Abstract
Epithelial to mesenchymal transition (EMT) is an extreme example of cell plasticity, important for normal development, injury repair, and malignant progression. Widespread epigenetic reprogramming occurs during stem cell differentiation and malignant transformation, but EMT-related epigenetic reprogramming is poorly understood. Here we investigated epigenetic modifications during TGF-β-mediated EMT. While DNA methylation was unchanged during EMT, we found global reduction of the heterochromatin mark H3-lys9 dimethylation (H3K9Me2), increase of the euchromatin mark H3-lys4 trimethylation (H3K4Me3), and increase of the transcriptional mark H3-lys36 trimethylation (H3K36Me3). These changes were largely dependent on lysine-specific deaminase-1 (Lsd1), and Lsd1 loss-of-function experiments showed marked effects on EMT-driven cell migration and chemoresistance. Genome-scale mapping revealed that chromatin changes were largely specific to large organized heterochromatin K9-modifications (LOCKs), suggesting that EMT is characterized by reprogramming of specific chromatin domains across the genome.
Introduction
Epithelial-mesenchymal transition (EMT) is a paradigm of cell plasticity characterized by reversible loss of epithelial characteristics coupled with gain of mesenchymal properties1,2. Recent evidence suggests that cells undergoing EMT also acquire stem cell traits3, and neoplastic epithelial cells undergoing EMT might represent the fraction of cancer “stem” cells thought to be present in many carcinomas4.
Epigenetic modifications are potentially reversible alterations in DNA methylation or chromatin not associated with changes in DNA sequence, which specify functional outputs from the DNA template and are often heritable through cell division5,6. Genome-wide reprogramming of epigenetic modifications occurs during both germ cell development and differentiation of stem cells into somatic cells7, and the pathologic reprogramming of these modifications contributes to carcinogenesis and other human diseases5,8. Because EMT is characterized by reversible changes in cell type with acquisition of both stem cell and malignant-type traits, and is important for normal development, injury repair, cancer, and other human diseases9, we hypothesized that epigenetic changes across the genome might also occur during EMT.
Results
TGF-β-mediated EMT induces changes in chromatin morphology
We induced EMT with transforming growth factor beta (TGF-β), a pleiotropic signaling molecule widely used to induce EMT in numerous lineages of cultured epithelial cells10,11. We utilized AML12 mouse hepatocytes, since these cells preferentially undergo EMT when growth arrested, confluent cultures are treated with TGF-β12–15. These cells are also not a transformed cell line and should be relatively free of epigenetic changes that might arise secondary to neoplastic transformation16.
EMT was confirmed by loss of the epithelial adhesion protein E-cadherin and gain of the mesenchymal marker vimentin, both hallmarks of EMT (Supplementary Fig. 1a). These changes were reversible upon removing TGF-β from the media and allowing cells to redifferentiate (Supplementary Fig. 1a). AML12 cells also displayed the EMT-characteristic pattern of reduced cytoplasmic and membranous E-cadherin immunostaining in response to TGF-β (Supplementary Fig. 1b). Immunostains with Ki67 confirmed that nearly all of the cells were growth arrested and not entering the cell cycle at the beginning of the experiments and during the TGF-β treatments (Supplementary Fig. 1c), and bromodeoxyuridine (BrDU) confirmed that cell proliferation was minimal during these experiments (Supplementary Fig. 1d). We also stained the cells with hematoxylin and eosin (H&E), since this procedure is the gold standard by which pathologists visualize changes in cellular morphology17 (Fig. 1a). As expected, TGF-β treatment resulted in reversion from the epithelioid shape of fully differentiated AML12 cells to the classic stellate shape observed during EMT. However, we were surprised to find that TGF-β also provoked striking changes in nuclear morphology, with nuclear enlargement, oval shape, and transition to “open”, hypochromatic chromatin with scattered punctate nucleoli (Fig. 1a). Examination of nuclear ultrastructure by transmission electron microscopy showed a smooth, uniform euchromatin pattern with scattered nucleoli in cells treated with TGF-β, consistent the H&E stains. Strikingly, TGF-β treated cells showed loss of the electron-dense heterochromatin areas that were observed invaginating from the nuclear periphery in differentiated cells (Fig. 1b). These observations are reminiscent of nuclear features seen in cells reacting to injury in histopathology specimens, and suggested to us that global epigenetic changes in DNA methylation or chromatin modifications might occur during EMT.
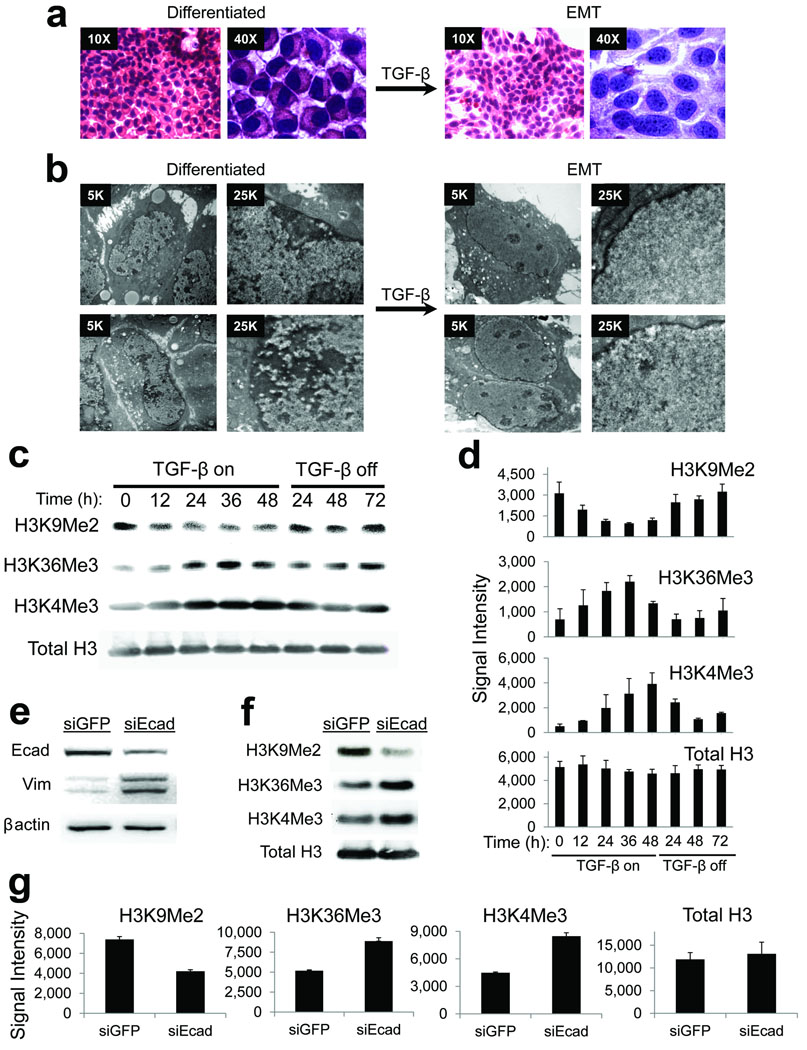
Figure 1. Global changes in bulk chromatin modifications during EMT.
(a) Hematoxylin and eosin (H&E) stains of differentiated AML12 cells and those treated with TGF-β, highlighting stellate morphology, nuclear enlargement, and altered chromatin texture during EMT.
(b) Electron microscopy of differentiated AML12 cells and those treated with TGF-β, highlighting smooth chromatin, scattered nucleoli, and loss of peripheral heterochromatin during EMT.
(c) Changes in bulk chromatin modifications during EMT. Representative western blots for the indicated histone H3 modifications of bulk histones from AML12 cells treated with TGF-β for the indicated timepoints. “TGF-β on” refers to timepoints during which cells were treated with TGF-β, which “TGF-β off” refers to timepoints after which TGF-β was removed from the media, allowing termination of EMT and re-differentiation to hepatocytes (as in Supplementary Figure 1). There is a reversible reduction in H3K9Me2 and increase in H3K4Me3 and H3K36Me3 in response to TGF-β. Total levels of histone H3 remain relatively constant during the treatments.
(d) Densitometry quantification summarizing western blots of histone modifications (including those depicted in Fig. 1B, n=2–3 biological replicates) for indicated times of TGF-β treatments. Data is plotted as raw signal intensities indicated on the x-axis. Error bars represent S.E.M.
(e) Western blots of whole cells extracts from AML12 cells treated with siRNA against E-cadherin (siEcad) or control siRNA against GFP (siGFP) shows reduced expression of E-cadherin and increased vimentin in response to siEcad, consistent with induction of EMT.
(f) Changes in bulk chromatin modifications in response to siEcad. Representative western blots for the indicated H3 modifications of bulk histones treated as indicated show similar changes as seen if Fig. 1c.
(g) Densitometry quantification summarizing western blots of histone modifications (including those depicted in Fig. 1F, n=2 biological replicates) for indicated siRNA treatments. Data is plotted as raw signal intensities indicated on the x-axis. Error bars represent S.E.M.
DNA methylation is preserved across the genome during EMT
Based on the changes in chromatin morphology, we first looked for genome-scale alterations in DNA methylation during EMT. Prolonged culture of proliferative epithelial cells undergoing oncogene-induced, irreversible EMT can eventually result in DNA methylation alterations at some genes18, and transformation-induced changes in DNA methylation might predispose carcinoma cells to undergo EMT9. However, there have been no comprehensive investigations into natural changes in DNA methylation during a round of EMT in a non-neoplastic setting, free of transformation-associated epigenetic instability. We investigated genome-wide DNA methylation during EMT in AML12 cells using comprehensive high throughput arrays for relative methylation (CHARM)19,20. This approach examines approximately 4.6 million CpG sites in the genome including almost all CpG islands as well as lower CpG density regions. These experiments were strongly negative, as pairwise comparisons of different timepoints post-TGF-β stimulation showed no observable changes in DNA methylation during EMT (e.g., Supplementary Fig. 2). Thus, DNA methylation patterns are faithfully retained during TGF-β treatments and may not strongly contribute to epigenetic plasticity during EMT in this system. These findings could have implications regarding preservation of epithelial cell fate during reversible mesenchymal transitions.
Bulk chromatin modifications are globally reprogrammed during EMT
We then asked whether chromatin modifications change during EMT. We began by investigating bulk levels of histone H3K9Me2, which seemed an appropriate target as it is found within large, non-repetitive facultative heterochromatin domains21–24. Western blotting showed a reduction in bulk H3K9Me2 by TGF-β, which was reversible upon removing TGF-β from the media (Fig. 1c, d). Immunostaining by immunofluorescence and immunohistochemistry showed that these changes were nuclear (Supplementary Fig. 3a), consistent with alterations in chromatin. Double immunostaining with H3K9Me2 and E-cadherin confirmed that loss of nuclear H3K9Me2 occurred in cells that also lost membranous staining of E-cadherin (Supplementary Fig. 3b). Levels of total H3 remained relatively constant throughout TGF-β treatments (Fig. 1c, d), and vehicle treated control cells did not display loss of H3K9Me2 (Supplementary Fig. 4). Thus, in contrast to DNA methylation, H3K9Me2, at least bulk levels, appeared to diminish during TGF-β mediated EMT.
Outside of S-phase, heterochromatin inhibits transcription25, and genes located within heterochromatin marked by H3K9Me2 are silent21,22,24. We therefore tested whether chromatin modifications associated with transcription might also change during EMT, in parallel with loss of H3K9Me2. Because histone H3K36Me3 within gene bodies is stimulated by active transcription26–28, we assayed for changes in bulk levels of H3K36Me3 during EMT. Western blot assays showed that bulk levels of H3K36Me3 increased in response to TGF-β (Fig. 1c, d), and this was reversible upon removal of TGF-β from the media although levels had not fallen completely back to baseline by the end of the time course. Gain of H3K36Me3 during EMT was also visible within nuclei by immunostaining (Supplementary Fig. 5). Despite loss of H3K9Me2, we did not observe an increase in H3K9 acetylation (Supplementary Fig. 6), a modification sometimes found on demethylated H3 lys9 residues within euchromatin. This observation could have implications regarding re-establishment of H3K9Me2 upon termination of the EMT program, since H3K9 acetylation inhibits H3K9 methylation. It would thus be interesting to investigate whether repeated rounds of EMT might result in replacement of H3K9Me2 with H3K9 acetylation in future studies. We next tested whether bulk levels of histone H3K4Me3 might increase during EMT in parallel to H3K36Me3, since H3K4Me3 is typically deposited around transcription start sites to attract protein complexes that remodel chromatin into an open, euchromatic state27,28. Indeed, H3K4Me3 displayed reversible increases in bulk levels upon TGF-β stimulation by western blot (Fig. 1c, d), similar to changes seen for H3K36Me3. Vehicle treated cells did not display similar changes in any of these modifications (Supplementary Fig. 4). Collectively, these results indicate that levels of chromatin modifications associated with active transcription and euchromatin increase during EMT in this system, in parallel with loss of heterochromatin.
We also wanted to confirm that reprogramming of bulk chromatin modifications was due to AML12 cells undergoing EMT, rather than a nonspecific effect of TGF-β treatments. Loss of E-cadherin expression by siRNA knockdown can trigger EMT in the absence of exogenous signaling molecules such as TGF-β 9,29,30. We therefore tested whether siRNAs directed against E-cadherin (siEcad) might induce similar changes in bulk chromatin modifications as TGF-β treatments. Incubation of AML12 cultures with siEcad resulted in loss of E-cadherin expression and increases in vimentin by western blot, consistent with induction of EMT (Fig. 1e). siEcad treatments also resulted in reduced H3K9Me2 with increased H3K4Me3 and H3K36Me3 (Fig. 1f, g), similar to induction of EMT with TGF-β. These findings support our conclusion that global reprogramming of chromatin modifications is due to AML12 cells undergoing EMT, rather than a nonspecific byproduct of TGF-β signaling.
EMT chromatin reprogramming is Lsd1-dependent
We next sought to identify proteins that might regulate the bulk changes in histone modifications during EMT. Loss of H3K9Me2 in growth arrested cells suggests this may be due to enzymatic removal, rather than a dilutional effect secondary to chromatin replication. Indeed, we found that protein expression of Lsd1, a dual histone H3 K4 and K9 demethylase31,32, reversibly increased during EMT (Fig. 2a). Normally, Lsd1 is complexed with co-repressors that direct H3K4 demethylation and heterochromatin assembly31,33. However, under some circumstances, Lsd1 can also complex with co-activators that direct H3K9 demethylation and euchromatin assembly32,34,35. We therefore screened for Lsd1 interactions by large-scale immunoprecipitation of endogenous Lsd1 from nuclear extracts of AML12 cells treated with either vehicle or TGF-β, followed by mass spectrometry (Fig. 2b, Supplementary Table 1). Although some known Lsd1 interacting proteins were common to both states (e.g. Bhc80, Ctbp, Hmg20), vehicle-treated extracts yielded other proteins including known Lsd1 co-repressors (e.g. Rest, CoRest). TGF-β-treated extracts instead yielded a distinct set of proteins, many of which were transcriptional coactivators and have been implicated in regulation of EMT, chromatin structure, and oncogenesis, including several catenins. Thus, we observed increased Lsd1 protein expression in response to TGF-β. Lsd1 in turn interacted with distinct sets of proteins in TGF-β and vehicle-treated AML12 cells, suggesting that it might function differently in cells undergoing EMT as opposed to differentiated cells.
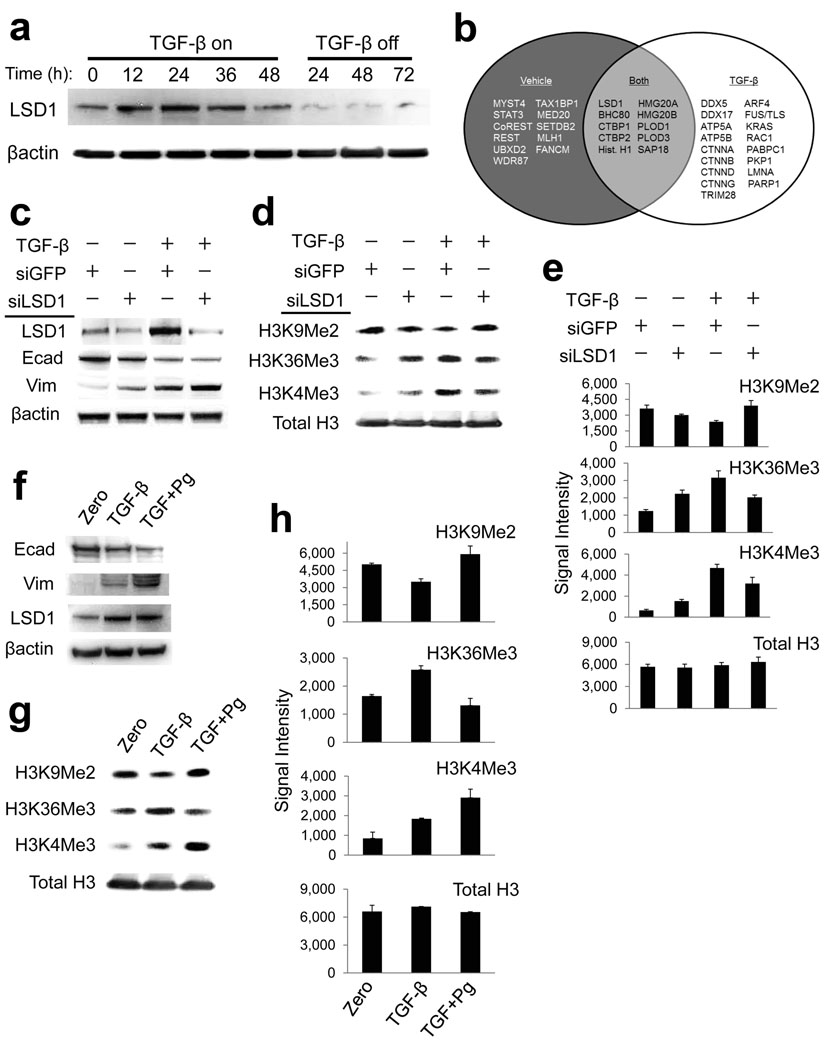
Figure 2. Lsd1 regulates bulk changes in chromatin modifications during EMT.
(a) Western blot of whole cell extracts from AML12 cells treated with TGF-β for the indicated time points, as in Fig. 1. Lsd1 protein expression is increased by TGF-β treatment.
(b) Venn diagram summarizing proteins that preferentially immunoprecipitated with Lsd1 from differentiated AML12 cells (vehicle) and those undergoing EMT (TGF-β). Proteins were identified by large-scale IP of endogenous Lsd1 from nuclear extracts followed by mass spectrometry of immunoprecipitates that had been separated on a polyacrylamide gel. “Both” refers to proteins that immunoprecipitated with Lsd1 under both conditions. We detected several known Lsd1 interactions during both conditions (e.g. BHC80, CTBP1, HMG20A). In contrast, several other interactions were specific for vehicle-treated cells including proteins involved in heterochromatin assembly (e.g. CoREST, STAT3, UBXD2). TGF-β treated cells instead yielded several proteins important for regulation of chromatin, EMT, and oncogenesis (e.g. several catenins, DEAD box RNA helicases, Rac1, PARP1).
(c) Western blots of the indicated proteins from AML12 cells treated with or without (vehicle-only) TGF-β, and incubated with the siRNAs listed in the panel above the blots. “+” denotes cells that received the indicated treatments, “−“ denotes cells that did not. siRNAs against Lsd1 but not GFP (control) knocks down Lsd1 expression but does not prevent upstream events that initiate EMT (loss of E-cadherin, gain of vimentin), as shown by western blot.
(d) Western blots of H3K9Me2, H3K36Me3, and H3K4Me3, as in Fig. 2C. siLsd1 prevented loss of H3K9Me2, and partially interfered with gain H3K36Me3 and H34Me3 in response to TGF-β. Total levels of histone H3 remain relatively constant.
(e) Densitometry quantification plots summarizing western blot data for the indicated siRNA treatments (including those depicted in Fig. 2D, n=2 biological replicates). Data is plotted as raw signal intensities indicated on the x-axis. Error bars represent S.E.M.
(f) Pargyline treatment does not affect E-cadherin, vimentin or Lsd1. AML12 cells were incubated with either TGF-β alone or co-incubated with TGF-β (TGF) and pargyline (Pg) as indicated. Pargyline did not prevent induction of upstream aspects of EMT (E-cadherin, vimentin) and did not affect expression of Lsd1, shown by western blots.
(g) Pargyline treatment blocks chromatin changes during EMT. Pargyline prevents loss of bulk H3K9Me2 and gain of H3K36Me3 in response to TGF-β, while total levels of histone H3 remain relatively constant. H3K4Me3 levels rise even further.
(h) Densitometry quantification summarizing western blots for pargyline experiments (including those depicted in Fig. 2G, n=2 biological replicates). Data is plotted as raw signal intensities indicated on the x-axis. Error bars represent S.E.M.
We therefore conducted Lsd1 loss of function experiments to test whether Lsd1 might direct the EMT-associated changes in bulk chromatin modifications. siRNAs directed against Lsd1 (siLsd1) knocked down protein expression of Lsd1 without interfering with the normal decrease in E-cadherin or normal increase in vimentin in response to TGF-β (Fig. 2c). Although siLsd1 had no effect upon initiation of these upstream aspects of EMT (E-cadherin and vimentin), siLsd1 blocked TGF-β-mediated loss of H3K9Me2, and siLsd1 partially blocked the TGF-β-mediated increases in bulk H3K36Me3 and H3K4Me3 (Fig. 2d, e), suggesting that Lsd1 regulates chromatin reprogramming downstream of events (E-cadherin and vimentin) that initate EMT. We also conducted Lsd1 loss of function experiments with pargyline, a monoamine oxidase inhibitor that inhibits the ability of Lsd1 to demethylate H3K9Me2 within chromatin32, with the caveat that pargyline, like most inhibitors, could have off-target effects that influence EMT independent of Lsd1. Similar to siLsd1, treatment with pargyline did not interfere with normal loss of E-cadherin or gain of vimentin in response to TGF-β (Fig. 2f) and actually appeared to accelerate these changes. The normal rise in Lsd1 expression was unaffected (Fig. 2f). Strikingly, despite enhanced loss of E-cadherin and vimentin, pargyline treatments completely blocked loss of H3K9Me2 and gain of H3K36Me3 in cells treated with TGF-β (Fig. 2g, h), again suggesting that Lsd1 globally reprograms these chromatin modifications downstream of changes in E-cadherin and vimentin. In contrast to siLsd1, pargyline treatments resulted in further increases in H3K4Me3 in response to TGF-β (Fig. 2g, h), possibly due to in vivo inhibition of Lsd1 or other demethylases toward H3K4 (discussed further in subsequent sections). We conclude that loss of H3K9Me2 and gain of H3K4Me3 and H3K36Me3 during EMT is in part dependent on Lsd1, and that pargyline abrogates Lsd1-dependent loss of H3K9Me2 and gain of H3K36Me3.
Lsd1 regulates EMT-driven cell motility and chemoresistance
We then wished to functionally test the role of Lsd1-mediated chromatin changes during EMT. We hypothesized that Lsd1-dependent changes in bulk chromatin might regulate phenotypic aspects of EMT downstream of events (e.g. loss of E-cadherin) that help initiate EMT in response to TGF-β. We tested this hypothesis with Lsd1 loss-of-function assays as above. A hallmark EMT phenotype is increased cell motility, which confers migratory capabilities. We therefore conducted scratch assays to test whether Lsd1 loss-of-function might affect migration of AML12 cells treated with TGF-β. Cultures treated with TGF-β displayed enhanced migration of individual cells into the scratched area relative to vehicle treated cells (Fig. 3a–d), consistent with increased motility during EMT. Interestingly, although siLsd1 partially increased migration in vehicle control cells, siLsd1 partially inhibited cell migration in cells treated with TGF-β (Fig. 3a, b), consistent with divergent functions for Lsd1 in differentiated cells verses those undergoing EMT. Strikingly, pargyline completely inhibited migration of cells treated with TGF-β, and cells regained migratory capacity upon removal of pargyline from the media (Fig. 3c, d). The total abolishment of EMT-driven cell migration by pargyline is impressive and raises the possibility that off-target effects of this inhibitor on other proteins that regulate cell motility might synergize with inhibition of Lsd1 to completely block cell migration during EMT. Thus, loss of Lsd1 function by both siRNA knockdown and pargyline interfered with cell migration during EMT, suggesting that Lsd1-mediated changes in chromatin modifications may be important for execution of this EMT phenotype.
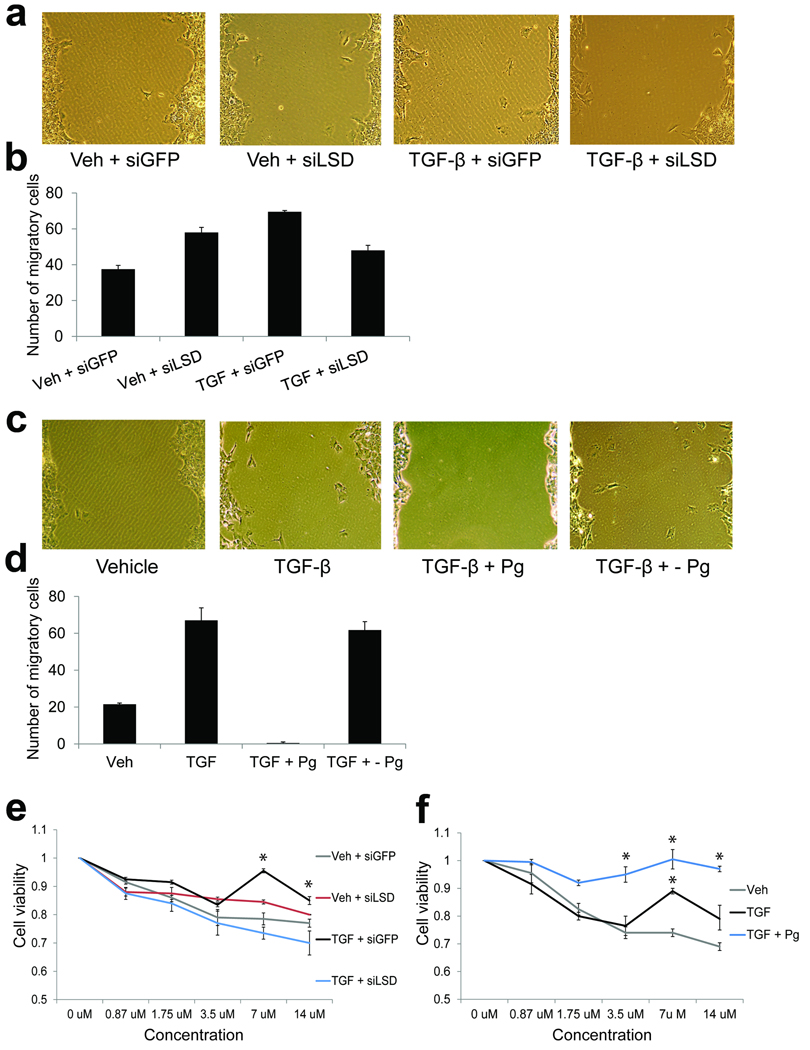
Figure 3. Lsd1 regulates certain EMT phenotypes.
(a) Light microscopy of representative areas of scratch assays performed on cultures of AML12 cells treated as indicated. siLsd1 partially inhibited cell migration as evidenced by reduced numbers of individual cells that have migrated into the scratched area. Interestingly, siLsd1 conveyed increased migration in vehicle-treated cells.
(b) Quantification of representative scratch assays (n=2) as above. Individual cells within the scratch area were counted over 10 microscopic fields for each condition. The average of total number of migratory cells are plotted, error bars represent the standard deviation. siLsd1 partially blocked cell migration in TGF-β-treated cells.
(c) Light microscopy of representative areas of scratch assays performed on cultures of AML12 cells treated as indicated. + - refers to cells that were treated with TGF-β and pargyline (Pg) for 36hrs, followed by removal of pargyline from the media for 36hrs. Pargyline inhibited cell migration as evidenced by the lack of individual cells that have migrated into the scratched area.
(d) Quantification of representative scratch assays as in (b). Individual cells within the scratch area were counted over 10 microscopic fields for each condition. The average of total number of migratory cells are plotted, error bars represent the standard deviation. Pargyline completely blocked cell migration.
(e) Representative dose response curves of AML12 cells treated as indicated, exposed to increasing concentrations of doxorubicin. Plots are absorbance values normalized to no drug (DMSO-only), and indicate cell viability. siLsd1 abolished resistance to doxorubicin (n=2, error bars represent the standard deviation, asterisks represents values with p < 0.5 relative to vehicle + siGFP controls).
(f) Representative dose response curves of AML12 cells treated as indicated, exposed to increasing concentrations of doxorubicin. Plots are absorbance values normalized to no drug (DMSO-only), and indicate cell viability. Pargyline conferred enhanced resistance to doxorubicin (n=2, error bars represent the standard deviation, asterisks represents values with p < 0.5 relative to vehicle controls).
We next examined chemoresistance, another phenotype acquired by cells undergoing EMT. We utilized the topoisomerase inhibitor doxorubicin, which causes cytotoxicity via induction of DNA double strand breaks (DSBs) leading to cell death. Relative to vehicle-treated controls, AML12 cultures treated with TGF-β displayed statistically significant resistance to cell death at high concentrations of doxorubicin (p=0.037 and 0.030 for 7uM and 14uM doxorubicin in siRNA experiments, p=0.042 for 7uM doxorubicin in pargyline experiments), as measured by calorimetric assays (Fig. 3e, f). Strikingly, cultures co-incubated with TGF-β and siLsd1 failed to show a similar statistically significant resistance to cell death (p=0.142 and 0.232 at 7um and 14uM doxorubicin), indicating that siLsd1 completely abolished chemoresistance in cells undergoing TGF-β-mediated EMT (Fig. 3e). In stark contrast, co-treatment with TGF-β and pargyline showed markedly enhanced chemoresistance, with nearly complete resistance even at concentrations where TGF-β alone had no effect (p=0.016 and 0.023 at 3.5uM and 14uM doxorubicin, Fig. 3f). These intriguing results suggest that protein expression of Lsd1 is required to confer EMT chemoresistance whereas Lsd1 demethylase activity may antagonize this phenotype, and highlight how different loss-of-function strategies can sometimes interfere with different aspects of enzyme function, leading to remarkably different effects on the same phenotype. We also note that siLsd1 and pargyline treatments resulted in opposite effects on H3K4Me3 during EMT (Fig. 2d–h). Deposition of H3K4Me3 at sites of programmed DSBs is required for V(D)J36,37 and meiotic recombination38, and H3K4Me3 can directly mediate repair of DSBs in response to exogenous genotoxic agents, independent of its role in transcription39. Because siLsd1 reduced EMT-driven H3K4me3 and chemoresistance while pargyline further enhanced these two events, we hypothesize that H3K4Me3 might contribute to Lsd1-regulated chemoresistance. We are currently investigating mechanisms whereby Lsd1-dependent chromatin reprogramming, including H3K4Me3, contributes to repair of DNA damage and chemoresistance during EMT in other studies (as outlined in the discussion).
Genome-scale mapping reveals that chromatin reprogramming is targeted to LOCKs
To gain further insight into whether the changes in bulk chromatin were important events during EMT, we wished to identify regions of the genome where these changes were located. LOCKs are large (100Kb–5Mb) non-repetitive heterochromatin domains enriched for H3K9Me2, which overlap with nuclear lamina-associated domains24,40. We first examined levels of H3K9Me2 within LOCKs by chromatin immunoprecipitation followed by genomic DNA microarray analysis (ChIP-chip) spanning mouse chromosomes 4–14. While LOCKs were visible in identical locations in TGF-β treated and untreated cells, we found that the quantitative levels of H3K9Me2 were reduced across 95.9% (821 of 856) of the LOCKs detected on the arrays (Supplementary Database 1, e.g. Fig. 4a–c), which was not observed for H3K9Me2 in non-LOCK regions (p=0, Supplementary Fig. 7). These results were verified by replicate ChIP-chip and replicate ChIP-qPCR analyses (Supplementary Fig. 8). These findings are reminiscent of the quantitative abatement of LOCKs seen in cultured embryonic stem cells and cancer cells24.
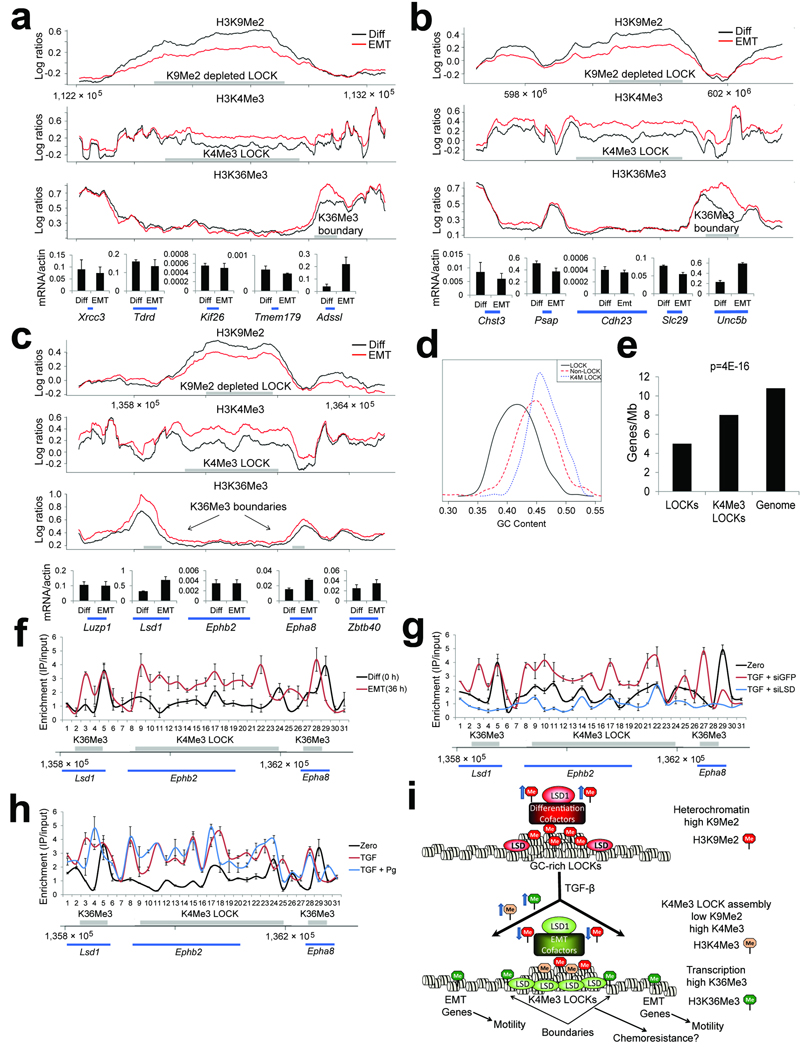
Figure 4. ChIP-chip analysis of EMT reveals alterations of chromatin in LOCK domains.
(a–c) Representative plots of ChIP-chip enrichments of histone modifications over loci from mouse chromosomes 12 (A), 10 (B) and 4 (C) during TGF-β induced EMT. Locations of genes that were assayed by RT-PCR are shown at the bottom. Regions where we detected LOCKs (top panel, H3K9Me2), K4Me3 LOCKs (middle panel, H3K4Me3), and peaks of H3K36Me3 (bottom panel, H3K36Me3) are displayed as gray bars. Chromatin was harvested from differentiated AML12 cells (time 0 hrs) and from AML12 cells undergoing EMT (TGF-β treated, 36 hrs). There is reduction in H3K9Me2 within LOCK regions, coupled with enrichment of H3K4Me3 in the same regions. Areas with enriched H3K36Me3 are present over genes at the boundaries of these LOCKs. See Fig. S8–9 for replicates of various regions, including region shown in Fig. 4C by ChIP-qPCR. Graphs underneath the ChIP-chip plots show RT-PCR results for the genes beneath them, demonstrating that genes at LOCK boundaries that acquire H3K36Me3 are upregulated during EMT.
(d) Histogram comparing GC content of LOCKs, K4Me3 LOCKs, and the whole genome. Whereas LOCKs are AT-rich overall, those that acquire H3K4Me3 during EMT are GC-rich.
(e) Gene content in all LOCKs, K4Me3 LOCKs, and the whole genome. Whereas LOCKs are gene-poor overall, the subset of LOCKs that acquire K4Me3 during EMT are gene-enriched over LOCKs that do not.
(f) ChIP assays for Lsd1 across the locus depicted in Fig. 4C. ChIP assays were performed for Lsd1 and quantitative real-time PCR was performed with 31 primers spaced across the locus. Gray bars represent where we detected a K4Me3 LOCK and H3K36Me3-enriched boundaries by ChIP-chip. Whereas Lsd1 binding is restricted to the 5’-end of the Lsd1 and Epha8 genes in differentiated cells, Lsd1 is enriched across the entire K4Me3 LOCK during EMT.
(g) ChIP assays for Lsd1 as in Fig. 4F show that chromatin isolated from AML12 cells co-treated with TGF-β and siLsd1 shows loss of Lsd1 enrichment within the K4Me3 LOCK during EMT.
(h) ChIP assays for Lsd1 as in Fig. 4F show that chromatin isolated from AML12 cells cotreated with TGF-β and pargyline shows retention of Lsd1 enrichment within the K4Me3 LOCK during EMT.
(i) Model of LOCK reprogramming during EMT. In differentiated AML12 cells, LOCKs have high levels of H3K9Me2. Lsd1 is complexed with proteins that may facilitate heterochromatin assembly at LOCKs. During TGF-β-mediated EMT, Lsd1 spreads from LOCK boundaries into the LOCKs and other proteins converge upon Lsd1, which may assist in directing demethylation of H3K9Me2 and recruitment of H3K4Me3. H3K9Me2 within all LOCKs is reduced, and specific, GC-rich LOCKs acquire H3K4Me3 adjacent to sites of transcription. These K4Me3 LOCKs might function in a surveillance DDR pathway, providing enhanced chemoresistance during EMT. H3K36Me3 is targeted to K4Me3 LOCK boundaries and numerous EMT-related genes located in non-LOCK regions across the genome, which are involved in conferring cell motility during EMT.
We next mapped the locations of H3K36Me3 by ChIP-chip. As expected27,28, we detected peaks of H3K36Me3 over gene coding regions, many of which were identical in differentiated AML12 cells and those treated with TGF-β. When we searched for regions with altered levels of H3K36Me3 during EMT, we observed that numerous regions harboring genes across the genome displayed elevated levels of H3K36Me3 in cells treated with TGF-β (1,736 total regions harboring 1,701 known genes, Supplementary Database 1, e.g. Fig. 4a–c). Interestingly, most of these genes were located in gene-rich regions between LOCKs, rather than within LOCKs themselves (1,231 of 1,701, 72.3%). GO analysis41 of genes with elevated H3K36Me3 during EMT revealed that these genes encode proteins with EMT-related functions, including cytoskeletal remodeling, cell migration, cell adhesion, Ras signaling, and various enzymatic activities (Supplementary Table 2). These findings are consistent with inhibition of cell migration by siLsd1 and pargyline, which block accumulation of bulk H3K36Me3 during EMT.
We next mapped H3K4Me3 enrichments by ChIP-chip. As expected28, most enrichments manifested as sharp peaks of H3K4Me3 directly over transcription start sites. Most of these were identical in differentiated and TGF-β treated cells and could not account for the increase in H3K4Me3 during EMT. Surprisingly, when we searched for the regions responsible for increased bulk H3K4Me3 during EMT, we identified 219 large (100kb–1.4Mb, avg. 260kb), discrete regions of the genome displaying broad enrichments (rather than sharp peaks) of newly acquired H3K4Me3. Strikingly, most (86.8%) of these new domains were distributed over a subset of 190 LOCKs (p=0, Supplementary Table 3 and Database 1, e.g. Fig. 4a–c), comprising 21.1% of all LOCKs detected on our arrays (p=0, Supplementary Table 3). Although LOCKs on average are AT-rich and gene poor, the K4Me3 LOCKs were GC- and gene-rich (Fig. 4d, e). We also noticed that one-third of K4Me3 LOCKs were flanked by regions with newly acquired H3K36Me3 at their boundaries (p=0, Fig. 4a–c, Supplementary Table 3), and that the majority (76%) of K4Me3 LOCKs were flanked by regions with either pre-existing or newly acquired H3K36Me3 (p=0, Supplementary Table 3). GO analysis of genes located within K4Me3 LOCKs revealed sets of genes that were distinct from those enriched with H3K36Me3, including those involved in glycoprotein activities, ion balance, plasma membrane signaling, extracellular functions, and development (Supplementary Table 4). Replicate ChIP-qPCR analysis over a region from chromosome 4 supported the ChIP-chip findings for H3K36Me3 and H3K4Me3 at this locus (Supplementary Fig. 9).
Although many genes situated at LOCK boundaries were enriched with H3K36Me3 during EMT, most known genes within K4Me3 LOCKs did not acquire this modification (18 of 262, 6.9%), suggesting that genes at the boundaries but not within the K4Me3 LOCKs themselves are transcriptionally active during EMT. Indeed, we observed elevated mRNA expression of genes situated at K4Me3 LOCK boundaries but not those within the LOCKs during EMT by RT-PCR, and absolute mRNA levels of genes located in the interior of K4Me3 LOCKs were very low relative to other genes (Fig. 4a–c). Further studies are needed to determine whether K4Me3 LOCKs serve to “poise” chromatin for transcriptional activation similar to what is proposed for bivalent domains harboring both H3K4Me3 and H3K27Me342 or whether they serve other functions during EMT, such as triggering release of LOCK subsets from the nuclear lamina or participating in a surveillance DNA damage response (DDR) as proposed for large-scale changes in chromatin structure43.
Lsd1 is enriched within a K4Me3 LOCK during EMT
We next selected a K4Me3 LOCK to map chromatin-bound Lsd1 in differentiated AML12 cells and in those undergoing TGF-β-mediated EMT. We investigated the region depicted in Fig. 4c, since we found it interesting that the Lsd1 gene itself was present at one of the LOCK boundaries. We therefore performed ChIP for Lsd1 with 31 PCR primers spaced across this 500Kb locus. In differentiated cells, we observed peaks of Lsd1 present over the 5’-regions of the genes situated at the LOCK boundaries (Lsd1 and Epha8). These findings are similar to other studies in lower eukaryotes that have reported peaks of Lsd1 enrichment at euchromatin-heterochromatin boundaries, where it functions to maintain boundaries between large heterochromatin and euchromatin domains44. Remarkably, Lsd1 was enriched across the entire K4Me3 LOCK in chromatin isolated from cells treated with TGF-β, in addition to new Lsd1 peaks within the gene bodies of Lsd1 and Epha8 (Fig. 4f). The appearance of Lsd1 across an entire LOCK depleted of H3K9Me2, in conjunction with blockade of H3K9Me2 demethylation by siLsd1 and pargyline, suggests that Lsd1 acquires the ability to demethylate H3K9Me2 substrates during EMT.
In contrast to the loss of Lsd1 within this K4Me3 LOCK induced by siLsd1 (Fig. 4g), pargyline did not affect Lsd1 enrichment across this LOCK during EMT (Fig. 4h). This is not unexpected since pargyline inhibits the demethylase activity but not protein expression of Lsd1. Given that the spectrum of proteins that interacted with Lsd1 shifted toward transcriptional co-activators during EMT (Fig. 2b), including several catenins which can recruit H3K4Me3 methyltransferase activity to chromatin45, the increase in EMT-driven H3K4Me3 during pargyline treatments noted earlier (Fig. 2g, h) is likely due to recruitment of such proteins by Lsd1 during EMT, coupled with inhibition of H3K4 demethylase activity by pargyline. Determination of the precise mechanisms whereby Lsd1 mediates epigenetic reprogramming of LOCKs and other regions of the genome during EMT will require extensive investigations into the roles of the various Lsd1 protein interactions identified in Fig. 2b, including genome-wide mapping of Lsd1 and its EMT interacting partners.
Discussion
In summary, our findings illustrate the dynamic nature of genome-scale epigenetic reprogramming in an experimental model of EMT, characterized by genome-wide reprogramming of large heterochromatin domains (LOCKs) to a state of reduced H3K9Me2, new LOCK-wide modifications of H3K4Me3 at specific GC-rich LOCKs, and enrichment of H3K36Me3 at LOCK boundaries and numerous EMT-related genes across the genome (Fig. 4i). This reprogramming may be critical for proper execution of EMT functions, as inhibition of bulk chromatin changes with Lsd1 loss-of-function experiments had marked effects on cell migration and chemoresistance. Finally, it is probable that many other chromatin modifications not examined here are also reprogrammed during EMT. Indeed, repression of epithelial-specific genes during EMT may be mediated by H3K27Me3 via a Snai1-dependent process, and forced overexpression of Snai1 can also recruit Lsd1 to epithelial genes to assist in repression46, suggesting that Lsd1 might play dual roles in repressing and activating areas of the genome important for EMT. In this regard, the balance of Lsd1-interacting factors identified in Fig. 2b may play a crucial role in directing Lsd1 toward activation or repression.
It has been proposed that cells undergoing EMT acquire stem cell traits3,4. Our findings of quantitative abatement of LOCKs and elevated levels of H3K36Me3 across the genome during EMT supports this notion at the level of chromatin, as previous studies have demonstrated similar results for cultured ESCs24,47. In contrast, other studies have reported that the reverse process, mesenchymal to epithelial transition (MET), is required to produce induced pluripotent stem cells (iPSCs) from fibroblasts 48,49. These findings raise the possibility that the reverse chromatin changes described here might occur during generation of iPSCs. Alternatively, the process described in the current work could be a general mechanism for increasing epigenetic plasticity and reprogramming, regardless of cell fate directionality.
EMT is also characterized by acquisition of malignant-type traits. Intriguingly, loss of LOCKs is seen in various malignant human cancer cell lines24, similar to our EMT results. We further observed H3K36Me3 enrichment at numerous genes involved in cell motility during EMT, another trait common to both EMT and malignancy. Indeed, Lsd1 loss-of-function experiments interfered with both H3K36Me3 and cell migration during EMT. We also consider that a shift in balance from heterochromatic (H3K9Me2) to euchromatic (H3K4Me3) and transcription-coupled (H3K36Me3) modifications may increase susceptibility to DNA damaging agents during stress, injury, or malignancy. Other cells with euchromatic genomes appear to activate a surveillance DDR to cope with this threat43, although this could generate chromosomal rearrangements at sites of hyperactive transcription50,51. Thus, if a surveillance DDR is activated concomitant with elevated transcription across the genome during EMT, this might confer robust repair of DNA damage at the expense of generating mutations.
This latter possibility would be compatible with a surveillance role for H3K4Me3 in repair of DSBs during EMT, consistent with the effects of siLsd1 and pargyline on H3K4Me3 and chemoresistance. Our ChIP-chip results revealed that K4Me3 LOCKs are specifically situated in large gene and CG-rich regions of the genome flanked by genes with high levels of transcription, which are prone to acquire DSBs in response to genotoxic agents such as doxorubicin50–52. Indeed, components of the DDR (such as γH2AX) must spread several hundred kilobases from a DSB to facilitate repair, and heterochromatin inhibits this process43. We therefore hypothesize that one function of K4Me3 LOCKs might be to provide a euchromatic scaffold in gene-rich regions of the genome, that allows efficient spreading of the DDR through large heterochromatin domains in response to DSBs that might occur during EMT, thereby conferring robust repair of DSBs and enhanced chemoresistance.
Like stem cell differentiation7,27, germ cell development53, and malignant transformation5,8, epigenetic reprogramming during EMT is mediated by widespread changes in chromatin modifications. Unlike reprogramming in these other cell types, DNA methylation patterns are faithfully preserved during EMT. Further studies in other cell systems are needed to determine whether our findings are a general property of EMT, and whether similar epigenetic reprogramming occurs in other physiological contexts.
Methods
Cell culture
AML12 cells were cultured as described16, supplemented with 10mM glutamine. Briefly, cells between passage 7 and 19 were grown to confluence in 10% serum. Cells were then growth arrested by serum starvation (0.5%) for 48hrs. Recombinant TGF-β1 (5ng mL−1, R&D systems) or vehicle was then added to the media for the indicated times. For redifferentiation, cells were treated for 36hrs followed by washes with PBS and replacement of the TGF or vehicle containing media with new serum starvation media (0.5% serum) and incubated for the allotted times. For siRNA experiments, cells were pretreated with oligofectamine (Invitrogen) containing siRNAs against GFP, Lsd1, or E-cadherin (silencer select, Ambion, s97505 and s63752) when cells were 70% confluent. Cells were subsequently taken through the TGF-β protocol upon reaching confluence the next day, as described above. For pargyline experiments, cells were co-incubated with TGF-β and 3mM Pargyline (Sigma). Cell viability was determined by absorbance with CellTiter96 (Promega). Doxorubicin was purchased from Sigma and dissolved in DMSO. P-values were obtained by running TGF-β-treated samples against vehicle-treated controls (vehicle+siGFP, Fig. 3e, vehicle alone, Fig. 3f) using two-tailed t-tests with Microsoft Excel software. Scratch assays were performed by making a single scratch down the middle of a well (in 6-well plates) with the sharp edge of a P1000 pipette tip, at the beginning of TGF-β and pargyline treatments. After 36hrs, individual cells within the scratch were counted as migratory cells. Cells still adhesed to other cells at the edge of the scratched areas were not counted. Cells treated with TGF-β + pargyline were then washed with PBS and the media replaced with TGF-β without pargyline, allowed to incubate, and counted after 36hrs.
Staining procedures
For immunofluorescence, cells were grown on coverslips and fixed with either paraformaldehyde (H3K9Me2, E-cadherin) at 37°C or methanol (H3K36Me3) at − 20°C. Images were visualized with ImageJ software. Cells harvested for electron microscopy were fixed in glutaraldehyde and further processed and imaged by the microscopy facility at Johns Hopkins. For immunohistochemistry, cells were grown on coverslips and fixed with acetone. Immunostains were detected with DAB substrate (Abcam). Cells grown on coverslips were fixed with ethanol for hematoxylin and eosin staining. Antibodies with dilutions are as follows: (Ecadherin, Cell Signaling, 1:200) (H3K9Me2, Abcam 1220, 1:500), (H3K36Me3, Abcam, 1:300).
CHARM analysis
CHARM assays were performed as described19. AML12 cells were harvested at various timepoints (zero, 24 h, 36 h) of TGF-β treatments. Cells were snap frozen in liquid nitrogen and stored at −80°C. DNA was later extracted and CHARM assays undertaken. Representative plots are shown in Supplementary Fig 2. The degree of DNA methylation is indicated by p (percent methylation) value versus genomic location, where the curve represents averaged smoothed p values. The location of CpG dinucleotide (black tick marks on x axis), CpG density (smoothed black line) calculated across the region using a standard density estimator, location of CpG islands (orange line), as well as gene annotation indicating the transcript (thin outer gray line), coding region (thin inner gray line), exons (filled gray box) and gene transcription directionality on the y axis (sense marked as +, antisense as −) are also shown. Examples are shown for Rab30 and Rplp1.
Western blots, ChIP, IP
Unless otherwise specified, cells taken for all assays were washed with ice cold PBS, scraped, snap frozen in liquid nitrogen, and stored at −80° C until further harvesting. Histones were extracted as described54. Whole cell extracts were prepared with RIPA buffer. All experiments with protein contained Complete EDTA-free protease inhibitor cocktails (Roche). Western blots were performed according to standard protocols. An iblot apparatus (Invitrogen) was used for protein transfer and a bio-imager for image analysis. Densitometry was performed with ImageJ software. Antibodies are as follows: (K9Me2, Abcam 1220), (K9Ac, Abcam ab12179), (K4Me3, Abcam 8580), (K36Me3, Abcam, ab9050), (Ecadherin, Cell Signaling), (Vimentin V9, Neomarkers), (β-actin, Sigma), (Lsd1, Abcam, ab17721).
Native ChIP with micrococcal nuclease digestion of native chromatin (to mononucleosomes) was performed as described24. ChIP experiments for Lsd1 were performed by similar methods except cells were fixed with 1% formaldehyde and chromatin was sonicated rather than digested with nuclease. For ChIP-chip, 1–2ug of whole genome amplified or straight genomic DNA from ChIP experiments was hybridized to Nimblegen 2.1M economy 2 and 3 mouse ChIP-chip genome arrays. These are whole-genome tiling arrays with probes spaced at 203bp intervals which cover the entirety of mouse chromosomes 4–14, excluding repetitive DNA regions. Data normalization is described in the statistical methods section. Plots were visualized with SignalMap software. Real-time PCR of ChIP samples with Fast Sybergreen (Applied Biosystems) was carried out on 10ng genomic DNA from input and IP samples with 31 primer pairs spaced over a 500Kb region on mouse chromosome 4. Enrichments of IP:Input were calculated as described55. RT-PCR was performed iScript RT-PCR kits (BioRad) followed by amplification of transcripts from cDNA by real-time PCR with Fast Sybergreen. Primer sequences for ChIP assays and RT-PCR are listed in Supplementary Table 5.
For large-scale IPs, 20mg of fresh nuclear extract isolated from 30 150mm plates of AML12 cells treated with either TGF-β or vehicle were incubated with Lsd1 polyclonal antibody (Abcam) under conditions that favor isolation of Lsd1 complexes56. Briefly, 20mg of extract was pre-cleared with rabbit IgG cross-linked to protein A dynabeads (Invitrogen) for 2hrs at 4°C. The extracts were then incubated overnight with 40ug Lsd1 antibody and captured by a 2hr incubation with Protein A dynabeads. Complexes were washed 4X in IP buffer and eluted with 200ug Lsd1 peptide (Abcam). Eluates were run on a 4–12% polyacrylamide gel and stained with simplyblue coomassie (Invitrogen). The entire lane of protein for each condition was excised at 2mm intervals (27 cut gel strips) and submitted to the University of Arkansas mass spec facility for mass spectrometry analysis. Data was analyzed with scaffold 3 software.
Microarray data analysis
For each microarray we first conducted within-array normalization to correct probe sequence effects and position biases. We then did partial quantile normalization to correct for bias across different arrays. LOCKs were defined in a similar way as in Wen et al. To identify enriched H3K36Me3 peaks and H3K4Me3 regions, we first smoothed the differences in log-ratios from 0- and 36-hour samples by moving average with different window sizes (10k base pairs for peaks and 100k base pairs for blocks). Enriched peaks or blocks are defined as the genomic regions where smoothed values are above 1.5 times median absolute value (MAD) of all the smoothed values. A detailed description is provided below. P-values were obtained by running 1000 permutation tests. Data was analyzed in R, and scripts are available upon request.
Normalization
Within-array normalization was performed to remove the spatial and probe sequence-dependent artifacts using an approach similar to previous studies24. Between-array normalization was performed using a partial quantile normalization algorithm as described24. As a result of data normalization, the log2 ratios between the ChIP and control intensities from all microarrays roughly follow the same probability distribution in the reference (non-methylated) regions.
Peak and block detection
LOCKs were identified using method similar to that previously described24. Peaks of H3K36Me3 were identified using a moving average based method. For each array, we first smoothed the log-ratios between the ChIP and control intensities using moving average with a window size of 10k base pairs (bp). Genomic regions with the smoothed values above 1.5 times median absolute value (MAD) of all the smoothed values were deemed peaks. We required the peaks to be at least 5000 bp long and contain at least 10 probes. Enriched peaks were detected using a similar way. We first computed the moving averages of the differences in log-ratios from 36- and 0-hour samples. Enriched peaks or blocks are defined as the genomic regions where smoothed values are above 1.5 times MAD of all the smoothed values.
The broadly enriched regions of H3K4Me3 were obtained in two steps. In the first step we excluded the genomic regions where there were peaks in data from 0-hour sample. That is because H3K4 are highly tri-methylated in promoter regions of many genes common to both the 0-hour and 36-hour arrays, so there are many sharp peaks in H3K4Me3 data. These peaks bring extra noise in broad region detection because they often have big impacts on the moving average values. We therefore excluded them. To do so we first detected the peaks at 0-hour sample using a moving average approach. Log2 ratios were smoothed with a 2000-bp window. The cutoff for peaks was determined so that the false discovery rate (FDR) for peaks is 10%. Genomic regions above the cutoff, with both ends extended by 2500bp are defined as H3K4Me3 peaks. In the second step, we computed the moving averages of the differences in log-ratios without the probes in the 0-hour H3K4Me3 peaks, using a window of 100k bp. Broadly enriched H3K4Me3 regions (e.g. K4Me3 LOCKs) were defined as genomic regions where smoothed values are above 1.5 times MAD of all the smoothed values.
Assessing the relationship among different regions
Throughout the paper, a genomic region A is “flanked by” another region B is defined as that either boundary of region A is within 100k bp of region B. All the statistical significances (p-values) were computed based on randomization tests. For example to assess the overlapping of a list of regions (A) and another list of regions (B), we first randomly sample the same number of regions from the genome as in region list A. The lengths of the random regions match the lengths of regions in list A. We than compute the percentage of the random regions overlapping list B. Repeating such process for many times, we will obtain the null distribution of the overlapping percentage. A p-value of the overlapping between list A and list B is computed as the percent of times when the random overlapping percentage is bigger than the observed overlapping percentage.
Data Processing
All original graphs, images, and text presented in the figures were generated by Microsoft Excel (graphs), Adobe Photoshop or Image J software (images), and Adobe Illustrator or Microsoft Powerpoint (text) and converted to publication quality images with Adobe Illustrator and Photoshop. Images were created in accordance with the guidelines of the Nature publishing group for image integrity.
Supplementary Material
Acknowledgements
This work was supported by NIH grant 5R37CA054358 to A.P.F. We thank Alan Tackett for expert assistance with mass spectrometry experiments and data analysis, Chad McCall for helpful advice on large-scale IP experiments, the microscopy facility at Johns Hopkins for electron microscopy, and Karen Reddy and Sean Taverna for helpful advice. OGM and APF conceived of this work, designed the experiments, and wrote the manuscript. HW performed the statistical analyses. WT performed the immunofluorescence. AD performed the CHARM.
Footnotes
Accession Codes
NCBI GEO: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE28581
Original ChIP-chip data are deposited under accession number GSE28581.
Original CHARM data are deposited under accession number GSE27968.
Competing Interests: The authors declare no competing conflicts of interests.
References
- 1.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of Clinical Investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Mani SA, et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta PB, et al. Identification of Selective Inhibitors of Cancer Stem Cells by High-Throughput Screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 6.Jenuwein T, Allis CD. Translating the Histone Code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 7.Hawkins RD, et al. Distinct Epigenomic Landscapes of Pluripotent and Lineage-Committed Human Cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. The Epigenomics of Cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roussos ET, et al. AACR Special Conference on Epithelial-Mesenchymal Transition and Cancer Progression and Treatment. Cancer Research. 2010;70:7360–7364. doi: 10.1158/0008-5472.CAN-10-1208. [DOI] [PubMed] [Google Scholar]
- 10.Rahimi RA, Leof EB. TGF-beta signaling: A tale of two responses. Journal of Cellular Biochemistry. 2007;102:593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- 11.Xu J, Lamouille S, Derynck R. TGF-[beta]-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dooley S, et al. Hepatocyte-Specific Smad7 Expression Attenuates TGF-β-Mediated Fibrogenesis and Protects Against Liver Damage. Gastroenterology. 2008;135:642–659. e46. doi: 10.1053/j.gastro.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Pan X, Lei W, Wang J, Song J. Transforming growth factor-beta1 induces epithelial-to-mesenchymal transition and apoptosis via a cell cycle-dependent mechanism. Oncogene. 2006;25:7235–7244. doi: 10.1038/sj.onc.1209712. [DOI] [PubMed] [Google Scholar]
- 14.Kaimori A, et al. Transforming Growth Factor-β Induces an Epithelial-to-Mesenchymal Transition State in Mouse Hepatocytes in Vitro. Journal of Biological Chemistry. 2007;282:22089–22101. doi: 10.1074/jbc.M700998200. [DOI] [PubMed] [Google Scholar]
- 15.Pan X, et al. Nitric oxide suppresses transforming growth factor-β1–induced epithelial-to-mesenchymal transition and apoptosis in mouse hepatocytes. Hepatology. 2009;50:1577–1587. doi: 10.1002/hep.23156. [DOI] [PubMed] [Google Scholar]
- 16.Wu JC, Merlino G, Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. PNAS. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills SE, C D, Greenson JK, Oberman HA, Reuter V, Stoler MH. Sternberg's Diagnostic Surgical Pathology. Philadelphia, PA: Lippincott Williams & Wilkins; 2004. pp. 1–2709. [Google Scholar]
- 18.Dumont N, et al. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. PNAS. 2008;105:14867–14872. doi: 10.1073/pnas.0807146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Research. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes & Development. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice JC, et al. Histone Methyltransferases Direct Different Degrees of Methylation to Define Distinct Chromatin Domains. Molecular Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 23.Peters AHFM, et al. Partitioning and Plasticity of Repressive Histone Methylation States in Mammalian Chromatin. Molecular Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 24.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 26.Kizer KO, et al. A Novel Domain in Set2 Mediates RNA Polymerase II Interaction and Couples Histone H3 K36 Methylation with Transcript Elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Onder TT, et al. Loss of E-Cadherin Promotes Metastasis via Multiple Downstream Transcriptional Pathways. Cancer Research. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 30.Zheng G, et al. Disruption of E-Cadherin by Matrix Metalloproteinase Directly Mediates Epithelial-Mesenchymal Transition Downstream of Transforming Growth Factor-β in Renal Tubular Epithelial Cells. The American journal of pathology. 2009;175:580–591. doi: 10.2353/ajpath.2009.080983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y, et al. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog Lsd1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Metzger E, et al. Lsd1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, et al. Lsd1 Is a Subunit of the NuRD Complex and Targets the Metastasis Programs in Breast Cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y-J, et al. Regulation of Lsd1 Histone Demethylase Activity by Its Associated Factors. Molecular Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, et al. Opposing Lsd1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A Plant Homeodomain in Rag-2 that Binds Hypermethylated Lysine 4 of Histone H3 Is Necessary for Efficient Antigen-Receptor-Gene Rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthews AGW, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faucher D, Wellinger RJ. Methylated H3K4, a Transcription-Associated Histone Modification, Is Involved in the DNA Damage Response Pathway. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001082. e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 41.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 42.Bernstein BE, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 43.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chosed R, Dent SYR. A Two-Way Street: Lsd1 Regulates Chromatin Boundary Formation in S. pombe and Drosophila. Molecular cell. 2007;26:160–162. doi: 10.1016/j.molcel.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes & Development. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase Lsd1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29:4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Efroni S, et al. Global Transcription in Pluripotent Embryonic Stem Cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samavarchi-Tehrani P, et al. Functional Genomics Reveals a BMP-Driven Mesenchymal-to-Epithelial Transition in the Initiation of Somatic Cell Reprogramming. Cell stem cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 49.Li R, et al. A Mesenchymal-to-Epithelial Transition Initiates and Is Required for the Nuclear Reprogramming of Mouse Fibroblasts. Cell stem cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Lin C, et al. Nuclear Receptor-Induced Chromosomal Proximity and DNA Breaks Underlie Specific Translocations in Cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haffner MC, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ju B-G, et al. A Topoisomerase II{beta}-Mediated dsDNA Break Required for Regulated Transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 53.Feng S, Jacobsen SE, Reik W. Epigenetic Reprogramming in Plant and Animal Development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat. Protocols. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 55.Litt MD, Simpson M, Recillas-Targa F, Prioleau M-N, Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee MG, Wynder C, Norman J, Shiekhattar R. Isolation and characterization of histone H3 lysine 4 demethylase-containing complexes. Methods. 2006;40:327–330. doi: 10.1016/j.ymeth.2006.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.