gene in DYT3 patients (Makino et al., 2007), DYT3 dystonia can be classified as an example of non-polyQ transcriptional dysregulation syndrome, as is DYT6 dystonia (Bressman et al., 2009). The cellular mechanisms by which the genes affected in these diseases contribute to disease-specific pathology have so far been difficult to determine in these transcription dysregulation syndromes, as the mutations occur in widely expressed genes and yet evoke tissue-specific illness (Goodchild et al., 2005). Thus localization of the transcripts of these genes is a crucial step in determining the pathogenetic basis of these diseases.
In DYT3 dystonia, the most striking neuropathology so far observed is a primary and progressive degeneration of striatal neurons in a cell type-specific and compartment-predominant pattern (Goto et al., 2005 and 2010). Our findings here demonstrate a striking similarity between this pattern of striatal degeneration and the distribution of N-TAF1 protein, as assayed with a novel N-TAF1-selective antibody in the rodent brain. In DYT3, medium spiny projection neurons are the most vulnerable, among striatal neurons, and the large cholinergic interneurons are spared, even in the late stage of disease progression (Goto et al., 2005). The striatal pathology at the early stage of the disease period is characterized by a more prominent loss of neurons in striosomes than of neurons in the matrix compartment (Goto et al., 2005). We show here that in the rodent brain, N-TAF1 protein likewise is preferentially located in striatal MSNs and enriched in the MSNs of striosomes, but is rare in striatal cholinergic neurons. Moreover, we found N-TAF1 immunostaining was in the nuclei of these immunostained striatal neurons, consonant with a nuclear function. Together these findings suggest the existence of cell type-specific actions of an alternative splicing isoform of the TAF1 gene within the striatum. Consistent with the hypothesis that the selectivity of neuronal death could be the consequence of a higher concentration of the affected protein in the targeted cells in neurodegenerative disorders (Trorrier et al., 1995), our findings raise the possibility that both at the cellular level and at the compartmental level, the distribution of N-TAF1 protein in the striatum is correlated with the pattern of striatal neuropathology in DYT3. Among the transcription syndromes, DYT3 dystonia thus could be the first example with a demonstrated tight link of the affected gene product with the disease-specific histopathology.
How could the loss of N-TAF1 protein produce the death of striatal neurons in DYT3 dystonia? The essential function of TAF1 can be attributed to its broad requirement during RNAPII-dependent gene transcription. It has been shown that null mutations in Drosophila TAF1 result in lethality late in embryogenesis or early in larval development, suggesting that it functions in relation to cell proliferation and survival (Wassarman et al., 2000). Given the hypothesis that, in mature brain cells, N-TAF1 is critical for the transcription of most protein-coding genes as suggested for TAF1 (Chen and Hampsey, 2002; Matangkasombut et al., 2004; Kim et al., 2005), an impaired transcriptional machinery induced by loss of N-TAF1 protein could be a critical molecular event in the DYT3 pathogenesis. It is not clear, however, that the loss of N-TAF1 protein is sufficient for the induction of neurodegeneration that occurs preferentially in striatal MSNs in DYT3 dystonia. We also show that neurons with N-TAF1 immunostained nuclei are found not only in the striatum, but also in many other brain regions (Fig. 2) in which a definite pathology has not been reported. The preferential loss of striatal projection neurons with relative sparing of the large (cholinergic) interneurons of the striatum has also been documented in another adult-onset transcription dysregulation syndrome, HD (Ferrante et al., 1987), for which evidence suggests that glutamate-mediated (Gubellini et al., 2004; Lin and Beal, 2006) or dopamine-mediated (Jakel and Maragos, 2000) neurotoxicity may result in the highly patterned neuronal loss in the striatum. Certain cellular factors to which striatal MSNs might be highly susceptible also may contribute to producing the DYT3-specific histopathology.
This possibility places special value on the coordinate findings that both the DYT3 pathology, and, as demonstrated here, the distribution of N-TAF1 in normal brain, are preferentially high in the striosomal compartment. Differential neurodegeneration in striosomes has been documented in subtypes of HD, including patient populations in which mood disorders are predominant early symptoms (Hedreen and Folstein, 1995; Tippett et al., 2006). Our previous postmortem analyses on the DYT3 patients documented clinicopathological correlations that the severity of neostriatal lesions appears to depend on disease progression; the preceding severe loss of the striosomes is linked to dystonia manifestation (Goto et al., 2005). The regulation of movement by the striatum may depend not only on the balance of activity between the matrix-based direct and indirect pathways (DeLong 1990, Albin et al., 1995), but also on a balance in activity between these pathways and the striosomal pathway (Graybiel et al., 2000). For example, an imbalance between compartmentally organized basal ganglia circuits has been suggested to contribute to the generation of repetitive movement and behavioral disorders including motor stereotypies (Graybiel, 2008), levodopa induced dyskinesias (Crittenden et al., 2009) and dystonias (Sato et al., 2008; Goto et al., 2010). We suggest that in DYT3 dystonia, preceding and predominant degeneration of MSNs in the striosomes caused by loss of N-TAF1 may alter the functional activities of matrix-based striatal output systems, resulting in occurrence of dystonia. Information about differences between gene regulation in striosome and matrix compartments could be critical to identifying the specific role of N-TAF1 protein in nuclear functions and the precise pathophysiologic mechanisms underlying neuronal death in DYT3 dystonia.
Highlights.
We developed a specific monoclonal antibody against the N-TAF1 protein. Immunohistochemical localization of N-TAF1 in rat brain was studied. N-TAF1 protein appears as a neuronal nuclear protein in multiple brain regions. In the striatum, neurons possessing N-TAF1 are largely of the medium spiny types. A marked tendency of these neurons to locate in the striosomes is also found.
ACKNOWLEDGEMENTS
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (grant-in-aid for Scientific Research, 20591025, 2139026900), and the United States National Institutes of Health (P50 NS38372 and R37 HD028341).
Comprehensive list of abbreviations
- MSNs
medium spiny neurons
- mAb
monoclonal antibody
- N-TAF1
neuron-specific isoform of the TAF1 gene
- TBP
TATA box-binding protein
- TFIID
transcription factor IID complex
- C-TAF1
common form of the TAF1 gene
- DARPP-32
the dopamine and cAMP-regulated phosphoprotein of 32 kDa
- MOR
μ-opiate receptor
- KLH
keyhole limpet hemocyanin
- BSA
bovine serum albumin
- TFIID
transcription factor IID complex
- RNAPII
RNA polymerase II
- SO
stratum oriens
- SP
stratum pyramidale
- SR
stratum radiatum
- GCL
granule cell layer
- H
hilus dentata
- ML
molecular layer
- PCL
Purkinje cell layer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Web Resources: Accession numbers and URLs for data presented herein are as followed : GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for the mRNA sequence of the C-TAF1 [accession number NM_004606]) and N-TAF1 [accession number AB300418]).
REFERENCES
- Albin RL, Young AB, Penny JB. The functional anatomy of disorders of the basal ganglia. Trends Neurosci. 1995;18:63–64. [PubMed] [Google Scholar]
- Bressman SB, Raymond D, Fuchs T, Heiman GA, Ozelius L, Saunders-Pullman R. Mutations in THAP1 (DYT6) in early-onset dystonia: a genetic screening study. Lancet Neurol. 2009;8:441–446. doi: 10.1016/S1474-4422(09)70081-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales JJ, Graybiel AM. A measure of striatal function predicts motor stereotypy. Nat Neurosci. 2000;3:377–383. doi: 10.1038/73949. [DOI] [PubMed] [Google Scholar]
- Chen BS, Hampsey M. Transcription activation: unveiling the essential nature of TFIID. Curr Biol. 2002;12:R620–R622. doi: 10.1016/s0960-9822(02)01134-x. [DOI] [PubMed] [Google Scholar]
- Crittenden JR, Cantuti-Castelvetri I, Saka E, Keller-McGandy CE, Hernandez LF, Kett LR, Young AB, Standaert DG, Graybiel AM. Dysregulation of CalDAG-GEFI and CalDAG-GEFII predicts the severity of motor side-effects induced by anti-parkinsonian therapy. Proc Natl Acad Sci USA. 2009;106:2892–2896. doi: 10.1073/pnas.0812822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong MR. Primate models of movement disorders of basal ganglia origin. Trends Neurosci. 1990;13:281–289. doi: 10.1016/0166-2236(90)90110-v. [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Kowall NW, Beal MF, Martin JB, Bird ED, Richardson EP., Jr Morphologic and histochemical characteristics of a spared subset of striatal neurons in Huntington’s disease. J Neuropathol Exp Neurol. 1987;46:12–27. doi: 10.1097/00005072-198701000-00002. [DOI] [PubMed] [Google Scholar]
- Freman RN, Tjian R. Neurodegeneration: a glutamine-rich trail leads to transcription factors. Science. 2002;296:2149–2150. doi: 10.1126/science.1073845. [DOI] [PubMed] [Google Scholar]
- Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Goto S, Lee LV, Munoz EL, Tooyama I, Tamiya G, Makino S, Ando S, Dantes MB, Yamada K, Matsumoto S, Shimazu H, Kuratsu JI, Hirano A, Kaji R. Functional anatomy of the basal ganglia in X-linked recessive dystonia-parkinsonism. Ann Neurol. 2005;58:7–17. doi: 10.1002/ana.20513. [DOI] [PubMed] [Google Scholar]
- Goto S, Nagahiro S, Kaji R. Striosome-matrix pathology of dystonias: a new hypothesis for dystonia genesis. In: Kurstot J, Forsstrom M, editors. Dystonia: causes, symptoms and treatment. New York, NY: Nova Science Publishers; 2010. pp. 1–22. [Google Scholar]
- Graybiel AM. Habits, rituals, and the evaluative brain. Annu Rev Neurosci. 2008;31:359–387. doi: 10.1146/annurev.neuro.29.051605.112851. [DOI] [PubMed] [Google Scholar]
- Graybiel AM, Canales JJ, Capper-Loup C. Levodopa-induced dyskinesias and dopamine-dependent stereotypies: a new hypothesis. Trends Neurosci. 2000;23:571–577. doi: 10.1016/s1471-1931(00)00027-6. [DOI] [PubMed] [Google Scholar]
- Gubellini P, Pisani A, Centonze D, Bernardi G, Calabresi P. Metabotropic glutamate receptors and synaptic plasticity: implications for neurological diseases. Prog Neurobiol. 2004;74:271–300. doi: 10.1016/j.pneurobio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Haberhausen G, Schmitt I, Kohler A, Peters U, Rider S, Chelly J, Terwilliger JD, Monaco AP, Müller U. Assignment of the dystonia-parkinsonism syndrome locus, DYT3, to a small region within a 1.8-Mb YAC contig of Xq13.1. Am J Hum Genet. 1995;57:644–650. [PMC free article] [PubMed] [Google Scholar]
- Hedreen JC, Folstein SE. Early loss of neostriatal striosome neurons in Huntington’s disease. J Neuropathol Exp Neurol. 1995;54:105–120. doi: 10.1097/00005072-199501000-00013. [DOI] [PubMed] [Google Scholar]
- Jakel RJ, Maragos WF. Neuronal cell death in Huntington’s disease: a potential role for dopamine. Trends Neurosci. 2000;23:239–245. doi: 10.1016/s0166-2236(00)01568-x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Wilson CJ, Augood SJ, Emson PC. Striatal interneurones: chemical, physiological and morphological characterization. Trends Neurosci. 1995;18:527–535. doi: 10.1016/0166-2236(95)98374-8. [DOI] [PubMed] [Google Scholar]
- Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley KC, Bergson C, Greengard P, Ouimet CC. Co-localization of the D1 dopamine receptor in a subset of DARPP-32-containing neurons in rat caudate-putamen. Neuroscience. 1997;78:977–983. doi: 10.1016/s0306-4522(96)00583-0. [DOI] [PubMed] [Google Scholar]
- Lee LV, Maranon E, Demaisip C, Peralta O, Borres-Icasiano R, Arancillo J, Rivera C, Munoz E, Tan K, Reyes MT. The natural history of sex-linked recessive dystonia parkinsonism of Panay, Philippines (XDP) Parkinsonism Relat Disord. 2002;9:29–38. doi: 10.1016/s1353-8020(02)00042-1. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Makino S, Kaji R, Ando S, Tomizawa M, Yasuno Y, Goto S, Matsumoto S, Tabuena D, Maranon E, Dantes M, Lee LV, Ogasawara K, Akatsu H, Nishimura M, Tamiya G. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am J Hum Genet. 2007;80:393–406. doi: 10.1086/512129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut O, Auty R, Buratowski S. Structure and function of the TFIID complex. Adv Protein Chem. 2004;67:67–69. doi: 10.1016/S0065-3233(04)67003-3. [DOI] [PubMed] [Google Scholar]
- Müller U. The monogenic primary dystonias. Brain. 2009;132:2005–2025. doi: 10.1093/brain/awp172. [DOI] [PubMed] [Google Scholar]
- Nolte D, Niemann S, Müller U. Specific sequence changes in multiple transcript system DYT3 are associated with X-linked dystonia parkinsonism. Proc Natl Acad Sci USA. 2003;100:10347–10352. doi: 10.1073/pnas.1831949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotactic Coordinates. 2nd edn. San Diego: Academic Press; 1986. [Google Scholar]
- Pisani A, Bernardi G, Ding J, Surmeier J. Re-emergence of striatal cholinergic interneurons in movement disorders. Trends Neurosci. 2007;30:545–553. doi: 10.1016/j.tins.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Sako W, Morigaki R, Nagahiro S, Kaji R, Goto S. Olfactory type G-protein α subunit (Gαolf) in striosome-matrix dopamine systems in adult mice. Neuroscience. 2010;170:497–502. doi: 10.1016/j.neuroscience.2010.06.072. [DOI] [PubMed] [Google Scholar]
- Sato K, Sumi-Ichinose C, Kaji R, Ikemoto K, Nomura T, Nagatsu I, Ichinose H, Ito M, Sako W, Nagahiro S, Graybiel AM, Goto S. Differential involvement of striosome and matrix dopamine systems in a transgenic model of dopa-responsive dystonia. Proc Natl Acad Sci USA. 2008;105:12551–12556. doi: 10.1073/pnas.0806065105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippett LJ, Waldvogel HJ, Thomas SJ, Hogg VM, van Roon-Mom W, Synek BJ, Graybiel AM, Faull LM. Striosomes and mood dysfunction in Huntington’s disease. Brain. 2006;130:206–221. doi: 10.1093/brain/awl243. [DOI] [PubMed] [Google Scholar]
- Trorrier Y, Devys D, Imbert G, Saudou F, An I, Lutz Y, Weber C, Agid Y, Hirsch EC, Mandel JL. Cellular localization of the Huntington’s disease protein and discrimination of the normal and mutated form. Nat Genet. 1995;10:104–110. doi: 10.1038/ng0595-104. [DOI] [PubMed] [Google Scholar]
- Vermeulen W, et al. Three unusual repair deficiencies associated with transcription factor BTF2 (TFIIH): evidence for the existence of a transcription syndrome. Cold Spring Harb Symp Quant Biol. 1994;59:317–329. doi: 10.1101/sqb.1994.059.01.036. [DOI] [PubMed] [Google Scholar]
- Wassarman DA, Aoyagi N, Pile LA, Schlag EM. TAF250 is required for multiple developmental events in Drosophila. Proc Natl Acad Sci USA. 2000;97:1154–1159. doi: 10.1073/pnas.97.3.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman DA, Sauer F. TAFII250: a transcription toolbox. J Cell Sci. 2001;114:2895–2902. doi: 10.1242/jcs.114.16.2895. [DOI] [PubMed] [Google Scholar]