Abstract
Hutchinson-Gilford progeria syndrome (HGPS) is a premature aging syndrome caused by the expression and accumulation of a mutant form of lamin A, Δ50 lamin A. As a component of the cell's nucleoskeleton, lamin A plays an important role in the mechanical stabilization of the nuclear envelope and in other nuclear functions. It is largely unknown how the characteristic 50 amino acid deletion affects the conformation of the mostly intrinsically disordered tail domain of lamin A. Here we perform replica exchange molecular dynamics simulations of the tail domain and determine an ensemble of semi-stable structures. Based on these structures we show that the ZMPSTE 24 cleavage site on the precursor form of the lamin A tail domain orients itself in such a way as to facilitate cleavage during the maturation process. We confirm our simulated structures by comparing the thermodynamic properties of the ensemble structures to in vitro stability measurements. Using this combination of techniques, we compare the size, heterogeneity of size, thermodynamic stability of the Ig-fold, as well as the mechanisms of force-induced denaturation. Our data shows that the Δ50 lamin A tail domain is more compact and displays less heterogeneity than the mature lamin A tail domain. Altogether these results suggest that the altered structure and stability of the tail domain can explain changed protein-protein and protein-DNA interactions and may represent an etiology of the disease. Also, this study provides the first molecular structure(s) of the lamin A tail domain, which is confirmed by thermodynamic tests.
Keywords: Laminopathy, structure, mechanics, protein stability, nuclear mechanics
1. Introduction
1.1 Lamin A, laminopathies and HGPS
The nucleoskeleton of a cell, located at the inner face of the inner nuclear membrane, is composed mainly of intermediate filaments (IFs) made of either A-type and B-type lamin protein along with other accessory proteins (Wilson and Berk, 2010). A-type lamins, primarily lamin A and lamin C, are spliced from the LMNA gene. B-type lamins are encoded by LMNB1 and LMNB2. Lamin A is an A-type lamin of particular mechanical interest since the loss of lamin A appears to influence nuclear mechanics more than other A-type lamins or B-type lamins (Lammerding et al., 2006). Conversely, a loss of lamin A has no apparent defect in mice aside from some nuclear weakening as long as other A-type lamins are present (such as lamin C) (Fong et al., 2006). This `survivability' may be a reason that the LMNA gene has more than 100 disease-causing mutations (Worman et al., 2010). Mutations in different regions of the LMNA gene lead to alterations in different tissue types including fat, muscle, and brain as well as different aging disorders (Worman and Bonne, 2007). This group of diseases, collectively termed laminopathies, has led to a considerable interest in lamin A. Hutchison Gilford progeria syndrome (HGPS) is a segmented premature aging syndrome caused by a mutation in LMNA (Goldman et al., 2004).
1.2 Lamin A molecular structure and the HGPS Δ50 mutation
Lamin A is a characteristic type V IF protein that contains a globular N-terminal head, a segmented coiled-coil α-helical rod domain and a C-terminal tail containing an immunoglobulin (Ig)-fold (Herrmann et al., 2007). Lamin proteins are unique from other IFs as they feature an exceptionally long C-terminal tail domain (Herrmann, et al., 2007). The Ig-fold binds DNA and many other nuclear proteins (Zastrow et al., 2004). The C-terminus of the tail domain undergoes posttranslational processing, where the precursor form of lamin A is farnesylated, carboxymethylated localized to the inner nuclear membrane (Coffinier et al., 2010) and then the last 18 amino acids are cleaved by an endoprotease ZMPSTE-24 to produce mature wild-type lamin A (mwt LA) (Young et al., 2005). In HGPS, a single point mutation in the LMNA gene activates a cryptic splice site causing 50 amino acids encoded by exon 11 to be deleted, and the resulting mutant protein is called Δ50 lamin A (Δ50 LA) (De Sandre-Giovannoli et al., 2003). The deletion in Δ50 LA includes the ZMPSTE-24 cleavage site resulting in the retention of the C-terminal farnesylation, which is suggested to be responsible for the accumulation of Δ50 LA at the inner nuclear membrane. Similarly, the loss of the ZMPSTE-24 protease causes an accumulation of the precursor lamin A protein, prelamin A, at the inner nuclear membran (Navarro et al., 2004; Taimen et al., 2009). However, the retained farnesylation cannot explain all of the molecular changes in HGPS. Recently, in vitro binding assays have shown differential binding of Δ50 LA to nuclear proteins and chromatin (Bruston et al., 2010). Two transgenic mice models containing an unfarnesylated Δ50 LA showed varied, but present, clinical pathology (Yang et al., 2011; Davies et al., 2010; Leuba et al., 1994). These results suggest that the loss of 50 amino acids from the lamin A tail may alter the protein more than simply retaining a farnesylation (Young et al., 2006).
1.3 Lamin A tail is an intrinsically disordered protein
The tail domain of lamin A is mostly disordered and demonstrates the characteristic qualities of intrinsically disordered proteins (Rauscher and Pomes, 2010) including a promiscuity in protein binding (Schirmer and Foisner, 2007; Zastrow et al., 2004), tendency to aggregate (Linding et al., 2004) and a high proline and glycine content. It is difficult to predict how the removal of 50 amino acids in a region lacking secondary structure will affect the overall structure of the protein domain. Disordered regions are typically difficult to crystallize and take on many conformations, making traditional structural analyses such as X-ray crystallography and NMR challenging (Krimm et al., 2002). Instead, here we use Replica Exchange Molecular Dynamics (REMD) simulations to determine an array of semi-stable structures based on ordered secondary structures (Sugita and Okamoto, 1999). REMD has been shown to be successfully utilized as a computational method used to improve the dynamic properties of conventional Molecular Dynamics and Monte Carlo methods, aiming at obtaining global-minimum free energy states of polymers by overcoming kinetic trapping without giving any specific initial structures (Liu et al., 2005; Snow et al., 2005; Sugita and Okamoto, 1999; Zhou et al., 2001). Nevertheless, this method can reduce accuracy when it is applied to extremely large systems (Snow et al., 2005). We focus on the amino acid sequence of the tail domains and apply massively parallelized computational power to study their intrinsically disordered structures (Liu et al., 2005) (see Methods and Supplementary Material for detail). When taken together, these structures represent the spectrum of conformations of the mwt LA and Δ50 LA tail domains. Then, we computationally and experimentally probe the protein stability to determine global structural changes in the tail domains. These structural changes likely impact binding, filament formation as well as protein stability. Specifically, we test the stability of the structured Ig-fold, which has previously been examined in other laminopathies (Krimm et al., 2002; Shumaker et al., 2005). However, an altered stability has only been observed for point mutations which occur in the Ig-fold. By combining data obtained from experiment and simulation, we extract information on changes in the stability of both normal and mutant proteins and develop a mechanistic model of higher-level nuclear structural and mechanical changes in diseased and normal states.
2. Results
2.1 Structure of the Δ50 region of lamin A
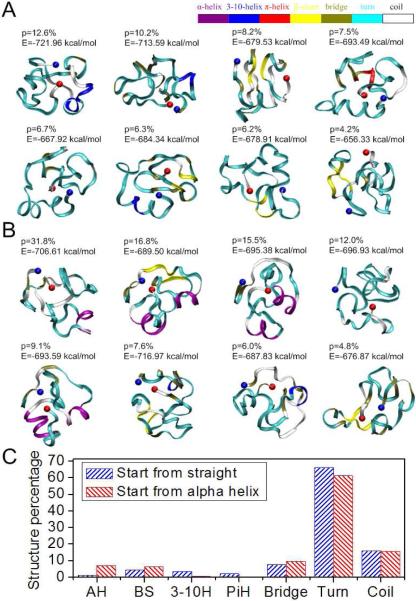
By applying REMD, we identify folded structures of different segments within the tail domain of LA. As shown in Figure 1, the segments include AA607–656 (Δ50) for the 50 amino acids coded in exon 11 that are missing in the Δ50 LA mutation (sequence in Figure 1A), AA428–646 for the mwt LA tail domain (sequence in Figure 1B) and AA428–614* for the Δ50 LA (sequence in Figure 1C). Since the Δ50 region from exon 11 is lost in Δ50 LA, we examine the folded conformation of that region. To increase the sampling space and to examine the dependence on the initial geometry, we consider two extreme initial conformations; a straight conformation and an α-helical conformation. This strategy has been effectively performed in earlier REMD studies applied to investigate protein structures (Liu et al., 2005). Likely structures of the Δ50 domain, that is, those associated with highest significance factors and lowest energies are listed in Figures 2A and B. There are no dominant conformations for the structure based on the energy calculations of the different structures. The average end-to-end length of this segment is determined to be 10 ± 3.9 Å (Figure S1C), obtained by statistical measurement of all the conformations generated. The flexibility of this segment is confirmed by comparing this end-to-end length with the contour length of 175 Å. These observations correlate with the experimental finding that the structure of this domain is amorphous and flexible (Kobayashi et al.; Krimm et al., 2002). We confirm that two sets of initially differently folded structures converged to similar conformations characterized by the secondary structure distribution as shown in Figure 2C. Figure 2C clearly shows that the generated structures have a secondary structure indicative of intrinsic disorder, with >80% turns and coil. This is consistent with our experimental CD measurements (Figure S2 and Table 1), where we find a high glycine content (20%) in this region that may be the source of the high flexibility of this segment. We observe a relatively high α-helix character for Δ50 segments that were initialized as a pure α-helix (Figure 2B). The α-helix character is retained near the C-terminal of this Δ50 domain around AA645 (8 AAs) and 623 (6 AAs), suggesting that they are more difficult to unfold rather than to maintain.
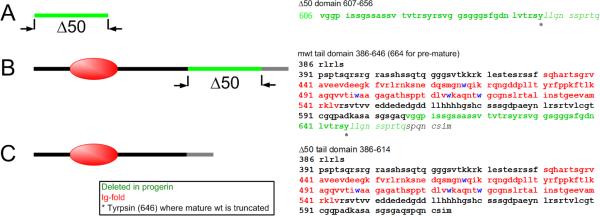
Figure 1. Schematic of the sequences of lamin tail domains investigated in this study.
(A) The segment of amino acids deleted in the D50 LA tail. (B) The mwt LA tail ends at tyrosine 646 (*), and the remaining residues (italics) are cleaved during posttranslational processing (these residues are included in our simulations). (C) Δ50 LA, also called progerin, has lost 50 amino acids (green) including the proteolytic site. Thus, Δ50 LA ends at a different 614, which we call 614*.
Figure 2. Conformations of the 50 amino acid segment of exon 11 with high significance factors.
Simulations of protein folding with initial conditions of (A) straight and (B) α-helix show similar final results, which are mostly disordered. Each amino acid is colored according to their secondary structure. The significance factor (p) as well as the potential energy (E) is marked for each conformation. The starting point (AA607) is in blue and the end point (AA656) is in red; (C) Secondary structure of the 50 amino acid segment shows little structure.
Table 1.
Fit CD data (Whitmore and Wallace, 2004; Whitmore and Wallace, 2008) of both tail domains shows a majority of disorder with ≈25% β-sheet structure.
| LA tail domain | Fraction of βsheet structure | Fraction of disorder |
|---|---|---|
| mwt | 0.27 ± 0.05 | 0.66 ± 0.05 |
| Δ50 | 0.23 ± 0.07 | 0.70 ± 0.07 |
Another intriguing character of the Δ50 region is that AA645–647 are always found on the outside of the structure with side-chains of the amino acids pointing outward (Figures S1A and B in red). AA645–647 are the site of endoproteolytic cleavage by the protease, ZMPSTE-24, cleaving prelamin A to mwt LA (shown in Figure 1A). Our computational results suggest that this “substrate” region faces outward possibly to facilitate interacting with the protease. This location may be critical to ensure the fast protease reaction kinetics of premature lamin A cleavage observed in cells (Liu et al., 2010).
2.2 Structures of tail domains of mwt LA and Δ50 LA
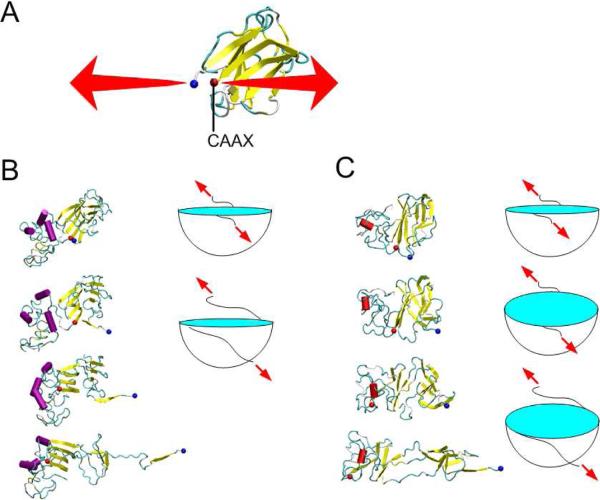
Experimentally, the tail domains of mwt LA and Δ50 LA show a β-sheet structure with random coil but no α-helix (Figure S2, Table 1). After purification, the proteins are estimated to be ≈4.0 +/− 0.3 nm in size, as measured by DLS (Figure S3). Using the same REMD technique as used above, we predict conformations of the full tail domains of mwt LA and Δ50 LA. As expected for a mostly intrinsically disordered protein, our simulations yield an ensemble of conformations for the tail domains of mwt LA and Δ50 LA. We present the eight conformations that are most frequently visited by the replica with the lowest temperature in the order of decreasing significance (Figures 3A and B). Initially, the structure of the s-type Ig-fold of the tail domain is modeled from the starting conformation available from the protein database (PDB with ID 1IVT (Krimm et al., 2002)), and the remaining tail domain is given a straight conformation. We observe that the Ig-fold maintains a majority of its β-sheet structure throughout the simulation, while other regions become disordered with a distribution of structures, but no single, dominant, equilibrium state. This intrinsic disorder is expected by the high proline and glycine content (3% P and 16% G for mwt LA tail; 4% P and 10% G for Δ50 LA tail).
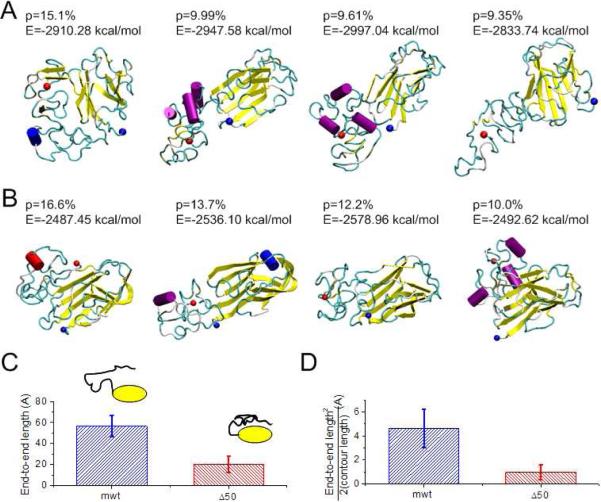
Figure 3. Conformations and structures of tail domains for normal and disease cases.
Conformations with high significance factors of the (A) mwt LA tail domain AA428–646 and (B) Δ50 LA tail domain, AA428–614* show different structures. The beginning amino acid is marked with a blue dot and the C-terminus is marked with a red dot. (C) The end-to-end length from the end of the Ig-fold domain (AA545) to the tail end (AA646 for mwt LA and AA614* for Δ50 LA) of all the conformations of the mwt LA and Δ50 LA tail domains, respectively. (D) Normalizing the square of the end-to-end length by the contour length reflects the persistence length of the peptide. We find that the Δ50 LA tail domain is significantly more compact, even when considering the loss of amino acids in the truncation.
We find that the folded structures of Δ50 LA tail (Figure 3B) are more compact than structures of mwt LA tail (Figure 3A). From these simulation results, we quantify the end-to-end length (ree) of this disordered domain (AA545–646 for mwt LA and AA545–614 for Δ50 LA in Figure 3C). The average ree for mwt LA tail was measured to be ree = 56.5 ± 10.2 Å, which is much larger than that of the Δ50 LA tail (ree = 20.4 ± 7.6 Å). Some of the reduction in the ree in the Δ50 LA tail, as compared to mwt LA tail counterpart, can be accounted for by the reduced contour length from the net loss of 32 amino acids. However, the contour length is only reduced by 30%, much less than their difference in ree. The persistence length of an isolated polypeptide chain is ≈4 Å, which is much smaller than the loss of contour length (357 Å for 102 AAs of mwt LA versus 245 Å for 70 AAs of Δ50 LA). As such, we can use the Worm-Like-Chain (WLC) model for the disordered regions (in the limit of l >>εp) (Leuba et al., 1994; Marko and Siggia, 1995)
| (1) |
to estimate the persistence lengths of the mwt LA tail and the Δ50 LA tail. We obtain that the persistence length of the Δ50 LA tail (1.0 ± 0.6 Å) is 78% shorter than that of the mwt LA tail (4.6 ± 1.6 Å), as shown in Figure 3D. This result indicates that the tail domain of Δ50 LA is more able to adopt a greater variety of conformations. We calculate the effective radius of the molecule, and find that the diameter of Δ50 LA tail closely agrees with the experimental hydrodynamic radius from DLS (Table 2; Figure S4 to compare with Figure S3). Interestingly, the deviation of the protein sizes, both by experiment and simulation, are the most striking features (Table 2); and the mwt LA has a more heterogeneous distribution of protein sizes.
Table 2.
Hydrodynamic radius of the tail domain measured by DLS and simulation
| LA tail domain | DLS experiment (nm) | Simulation (nm) |
|---|---|---|
| mwt | 4.5 ± 1.3 nm | 5.0 ± 1.4 nm |
| Δ50 | 4.0 ± 0.3 nm | 3.2 ± 0.3 nm |
We quantitatively measure the overall structural composition of the mwt LA and Δ50 LA for the tail domain, and we observe that >70% of the amino acids feature turn or coil structures, confirming that this domain is mostly disordered (Figure S5A). We count the secondary structures of each amino acid and plot the ratio (Figures S5B and C). Some positions in the mwt LA tail are seen to form α-helices including 562 (24 AAs), 584 (8 AAs) and 620 (20 AAs). In the Δ50 LA tail, α-helices are similar if not slightly longer for the mid segment but are missing the third segment (562 (24 AAs) and 582 (12 AAs)). Compared with random coils, a peptide with a helix structure has a much longer persistence length (≈100 Å since the backbone is stabilized by hydrogen bonds), which can explain why the mwt LA tail, with net 16 more AAs in helices, has a longer persistence length.
2.3 The thermal denaturation of the mature lamin A and and Δ50 LA tail domains
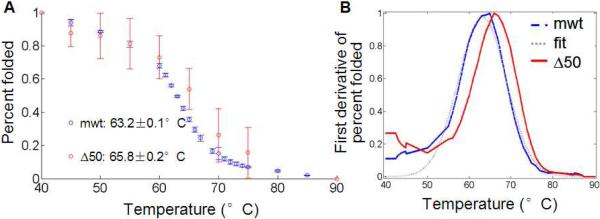
To experimentally measure the stability of the LA tails we utilize the presence of four hydrophobic tryptophan residues in the Ig-fold (Figure 1) as a way to monitor protein thermal unfolding (Eftink, 1994). We monitor the intensity of protein solutions at 342 nm, the peak wavelength of the emission spectra, as a function of temperature. The intensity is then normalized assuming a fully folded state at low temperature 37°C and a fully unfolded state at high temperature 95°C (see Methods and Figure S5). The resulting unfolding curve as a function of temperature is shown in Figure 4A. Measuring the thermal stability gives us insight into the tail domains' thermal equilibrium (kBT). Using this analysis, we find that the mwt LA tail shows little sample-to-sample variation and we determine a transitional melting temperature (TM,mwt) of 63.2 +/− 0.09 °C at which 50% of the protein is unfolded. The Δ50 LA tail domain shows an increased sample-to-sample variation and shows a TM,Δ50 of 65.8 +/− 0.23°C (Figure 4B, Table 3).
Figure 4. Thermal denaturation of Δ50 LA and mwt LA protein by measuring tryptophan fluorescence.
(A) Fluorescence of maximum tryptophan signal are normalized to signals corresponding to 100% folded (measured at 37°C) and 0% folded (at 95°C). The averages of two independent runs are shown of each protein. (B) To find the transition melting temperature, we take the first derivative of the signal. Interpolation, smoothing and fit to a Gaussian curve (see Figure S6) allow us to determine transition temperatures with confidence intervals (Table 3).
Table 3.
Experimentally measured TM of lamin A tails from thermal denaturation and tryptophan fluorescence.
| LA tail domain | Tm(°C) | r2 | σ (°C) | Tm (°C) | GTm (°C) |
|---|---|---|---|---|---|
| mwt | 63.13, 63.24 | 0.96, 0.94 | 0.070, 0.055 | 63.2 | 0.09 |
| Δ50 | 65.80, 65.82 | 0.32, 0.88 | 0.215, 0.085 | 65.8 | 0.23 |
We confirm the melting temperature of the Δ50 LA tail by examining changes in the secondary structure using CD with increasing temperature as well as by differential scanning calorimetry (DSC). We perform these analyses on Δ50 LA tail domain since the protein is less susceptible to aggregation and the baseline protein state was more regular (see Supplementary Material). The change in CD signal at 230 nm with increasing temperature normalized to room temperature and denatured points shows a characteristic denaturation when fit to a van't Hoff model
| (2) |
From this fit (Figure S6A), we observed a characteristic TM of 63°C based on secondary structure. We confirm this transition temperature using DSC, and observed a TM of 63°C (Figure S6B). These values are slightly below the TM proposed earlier, the similar range of temperatures for denaturation of secondary and tertiary structure suggests robustness of the thermal stability measures. However, changes in secondary structure are limited by low signal to noise ratio (small amounts of β-sheet compared to random coil).
2.4 Simulating the thermodynamic denaturation of tail domains under loading conditions
To complement the experimental data of protein stability described in the previous section, we use simulation to measure the mechanical stability of the mwt and Δ50 LA tail domains by unfolding the protein at 27°C and monitoring the conformation of the region containing the Ig-fold. We focus on the Ig-fold because: (a) the Ig-fold is the most ordered and probably most stabilized motif representing the limiting factor of mechanical strength; (b) the Ig-fold interacts with lamin binding proteins and DNA (Stierle et al., 2003; Zastrow et al., 2004); and (c) we can directly compare denaturation with experimental results based on the tryptophan residues in the Ig-fold. We apply the steered molecular dynamics (SMD) protocol to stretch the two regions flanking the Ig-fold domain (AA428 and 545) within the full tail domain to unfold the structure and record the force-extension curve during the stretching process (Figure 5A).
Figure 5. Results of SMD simulation of Ig-fold domain unfolding.
(A) The β-sheet structure of the Ig-fold domain and boundary conditions used to apply the mechanical load. While the force was applied to the Ig-fold, the other amino acids were in their pseudo-equilibrium states. (B) Snapshots of a mwt LA tail domain during the unfolding process, taken at extensions of 0, 15 Å, 38 Å and 75 Å, respectively show one β-strand at a time (yellow) unfolding from a central structure. (C) Snapshots of a Δ50 LA tail domain during the unfolding process, taken at extensions of 0, 15 Å, 38 Å and 75 Å, respectively. The images show an intermediate state with no “outlying” β-strands. The cartoons in panels B and C schematically describe the different deformation process of the mwt and Δ50 LA tail domains, respectively. Note the large blue binding pocket in panel C is larger for Δ50 LA tail domain, than that for mwt LA tail domain and present for larger deformations (and representing the intermediate state).
We find that the unfolding processes of the two protein segments reveal different deformation mechanisms under loading. The unfolding of the Ig-fold in the mwt LA tail shows a stepwise unwinding of the β-strands from the one end to the other end (Figure 5B). However, the unfolding of the Ig-fold of the Δ50 LA tail domain reveals a two-state process. In the first state, the Ig-fold maintained secondary β-sheet structure but shows a tertiary transition, similar as opening two halves of a sandwich along an edge. This unfolding pattern opens up a beta-sheet pocket. In the second stage, beta-strands are peeled away from the central structure in a similar fashion as mwt LA tail (Figure 5C). The two-state unfolding process of the Ig-fold domain appears to be unique to the Δ50 LA tail, which could qualitatively describe by a pocket-unwinding model as illustrated in Figure 5C. This result shows that the Ig-fold of the Δ50 LA tail transforms before unfolding under loading. These results suggest altered interactions within the Ig-fold and that the mutation makes unfolding more variable. Thus, the two-state unfolding complicates a normally continuous process and may allow for more stable intermediate states.
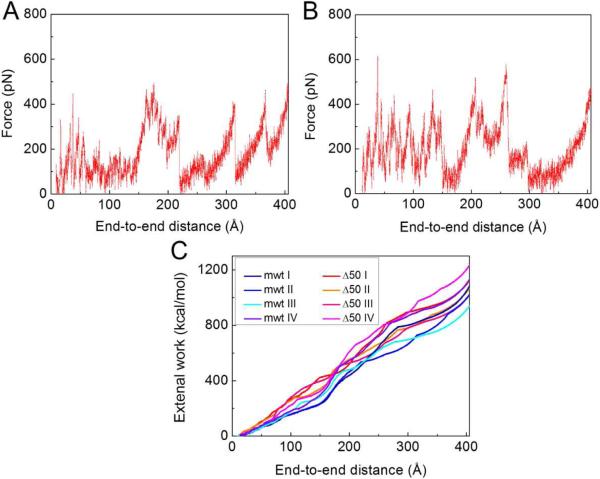
We repeat the loading process for other possible structures with high significance factor (structures in Figures 3A and B) and collect all force-extension curves as shown in Figures 6A and B from the initial conformation until the end-to-end distance equaled the contour length (≈403 Å). We model force-extension curves as worm-like-chain (WLC) models stretching at the end (Marko and Siggia, 1995). Initially, there were 4–5 transition peaks, each corresponding to an unfolding event of a β-strand, requiring the unfolding force of 357.8 ± 94.2 pN. After each unfolding event, the force drops and shifts rightward because the contour length has increased (Sotomayor and Schulten, 2007). We integrate the force-extension curves (Figure 6C) and find that the external work to fully unfold the Δ50 LA Ig-fold was 1,113.8 +/− 75.1 kcal/mol and 1057.3 +/− 70.2 kcal/mol for the mwt LA Ig-fold. To fully unfold the Ig-fold of the Δ50 LA tail domain, 56.5 kcal/mol more work is required than in the case of the mwt LA tail domain. We believe that this extra energy is required because the Δ50 LA tail is more compact, which stabilizes the Ig-fold and results in an increase of the unfolding energy. From Figure 6C, the energy difference at 75 Å extension, corresponding to the first-stage transition shown in Figure 5C, is 65 kcal/mol, suggesting that the additional energy to fully unfold the Ig-fold may primarily be required for the first-state, hinge-like transition.
Figure 6. Force-extension curves of the unfolding of Ig-fold domains.
(A) A force-extension curve as recorded during the unfolding process of the mwt LA Ig-fold as shown in Figure 5B. (B) A force-extension curve as recorded during the unfolding process of the Δ50 Ig-fold as shown in Figure 5C. (C) The external work on the Ig-fold domain during the unfolding process as a function of the end-to-end distance, which is computed by integrating of the force-extension curves obtained by unfolding each of the structures shown in Figures 3A and B.
We now qualitatively compare our in vitro result with the in silico results and find that the Δ50 LA tail is more stable in both experiment and simulation. Experimentally, the thermal transition midpoints are TM,Δ50 =65.8 +/− 0.23°C for the Δ50 LA tail domain and TM,mwt =63.2 +/− 0.09 °C for the mwt LA tail domain, indicating that the Δ50 LA tail domain is more difficult to unfold. This experimental result agrees with the in silico result that additional energy was required to fully unfold the Δ50 LA tail domain structure. According to a calculation based on the Bell-Evans model (Bell, 1978; Evans and Ritchie, 1997; Rief et al., 1998), we arrive at Δwexp = 37.0 +/− 7.0 kcal/mol (62 +/− 12 kBT for T=300 K). This corresponds to the additional energy of Δwsim = 69.8 +/− 17.4 kcal/mol (117 +/− 29 kBT for T=300 K) needed to unfold 50% of the Δ50 LA tail more than the mwt LA tail in our experimental studies. We find that the computational results are on the same order of magnitude as the experimental result. The difference between the experiment and simulation results could be caused by the intrinsic difference of the two methods. In the SMD simulation approach, the force is applied only at the two ends of the strand with a finite loading rate, the structure unfolded along the prescribed direction and the force required to overcome the energy barriers is applied only in this one prescribed direction. In temperature mediated changes in tryptophan fluorometry, the unfolded form has more degrees of freedom and the rate of temperature increase is also intrinsically different from the loading rate used in our simulation. Also, experiments could possibly include simultaneous kinetic aggregation, which was not included in the simulation (see Materials and Methods for detail).
3. Discussion
Using a combination of theoretical and experimental methods, we have studied atomic conformations and nanomechanical stabilities of the tail domain of the mwt LA tail and a mutant associated with HGPS (Δ50 LA tail). Our results demonstrate that the Δ50 LA tail is more compact than the mwt LA tail domain. Most importantly the unfolding of the Δ50 LA tail domain requires a higher energy barrier to overcome unfolding, likely due to transitioning through a pseudo-stable intermediate state (Figures 3–6 and Table 1). This phenomenon is in agreement with experimental measurements using both tryptophan fluorometry and circular dichroism methods (Figures 4 and S7).
These findings highlight the need to keep the protein geometry in mind when trying to understand the molecular mechanisms of the disease. Previously, experimental works of wild type and HGPS cells have shown a stiffening of nuclei from HGPS patients and resistance to mechanical pressure, and alteration in the mechanical properties is due to the presence and overaccumulation of the Δ50 LA protein and associated filaments at the inner nuclear membrane (Dahl et al., 2004; Goldman et al., 2004). The etiology of HGPS has long been thought to be a function of the retained farnesyl group alone (Capell et al., 2008; Leuba et al., 1994), but our results suggest that changes in protein stability may be contributing to the disease pathology. This finding is consistent with recent studies which show that the presence of unfarnesylated prelamin A and Δ50 LA still have cellular and organism effects whose mechanisms are not yet entirely clear (Yang et al., 2011; Davies et al., 2010; Verstraeten et al., 2008).
Furthermore, our experimental and computational results quantitatively and consistently show that the Δ50 LA tail is a more stable structure (by approximately 37–70 kcal/mol or 62–117 kBT), as well as a more compact molecule. This suggests an explanation for the increased compactness of in vitro reconstituted filaments of full length Δ50 LA than mwt LA (Taimen et al., 2009). This compact filament structure, likely caused by a smaller and more stable Δ50 LA tail, may also partly be responsible for reduced turnover of proteins from the nucleoskeleton (Dahl et al., 2004; Goldman et al., 2004) and is thus much more than just a farnesyl-related issue. We also observe an increased propensity for Δ50 LA tail domain to form dimers, suggesting an increase in dimer-dimer interaction strength and as a result, a reduced capacity for the Δ50 LA tail domain to be reorganized by thermal fluctuations (moderated temperatures or moderate force), consisted with the extra energy and higher temperature that is required to unfold the Δ50 LA tail domain seen here. Filaments that include the more stable Δ50 LA may more readily aggregate, show reduced turnover, and due to their stronger binding appear mechanically more rigid and therefore “brittle”, thereby directly explaining earlier experimental findings (Dahl et al., 2004; Dahl et al., 2006; Delbarre et al., 2006) based on the geometric and mechanistic insight developed in this study. Our results may also explain a recent experimental report that showed that the proliferation of HGPS cells is significantly impaired (Verstraeten et al., 2008).
From a mechanistic viewpoint we have shown that the mwt LA tail unfolds by individual beta-strands being pulled away from the central structure in a sequential process. In contrast, the Δ50 LA tail domain shows a two-state transition process with a tertiary opening of the Ig-fold into two halves, which we termed a “beta-sheet pocket”. Within this pocket there may exist other “hidden” binding sites similar to the internal structure of the Ig-folds of titin (Marszalek et al., 1999) or fibronectin (Klotzsch et al., 2009) that may alter the affinity to binding partners. The two stage transition process may also alter the unfolding process of the assembled filament, and we hypothesize that this mechanism could imply that the unfolding of Δ50 LA lamin filament features a greater structural variability, while the process in case of wild-type lamin filament is smoother. This could explain differential binding of Δ50 LA to proteins and DNA (Bruston et al., 2010). We suggest that there may be other differential protein-protein binding associated with Δ50 LA, but this binding may be energy dependent, such as with force or temperature.
The “structure” of intrinsically disordered proteins are described as an ensemble of conformations which may represent conformations allowing for protein-protein binding to different binding partners and have been shown to be necessary for specific mechanical functions of biological materials including spider silk and talin rods (Keten et al., 2010; del Rio et al., 2009). Given lamin A's proposed role in mechanotransduction, and the multiple binding partners of the tail domain, further study of the conformations of the tail domain relating to mechanical function should be explored in future work (Wang et al., 2009). Here, we have demonstrated the use of an integrated experimental-computational approach to determine a group of structures to gain molecular-level insight into pathological mechanisms.
4. Materials and Methods
4.1 Protein concentration, purity and size characterization
Protein concentration was measured using Coomaisse Plus (Bradford) Assay (Pierce). Purified protein size and purity was determined using 14% SDS-PAGE, according to standard protocols. Dynamic light scattering (Malvern Zeta Sizer Nano) was used to measure protein hydrodynamic radius and possible aggregation state.
4.2 Protein expression and purification
Lamin A tail domains were created using PCR from plasmids kindly provided by Misteli at NCI/NIH (Dahl et al., 2004) to create tail fragments of mwt LA (386–646; Figure 1B) and Δ50 LA (Figure 1C) using primers. For the mwt LA tail domain with encoded EcoR1, Xho1 and stop codon we used: 5'-GAA TTC GCT ACG CCT GTC CCC CA-3' and 5'-CTC GAG TTA GTA GGA GCG GGT GAC CAG-3' and for Δ50 LA tail domain with EcoR1, Xho1 and stop codon we used: 5'-GAA TTC CTA CGC CTG TCC CCC AGC CC-3' and 5'-CTC GAG TTA CAT GAT GCT GCA GTT CTG GGG GCT-3'. PCR products were ligated into pGEMTez vectors, amplified in DH5α, and subcloned into the pGST parallel vector for recombinant expression (Sheffield et al., 1999). GST-tagged lamin A tails were transformed into BL21-CodonPlus competent E. coli (Agilent Technologies), grown 2–3 hours at 37°C, and induced by IPTG at OD600 of 1.8 (mwt LA: 400 uM IPTG for 3 hrs; Δ50= 800 uM IPTG for 2 hrs). Δ50 LA tail was produced more easily and was less susceptible to degradation, possibly suggesting resistance to protease activity (Parsell and Sauer, 1989). Cells were harvested by centrifugation at 6000×g for 10 min, lysed with B-PER reagent (Pierce), diluted by 5× in binding/wash buffer: 125 mM Tris, 500 mM NaCl, pH 8.0 and 100 uM PMSF and added to glutathione magnetic beads overnight at 4°C (Pierce). Beads were washed with 20 volumes with binding/wash buffer and exposed to proTEV protease for 5 hours at 4°C according to manufacturer's instructions (Promega Madison, WI). Cleaved protein was collected in the supernatant after centrifugation (1,000 × g for 5 min) and exposed to glutathione agarose beads (Pierce) pre-equilibrated in proTEV buffer to remove excess GST overnight at 4°C. Proteins were ultracentrifuged at 100,000 × g for 30 minutes at 4°C to remove aggregates.
4.3 Tryptophan fluorometry
For tryptophan fluorescence tail domains were diluted to 0.1 mg/mL in 50 mM HEPES, 0.5 mM EDTA, pH 7.0. Sample was excited at 295 nm in a 1 cm pathlength polished cell in a Fluorolog-3 (Jobin Yvon-spex, Instruments S.A., Inc., Edison, NJ) and the emission at 342 nm was monitored as the temperature was increased from 40°C to 90°C in 1°C increments, ≈every 1 min. The temperature was manually controlled using a water bath (Neslab RTE-210, ThermoScientific) attached to a custom built sample holder, allowing for temperature regulation.
4.4 Molecular simulation approach and analysis
Replica Exchange Molecular Dynamics (REMD) and Steered Molecular Dynamics (SMD) simulations are carried out using the CHARMM19 all-atom energy function with an effective Gaussian model for the water solvent (additional details about the simulation approach and analysis methods are provided in Supplementary methods part). For the mechanical analysis the α-carbon atoms at the two ends of the Ig-fold are connected to SMD springs (spring stiffness 10 kcal/Å2mol) and they move against each other at a constant relative speed of 0.01 Å/ps (Qin and Buehler, 2010; Qin et al., 2009). The applied pulling force is recorded versus its displacement. We use Visual Molecular Dynamics (VMD) (Humphrey et al., 1996) for visualization of protein structures (Figures 2, 3 and 5). We assess secondary structures by analyzing both the hydrogen bond patterns and the backbone geometry with the STRIDE algorithm (Frishman and Argos, 1995).
4.5 Extra energy needed to unfold Ig fold of Δ50 LA tail
According to the Bell-Evans model (Bell, 1978; Evans and Ritchie, 1997; Rief et al., 1998), the unfolding probability of a structure stabilized by molecular bonds is proportional to exp (−Eb/kBT) where Eb is the energy barrier height and kBT is the thermal energy. Thereby, for Δ50 LA at 50% unfolding in simulation, we have
| (3) |
Where <wM,Δ50>= 551.5 +/− 40.1 kcal/mol is the averaged external energy applied to unfold 50% of the Δ50 LA as the end-to-end length equals to half of the fully unfolded length (Figure 6C) and T0=27°C is the room temperature. Eq. (3) means the increasing temperature has a same effect at the intermediate unfolding stage as the external energy as shown in Figure S7, and from this equation we obtain EΔ50 = 4815.7 kcal/mol. In experiments:
| (4) |
And since both of the two structures are at the intermediate unfolding stage with a same unfolding probability. We have similar conclusion for the simulations as
| (5) |
By solving Eqs. (4) and (5), we have Δwexp = EΔ50(TM,Δ50 − T>M,mwt)/TM,Δ50 = 37.0 +/− 7.0 kcal/mol (=62 +/− 12 kBT for T=300 K), which corresponds to the extra energy needed to unfold 50% of the Δ50 LA more than the mwt LA. Since <wM,mwt >= 481.7 +/− 46.1 kcal/mol is obtained by simulation from Figure 6C, we obtain the extra energy term from simulation as Δwsim = <wM,Δ50−wM,mwt>= 69.8 +/− 17.4 kcal/mol (=117 +/− 29 kBT for T=300 K), which is in the same order of magnitude as the experimental result.
Supplementary Material
Acknowledgements
ZQ and MJB acknowledge support from AFOSR and DOD-PECASE. AK and KND acknowledge: R. Tilton, T. Przybycien (Chemical Engineering), B. Armitage (Chemistry) and G. Rule (Biology) at Carnegie Mellon University for use of their shared equipment, as well as N. Kolluri, S. Chapman, P. Silberman and J. Wert for their assistances in experiments. Funding from the Progeria Research Foundation (to KND) and NIH (NRSA F30AG030905 from NIA to AK) is also acknowledged. We acknowledge helpful discussions with Tom Mistelli.
Abbreviations
- AA
amino acid
- CD
circular dichroism
- DLS
dynamic light scattering
- DSC
differential scanning calorimetry
- HGPS
Hutchinson–Gilford progeria syndrome
- Ig
immuno-globulin
- REMD
replica exchange method
- SMD
steered molecular dynamics
- WLC
worm-like chain
- mwt LA
mature wild type lamin A
- Δ50 LA
HGPS Δ50 mutant lamin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–27. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Bruston F, Delbarre E, Ostlund C, Worman HJ, Buendia B, et al. Loss of a DNA binding site within the tail of prelamin A contributes to altered heterochromatin anchorage by progerin. FEBS Lett. 2010;584:2999–3004. doi: 10.1016/j.febslet.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell BC, Olive M, Erdos MR, Cao K, Faddah DA, et al. A farnesyltransferase inhibitor prevents both the onset and late progression of cardiovascular disease in a progeria mouse model. Proc. Natl. Acad. Sci. USA. 2008;105:15902–7. doi: 10.1073/pnas.0807840105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, Jung HJ, Li Z, Nobumori C, Yun UJ, et al. Direct synthesis of lamin A, bypassing prelamin a processing, causes misshapen nuclei in fibroblasts but no detectable pathology in mice. J. Biol. Chem. 2010;285:20818–26. doi: 10.1074/jbc.M110.128835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 2004;117:4779–86. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- Dahl KN, Scaffidi P, Islam MF, Yodh AG, Wilson KL, et al. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2006;103:10271–10276. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BS, Barnes RH, 2nd, Tu Y, Ren S, Andres DA, et al. An accumulation of nonfarnesylated prelamin A causes cardiomyopathy but not progeria. Hum. Mol. Genet. 2010;19:2682–94. doi: 10.1093/hmg/ddq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- Del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre E, Tramier M, Coppey-Moisan M, Gaillard C, Courvalin JC, et al. The truncated prelamin A in Hutchinson-Gilford progeria syndrome alters segregation of A-type and B-type lamin homopolymers. Hum. Mol. Genet. 2006;15:1113–22. doi: 10.1093/hmg/ddl026. [DOI] [PubMed] [Google Scholar]
- Eftink MR. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys. J. 1994;66:482–501. doi: 10.1016/s0006-3495(94)80799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong LG, Ng JK, Lammerding J, Vickers TA, Meta M, et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J. Clin. Invest. 2006;116:743–52. doi: 10.1172/JCI27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins-Struct. Funct. Gene. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2004;101:8963–8. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat. Rev. Mol. Cell Biol. 2007;8:562–73. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Keten S, Xu ZP, Ihle B, Buehler MJ. Nanoconfinement controls stiffness, strength and mechanical toughness of beta-sheet crystals in silk. Nat. Mat. 2010;9:359–367. doi: 10.1038/nmat2704. [DOI] [PubMed] [Google Scholar]
- Klotzsch E, Smith ML, Kubow KE, Muntwyler S, Little WC, et al. Fibronectin forms the most extensible biological fibers displaying switchable force-exposed cryptic binding sites. Proc. Natl. Acad. Sci. USA. 2009;106:18267–72. doi: 10.1073/pnas.0907518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Kigawa T, Koshiba S, Inoue M, Yokoyama S. Solution structure of immunoglobulin like domain of mouse nuclear lamin. RCSB Protein Database. DOI:10.2210/pdb1ufg/pdb. [Google Scholar]
- Krimm I, Ostlund C, Gilquin B, Couprie J, Hossenlopp P, et al. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure. 2002;10:811–23. doi: 10.1016/s0969-2126(02)00777-3. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, et al. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006;281:25768–80. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- Leuba SH, Yang G, Robert C, Samori B, van Holde K, et al. Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc. Natl. Acad. Sci. USA. 1994;91:11621–5. doi: 10.1073/pnas.91.24.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Schymkowitz J, Rousseau F, Diella F, Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 2004;342:345–53. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- Liu P, Kim B, Friesner RA, Berne BJ. Replica exchange with solute tempering: a method for sampling biological systems in explicit water. Proc. Natl. Acad. Sci. USA. 2005;102:13749–54. doi: 10.1073/pnas.0506346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kim DI, Syme J, LuValle P, Burke B, et al. Dynamics of lamin-A processing following precursor accumulation. PLoS One. 2010;5:e10874. doi: 10.1371/journal.pone.0010874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- Marszalek PE, Lu H, Li H, Carrion-Vazquez M, Oberhauser AF, et al. Mechanical unfolding intermediates in titin modules. Nature. 1999;402:100–3. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- Navarro CL, De Sandre-Giovannoli A, Bernard R, Boccaccio I, Boyer A, et al. Lamin A and ZMPSTE24 (FACE-1) defects cause nuclear disorganization and identify restrictive dermopathy as a lethal neonatal laminopathy. Hum. Mol. Genet. 2004;13:2493–503. doi: 10.1093/hmg/ddh265. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Sauer RT. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J. Biol. Chem. 1989;264:7590–5. [PubMed] [Google Scholar]
- Qin Z, Buehler MJ. Molecular dynamics simulation of the alpha-helix to beta-sheet transition in coiled protein filaments: evidence for a critical filament length scale. Phys. Rev. Lett. 2010;104:198304. doi: 10.1103/PhysRevLett.104.198304. [DOI] [PubMed] [Google Scholar]
- Qin Z, Kreplak L, Buehler MJ. Hierarchical structure controls nanomechanical properties of vimentin intermediate filaments. PLoS One. 2009;4:e7294. doi: 10.1371/journal.pone.0007294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher S, Pomes R. Molecular simulations of protein disorder. Biochem. Cell. Biol. 2010;88:269–90. doi: 10.1139/o09-169. [DOI] [PubMed] [Google Scholar]
- Rief M, Fernandez JM, Gaub HE. Elastically coupled two-level systems as a model for biopolymer extensibility. Phys. Rev. Lett. 1998;81:4764–4767. [Google Scholar]
- Schirmer EC, Foisner R. Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 2007;313:2167–79. doi: 10.1016/j.yexcr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 1999;15:34–9. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Shumaker DK, Lopez-Soler RI, Adam SA, Herrmann H, Moir RD, et al. Functions and dysfunctions of the nuclear lamin Ig-fold domain in nuclear assembly, growth, and Emery-Dreifuss muscular dystrophy. Proc. Natl. Acad. Sci. USA. 2005;102:15494–9. doi: 10.1073/pnas.0507612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow CD, Sorin EJ, Rhee YM, Pande VS. How well can simulation predict protein folding kinetics and thermodynamics? Ann. Rev. Biophys. Biomol. Struct. 2005;34:43–69. doi: 10.1146/annurev.biophys.34.040204.144447. [DOI] [PubMed] [Google Scholar]
- Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- Stierle V, Couprie J, Ostlund C, Krimm I, Zinn-Justin S, et al. The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochem. 2003;42:4819–28. doi: 10.1021/bi020704g. [DOI] [PubMed] [Google Scholar]
- Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- Taimen P, Pfleghaar K, Shimi T, Moller D, Ben-Harush K, et al. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc. Natl. Acad. Sci. USA. 2009;106:20788–93. doi: 10.1073/pnas.0911895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten VL, Ji JY, Cummings KS, Lee RT, Lammerding J. Increased mechanosensitivity and nuclear stiffness in Hutchinson-Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell. 2008;7:383–93. doi: 10.1111/j.1474-9726.2008.00382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Bio. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. Analysis of peptaibol sequence composition: implications for in vivo synthesis and channel formation. Eur. Biophys. J. 2004;33:233–7. doi: 10.1007/s00249-003-0348-1. [DOI] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- Wilson KL, Berk JM. The nuclear envelope at a glance. J Cell Sci. 2010;123:1973–8. doi: 10.1242/jcs.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp. Cell Res. 2007;313:2121–33. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Ostlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2010;2:a000760. doi: 10.1101/cshperspect.a000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Chang SY, Ren S, Wang Y, Andres DA, Spielmann HP, Fong LG, Young SG. Absence of progeria-like disease phenotypes in knock-in mice expressing a nonfarnesylated version of progerin. Hum. Mol. Gen. 2011;20:436–444. doi: 10.1093/hmg/ddq490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SG, Fong LG, Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria--new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 2005;46:2531–58. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- Young SG, Meta M, Yang SH, Fong LG. Prelamin A farnesylation and progeroid syndromes. J. Biol. Chem. 2006;281:39741–5. doi: 10.1074/jbc.R600033200. [DOI] [PubMed] [Google Scholar]
- Zastrow MS, Vlcek S, Wilson KL. Proteins that bind A-type lamins: integrating isolated clues. J. Cell Sci. 2004;117:979–87. doi: 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- Zhang S, Iwata K, Lachenmann MJ, Peng JW, Li S, et al. The Alzheimer's peptide a beta adopts a collapsed coil structure in water. J. Struct. Biol. 2000;130:130–41. doi: 10.1006/jsbi.2000.4288. [DOI] [PubMed] [Google Scholar]
- Zhou RH, Berne BJ, Germain R. The free energy landscape for beta hairpin folding in explicit water. Proc. Natl. Acad. Sci. USA. 2001;98:14931–14936. doi: 10.1073/pnas.201543998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.