Abstract
ATOH8 is a bHLH domain transcription factor implicated in the development of the nervous system, kidney, pancreas, retina and muscle. In the present study, we collected sequence of ATOH8 orthologues from 18 vertebrate species and 24 invertebrate species. The reconstruction of ATOH8 phylogeny and sequence analysis showed that this gene underwent notable divergences during evolution. For those vertebrate species investigated, we analyzed the gene structure and regulatory elements of ATOH8. We found that the bHLH domain of vertebrate ATOH8 was highly conserved. Mammals retained some specific amino acids in contrast to the non-mammalian orthologues. Mammals also developed another potential isoform, verified by a human expressed sequence tag (EST). Comparative genomic analyses of the regulatory elements revealed a replacement of the ancestral TATA box by CpG-islands in the eutherian mammals and an evolutionary tendency for TATA box reduction in vertebrates in general. We furthermore identified the region of the effective promoter of human ATOH8 which could drive the expression of EGFP reporter in the chicken embryo. In the opossum, both the coding region and regulatory elements of ATOH8 have some special features, such as the unique extended C-terminus encoded by the third exon and absence of both CpG islands and TATA elements in the regulatory region. Our gene mapping data showed that in human, ATOH8 was hosted in one chromosome which is a fusion product of two orthologous chromosomes in non-human primates. This unique chromosomal environment of human ATOH8 probably subjects its expression to the regulation at chromosomal level. We deduce that the great interspecific differences found in both ATOH8 gene sequence and its regulatory elements might be significant for the fine regulation of its spatiotemporal expression and roles of ATOH8, thus orchestrating its function in different tissues and organisms.
Introduction
bHLH transcription factors play very important regulatory roles during embryonic development, e.g. in neurogenesis, myogenesis, hematopoiesis, sex determination, and gut development [1]. In animals, bHLH proteins have been classified into six groups, named A, B, C, D, E, and F, based on their phylogenetic relationships and different biochemical properties [2]. ATOH8 belongs to group A of bHLH transcription factors [1]. Specifically, it is classified as a member of NET family within the atonal superfamily which includes families of NeuroD, Neurogenin, Atonal, Oligo, Beta3, Delilah, Mist and NET [2], [3]. In general, proteins of the atonal family are encoded by one single exon and are involved in neurogenesis [4]. Exceptionally, ATOH8 is encoded by 3 exons and is implicated in multiple developmental events in addition to neurogenesis. In the fruit fly, the ATOH8 orthologue, NET, is involved in the wing vein morphogenesis [5]. In the mouse, ATOH8 (MATH6) induces neurogenesis but inhibits gliogenesis in the developing retina [4]. Mouse ATOH8 is also involved in podocyte differentiation during kidney development [6] and endocrine pancreas development [7]. Inactivation of ATOH8 results in embryonic lethality in mice [7]. In the zebrafish, ATOH8 is expressed in the developing retina and somites, and knockdown of ATOH8 results in malformation of the retina and skeletal muscles [8]. A recent study shows that ATOH8 inhibits neuronal differentiation in the developing retina in the chicken [9]. Besides, the level of ATOH8 expression increases in U2OS cells transfected with Cyclin-B1-EGFP fusion gene [10], and altered expression levels of ATOH8 are detected in human patients who suffer from oligodendrogliomas [11]. An altered ATOH8 expression level is also reported in glioblastoma multiforme [12]. Considering these multiple implications of ATOH8 in different biological, developmental and pathological processes, we were intrigued to know if the function and regulation of expression of ATOH8 are conserved across different evolutionary lineages.
In the present study, we assembled ATOH8 sequence of different species from the GenBank and our re-sequencing data, analyzed the phylogeny of ATOH8 and performed multiple sequence comparisons. The results show that among metazoans, ATOH8 has experienced a high sequence divergence which makes inferences on basal metazoan relationships difficult. Within the analyzed vertebrate species, the evolutionary relationship of ATOH8 gene is mostly compliant with the accepted classification of the analyzed species. The bHLH domain of vertebrate ATOH8 was highly conserved. Mammals developed another potential isoform during evolution. Some amino acids are absent in zebrafish, frog and chicken ATOH8 compared to the mammalian orthologues. Vertebrates had a TATA-box type element that secondarily shifted to CpG-island type in mammals except for the opossum. The opossum ATOH8 displayed an evolved structure with an extended C-terminus, accompanied with an absence of CpG islands and TATA elements in the regulatory region. Gene mapping showed that the ATOH8 host chromosome, chromosome 2 in human, has two orthologous chromosomes in non-human primates. Experimentally, we identified the effective promoter of human ATOH8 which could drive the expression of reporter genes in the chicken embryo. To summarize, while ATOH8 maintains the conserved bHLH domain, it shows a high sequence diversification among different evolutionary lineages, in particular when comparing orthologous loci in invertebrate species. This great interspecific diversity may contribute to the functional and regulatory diversification of this gene. Our data demonstrate an example of the diversification pattern of a gene encoding a transcription factor among different species and bring insight into the functional study of ATOH8 in different animal models.
Results
Gene structure of vertebrate ATOH8
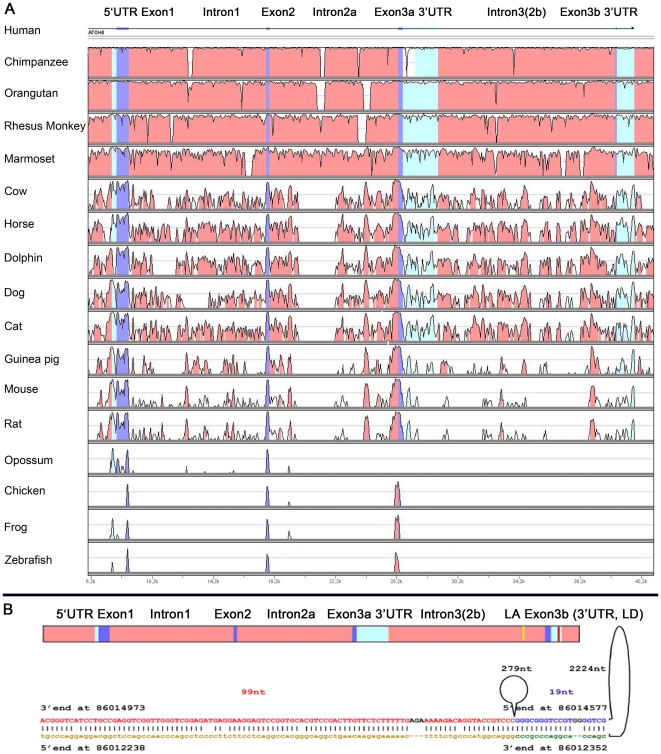
To determine the gene structure of ATOH8 in vertebrates, we filled the sequence gaps of ATOH8 gene loci of seven species (Accession No of Chimpanzee: FN868887.1; cat: FN868891.1; dog: FN868890.1; horse: FN868888.1; dolphin: FN868889.1; rhesus monkey FN868886.1 and chicken: FN868883). With these data supplementing the ATOH8 genomic sequences from ensemble genome browser (http://www.ensembl.org/index.html), we carried out the genomic sequence analysis among 17 vertebrate species, using the human ATOH8 genomic sequence as reference (Fig 1A). Our analysis showed that four exons which we termed as exon1, exon2, exon3a and exon3b were present within ATOH8 gene locus of eutherian mammals. ATOH8 consists of exon1, exon2 and exon3b as annotated in GenBank. Exon3a was verified by an EST (AL831857.1) from the amygdala of the human brain, which with exon1 and exon2 may form another isoform of ATOH8. The ATOH8 gene locus in opossum, chicken, frog and zebrafish contains three exons as well (exon1, exon2 and exon3b), although exon3b shows no significant homology to that of eutherian mammals. We do not know if exon3a exists in these four species or not. In all analyzed species except the opossum, exon3b encodes only one amino acid, the glutamic acid. Instead, exon 3b of the opossum encodes a dozen of amino acids resulting in an extended C-terminus.
Figure 1. Gene structure of ATOH8.
(A) Interspecies comparisons of ATOH8 gene loci. In the plot, the horizontal axis represents the sequence of the base genome, and the amplitude represents the degree of sequence conservation. With the hATOH8 gene locus as a standard reference sequence, the ATOH8 gene locus is divided into several regions, the exons (blue for protein coding regions and light cyan for UTRs) and the introns (pink). In vertebrates, ATOH8 consists of three exons (exon1, exon2 and exon3b shown in blue color) and an exon3a is predicted to encode a novel C-terminus of hATOH8 isoform 2. (B) A potential DNA loop in primate ATOH8 gene loci. One fragment is located in the intron between exon3a and exon3b as a potential loop acceptor (LA, marked with yellow); and the second one is located within the exon3b as a loop donor (LD, marked with red). The pink color marks introns, cyan the UTRs and blue the exons. The sketch of potential DNA loop derived from human ATOH8 is presented in the lower part of the figure.
With closer analysis we identified a pair of highly complementary sequences in primates, of which one was located in the intron between exon3a and exon3b referred to as loop acceptor (LA), and the complementary sequence was located in exon3b named as loop donor (LD) (Fig 1B and Table S1). This pair of complementary sequences can base-pair each other to potentially introduce a DNA loop (Fig 1B), which might provide additional regulatory facet for ATOH8 gene in primates.
Phylogenetic analysis of ATOH8 gene
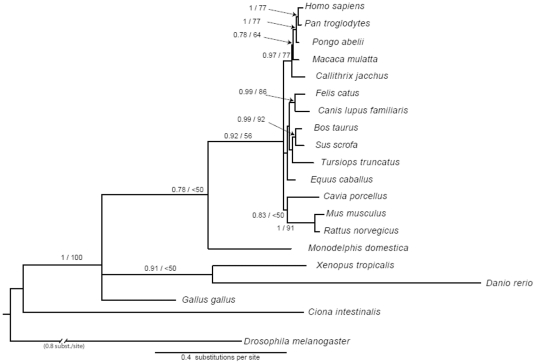
To study the evolutionary history of the ATOH8 gene, we first constructed the phylogenetic tree for the vertebrate ATOH8 (Fig 2), on the basis of the complete coding sequences of 20 species (Table S2). The translation alignment of the vertebrate ATOH8 sequences using MAFFT was trivial and resulted in a compact 1,188 bp alignment. When adding the outgroup sequences of the ATOH8 orthologue, NET, from the insect Drosophila melanogaster and the sea squirt Ciona intestinalis, the alignment was expanded to 2713 bp due to long insertions mainly in C. intestinalis. For the nucleotide multiple sequence alignment, jModeltest determined the GTR+G nucleotide substitution model as the most appropriate for the phylogenetic reconstruction.
Figure 2. Bayesian phylogenetic tree of ATOH8 gene.
The gene tree was built on the basis of complete coding DNA sequence alignment (2713 bp including gaps) of 20 species. The fruit fly NET gene was used as outgroup. Only branches with a posterior probability of at least 0.75 are shown. Branch support indicates the posterior probabilities and Maximum Likelihood Bootstrap.
Bayesian and Maximum Likelihood phylogenetic trees had similar topologies with the Bayesian trees having generally higher support values for the internal nodes. Fig. 2 displays the Bayesian ATOH8 phylogenetic tree with support values the two different reconstruction methods mapped. For comparison, the Ensembl species tree is also shown (Fig S1 and http://www.phylowidget.org/full/index.html?tree=http://tinyurl.com/ensembltree&useBranchLengths=true&minTextSize=7). The phylogenetic analysis of ATOH8 resolves a large well-supported vertebrate clade with the major vertebrate taxa only partly resolved as monophyla (e.g. mammals only with posterior probability of 0.78, ML Bootstrap <50, MP Bootstrap 66). Zebrafish as the fish representative is not reported as the most basal vertebrate but rather all major groups (fish, amphibians, mammals, birds) resemble one polyphyletic group in this analysis. Within the mammalian tree, opossum represents the basal group, which is consistent with the species tree. Tip-level phylogenetic relationships are mostly in accordance with the known classification (Primates, Carnivora, Cetartiodactyla, Rodentia) and mostly well-supported by the two methods. The urochordate Ciona intestinalis (sea squirt) clusters basal to the vertebrate in group and shows highly-supported affinities to the fruit fly orthologue NET.
Extended details regarding the evolution of the ATOH8 gene was reflected by the Ensembl ATOH8 gene tree (Fig S2) [13]. Searching in Ensembl genome browser (http://www.ensembl.org/Multi/Search/Results?species=allidx=q=ATOH8), we found the ATOH8 gene to be present in 36 species which could represent all extant classes of vertebrates. Based on the presently available genomic sequences of ATOH8 from these species, the ATOH8 gene tree was generated with the pipeline of gene orthology/paralogy prediction method of Ensembl using only the most conserved regions for analysis (Fig S2). Here, related species are clearly clustered into their respective higher-order taxa in the gene tree, e.g. higher fishes (teleosts) are monophyletic and are resolved as basal vertebrates followed by more derived ‘Amphibia’, ‘Reptilia/Aves’ and ‘Mammalia’; ATOH8 of chicken and zebrafinch share a common ancestor.
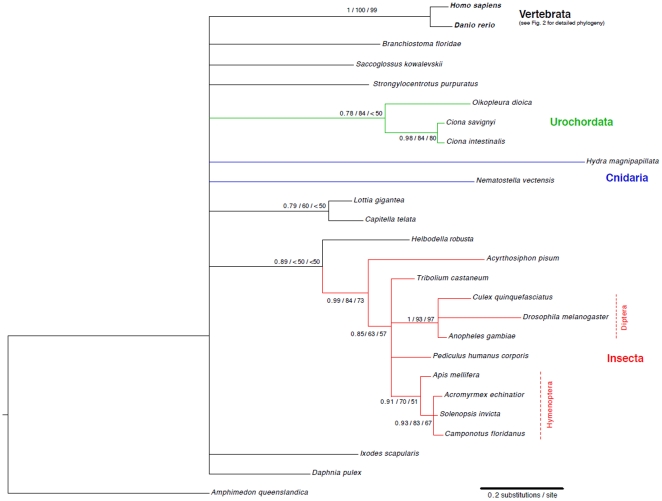
Using Drosophila melanogaster NET or Homo sapiens ATOH8 as queries, we searched in GenBank and published genome projects to identify possible ATOH8/NET orthologues in invertebrates. Sequences from 22 invertebrate species with a high similarity (Blastp e-values of < = 10−18) were collected (Table S3). We did not use sequences from invertebrate species of a genus twice (e.g. just one Drosophila species). To reconstruct the phylogenetic relationship of the NET/ATOH8 orthologues among metazoans, we chose the Amq1 sequence (GenBank: ABZ79673.1, Table S3) of the sponge Amphimedon queenslandica as outgroup, which resembles the most ancestral protein of Atonal and Twist superfamilies [2]. Human and zebrafish ATOH8 protein sequences were used as vertebrate representatives for this second phylogenetic analysis. The alignments of the 26 different protein sequences using the E-INS-I and the L-INS-I alignment algorithm in MAFFT were different and consisted of many gaps. Both alignments were analyzed using the software gBlocks to search for conserved and conveniently aligned blocks. For both alignments, the same 82 positions were reported from two blocks in the alignments (Table S3) and extracted. The L-INS-I alignment had five additional positions at the N-terminus. However, since no information was available for these positions for some taxa, we used the 82 positions reported from both alignments for subsequent analyses only. Within the 82 bp alignment 20 positions were invariant. Prottest reported the LG+G model as the most appropriate using the BIC and AIC criterion. For the ML calculations using PhyML and RAxML, we used the LG+G model. For Bayesian inferences using MrBayes (which does have the LG model), we used the JTT+G model, which was reported as the best model also implemented in MrBayes. The results of all calculations showed a large basal polytomy from which few well-supported groups branch off (Fig 3). The results of the Bayesian and the two ML analyses were mostly congruent. The vertebrates, urochordates, and insects were correctly resolved as monophyletic groups. Within the insects, the hymenopteran groups ((Camponotus, Solenopsis, Acromyrmex) and Apis) and the dipterans (Drosophila, Anopheles, Culex) were correctly resolved. Also, the holometabolous insects (hymenopterans + Tribolium + Pediculus + dipterans in the tree) were found as the sister group to the hemimetabolous insect representative Acrythosiphon, with low support although. In the Bayesian analyses, the two cnidarian species formed a clade (posterior probability of 0.68 only). However, in the ML analyses the two species clustered at two different positions within the tree with very low support although.
Figure 3. Bayesian phylogenetic tree based on the conserved 82 amino acid alignment of the bHLH domain region.
Branches with a posterior probability below 0.75 were collapsed. Support values on the branches resemble the posterior probability values, the PhyML bootstrap, and the RaxML bootstrap values, respectively.
Sequence analysis of the vertebrate ATOH8 protein
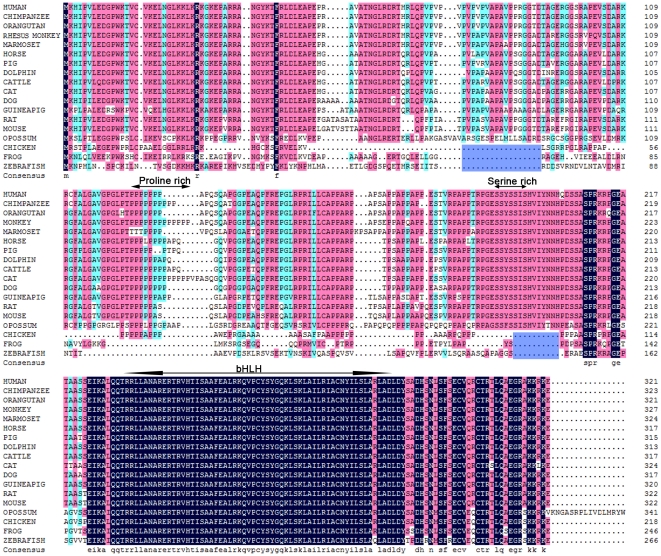
To understand further if ATOH8 varies among vertebrates, we analyzed the sequence alignments of the ATOH8 protein among the 18 vertebrate species that were collected from GenBank and our re-sequencing data (Table S2). The alignment of protein sequences in vertebrates showed a highly conserved bHLH domain among all species but there were two variable motifs between mammals and non-mammals (Fig 4). The frog and zebrafish lacked the proline-rich motif between the N-terminus and bHLH domain of the protein, and the serine-rich motif was only found in mammals. Among the vertebrates, chicken ATOH8 was the shortest, consisting of 218 amino acids. Dozens of amino acid were absent in the regions approximately 50 amino acid downstream of the N-terminus compared to its vertebrate orthologues. Shorter protein sequence was also observed in zebrafish and frog ATOH8, although not to the same extent as in the chicken. For the opossum, the ATOH8 protein showed a higher similarity to its mammalian orthologues than to the non-mammalian vertebrates. However, it had an extended C-terminus which was encoded by its third exon and was unique among vertebrates.
Figure 4. Comparative analysis of 18 vertebrate ATOH8 proteins.
The identical amino acids are labeled with black background. Sequences more than 80% conserved are highlighted with carmine. Sequences more than 50% conserved are highlighted with cyan. Dots represent the missing amino acids. The bHLH domain, proline-rich and serine-rich regions are defined with bi-directional arrowheads. The bHLH domain was defined by comparison of the ATOH8 sequence (UniProtKB/Swiss-Prot: Q96SQ7, ATOH8_HUMAN) to data from protein database according to the calculation in protein database and protein structure prediction (http://www.compbio.dundee.ac.uk/www-jpred/). The two commonly missing regions in zebrafish, frog and chicken in contrast to mammalian ATOH8 are indicated by blue.
In silico analysis of regulatory elements of ATOH8
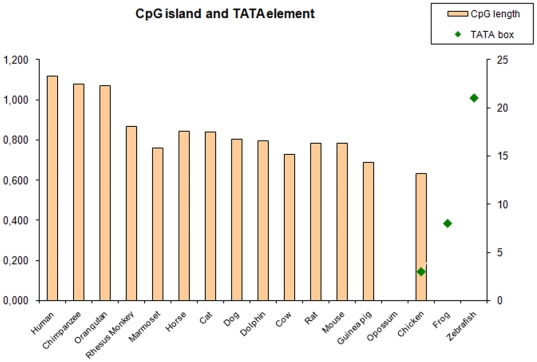
To characterize possible regulatory elements in the upstream region of the ATOH8 gene, we analyzed regions upstream of the ATOH8 gene in 17 vertebrate species. The results demonstrated a trend of increased CpG islands size upstream of the ATOH8 gene in eutherian mammals, where the GC-content in all examined islands was more than 65% (Fig 5). No CpG islands were present in the upstream region of zebrafish and frog ATOH8. Instead, we found an abundance of TATA elements in these species (Fig 5, Fig S3). In chicken, both CpG islands and TATA elements were present in the upstream region and the number of TATA elements decreased from zebrafish to frog and chicken (Fig 5). Uniquely, neither CpG islands nor TATA elements were identified in the upstream region of the opossum ATOH8.
Figure 5. CpG islands and TATA elements in the upstream region of ATOH8 start codon.
In mammals, the upstream region of ATOH8 coding region is characteristic of CpG islands, of which the human ATOH8 has the longest CpG islands. No TATA elements (green diamonds) are found. In chicken, both CpG islands and TATA elements are found in ATOH8 upstream region. In frog and zebrafish, there are no CpG islands but TATA elements present in the upstream region. From chicken, frog to zebrafish, the number of TATA elements increases.
Identification of functional regulatory elements of human ATOH8
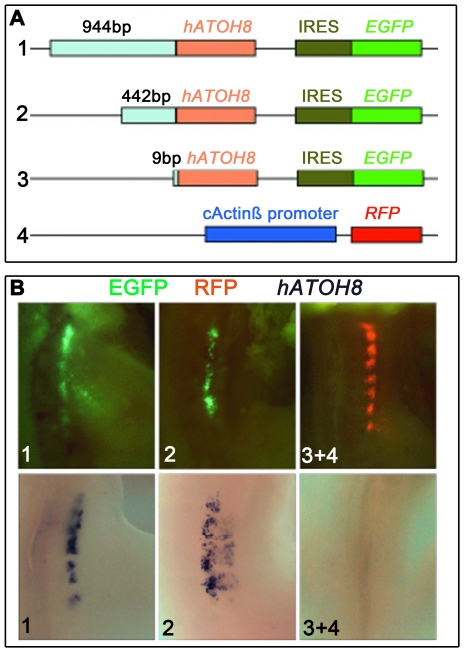
As ATOH8 has been found to be deregulated in a number of human diseases [11], [12], we were interested in analyzing the regulatory region of human ATOH8. To identify the potential promoter of human ATOH8 (hATOH8), we isolated a 944 bp and a 442 bp fragment upstream of the hATOH8 start codon along with the hATOH8 coding sequence and integrated them into pIRES2-EGFP vector by replacing its CMV promoter (Fig 6A). We transfected these two constructs into the somites of chicken embryos where the chicken ATOH8 is natively expressed (data not shown), and found that both fragments could drive the expression of EGFP (Fig 6B). To further verify the effect of expression, we analyzed those transfected embryos with a human ATOH8 probe by in situ hybridization and found that human ATOH8 transcripts were indeed present in the transfected region of the embryos (Fig 6B). In contrast, the control plasmid in which a 46 bp adaptor replaced the CMV promoter, no expression of EGFP or human ATOH8 transcripts was seen (Fig 6B).
Figure 6. Fragments upstream of human ATOH8 could drive gene expression in the chicken embryo.
(A) Diagram of constructed plasmids. 944 nt (1), 442 nt (2) and 9 nt (3) fragments (light cyan) upstream of the hATOH8 translation start codon, together with hATOH8 coding sequence (pink) are integrated into pIRES2-EGFP vector by replacement of its CMV promoter, respectively. The cActin-beta-RFP plasmid (4) is used as an additional indicator for effective transfection. (B) Exogenous gene expression after transfection of these plasmids into chicken embryos. Both 944 nt (1) and 442 nt (2) fragments could drive the expression of their downstream genes shown by, EGFP (green), and hATOH8 as shown here after in situ hybridization. In contrast, the embryo co-transfected with the control plasmid and cActin-beta-RFP plasmid shows no expression of EGFP or hATOH8 (3+4).
ATOH8 gene mapping in primates
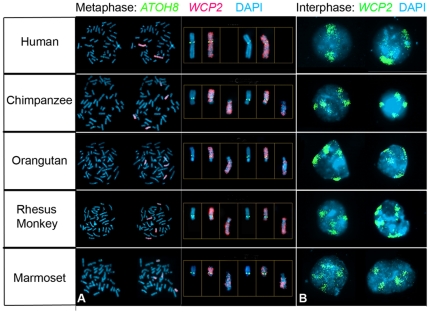
The hATOH8 gene is located in the human chromosome 2, which is a unique human ‘fused’ chromosome. To characterize the orthologues of hATOH8 host chromosomes and their territories in different primates, we performed fluorescent in situ hybridization (FISH) experiments with a human whole chromosome 2 painting probe (WCP2) and the hATOH8 gene-specific probe on fixed cells from human, chimpanzee, orangutan, rhesus monkey and marmoset. In metaphase cells, a pair of the human chromosomes was identified from the WCP2 hybridization signal and the ATOH8 gene was located in a region of the short arm, which was relatively near the centromere of the painted human chromosomes 2. However, the same probe WCP2 labelled two pairs of the intermediate-sized ‘non-fused’ chromosomes in non-human primates, and the hATOH8 signal was only located in the short arm of one pair of the chromosomes which were regarded to be the orthologue of the short arm of human chromosomes 2 (Fig 7a). In the human interphase cell, WCP2 labeled two chromatin territories, while in the non-human primates four separate territories were visible in the cell nuclei (Fig 7b). The results demonstrate that the short and long arms of human chromosome 2 are restricted to one single chromatin territory, whereas their orthologous chromosomes in non-human primates are separated into two territories.
Figure 7. Fish and chromosome painting show the host chromosome of ATOH8 gene in primates.
(A) Demonstration of host chromosomes of ATOH8 in metaphase cells. In human, ATOH8 (green) and WCP2 (red) staining signals are located on the same two chromosomes. ATOH8-positive signal is present on the short arm of WCP2-painted chromosomes. In other primates, ATOH8 signals are positive in two chromosomes, while WCP2 probe marked four chromosomes, out of which two chromosomes are ATOH8 positive. (B) The territories of WCP2-marked chromosomes in interphase cells. In human, WCP2-positive chromosomes (green) have two territories in the nucleus, while four separate territories are clearly visible in non-human primates.
Discussion
The phylogeny of ATOH8
The evolution of the vertebrate ATOH8 gene is mostly in accordance with the known phylogeny of the species analyzed. NET of the fruit fly is regarded as an orthologue of vertebrate ATOH8 [3] and was used together with the ATOH8 orthologue of Ciona intestinalis to root the topology. In the phylogenetic analysis, however, it was not possible to unambiguously align the two orthologues from Drosophila melanogaster and Ciona intestinalis to the vertebrate ingroup sequences at both nucleotide and protein level. The differences in the alignment had minor effects on branch lengths and support values for the two outgroup taxa relative to another and to the ingroup (results not shown). However, for the vertebrate ingroup which could be aligned consistently, this had no effect. In the phylogenetic trees (Fig. 3) calculated from the 82 amino acid alignment of mainly the bHLH sequences of invertebrates and two vertebrate representatives, the resolution to discriminate between major metazoan lineages (sponges, radialia, protostomes and deuterostomes) is very low. The tree shows many basal polytomies. Even the different chordate lineages do not group together in one monophylum. In other phylogenetic studies based on individual genes, the urochordates were resolved as the sister group to vertebrates, while the cephalochordates represented the most basal chordate lineage [14]. The results of our study hence add no support for either scenario. With this gene it is possible, however, to infer more recent evolutionary events such as the divergence within the vertebrates (Fig. 2) or also within the insects (Fig. 3). Interestingly, the crustacean Daphnia pulex NET candidate gene was not resolved basal to the insects, forming a pancrustacean clade, but showed affinities to the sponge sequence. Since about 20% of the genome sequence in Daphnia pulex has not been sequenced yet, and prominent gene duplication has been found in this species [15], it is likely, that the NET candidate gene found for this species may not be the true NET orthologue gene. Helobdella robusta clustered basal to the insect group. Due to the very low support value we regard this as an artifactual signal (homoplasy) only. The same seems to be the case for the clade consisting of Lottia gigantea and Capitella telata, which is also weakly supported. In general, our results show that within the different evolutionary lineages, NET/ATOH8 has experienced a high sequence divergence which makes inferences on basal metazoan relationships difficult, and probably correlates to the notable functional diversification of NET/ATOH8. For instance, NET of fruit fly plays a role in the morphogenesis of wing vein [5], while its vertebrate orthologue ATOH8 is engaged in multiple developmental processes such as neurogenesis [4], [8], development of pancreas [7] and so forth.
It is noteworthy that we did not find any strong support for homologous sequences of NET/ATOH8 in the released genome of C. elegans and the other two species of nematodes with tblastn. However, NET of C. elegans has been reported previously and referred to as ZK682.4 [2] and T05G5.2 [3] respectively. ZK682.4 has been classified into MyoR family [3] and remains doubtful if it belongs to NET [2], while T05G5.2 is annotated as Achaete-scute transcription factor-related in WormBase (http://www.wormbase.org/; WBGene00001951). Due to such an uncertainty, we excluded the nematodes in our phylogenetic analysis.
Gene structure of ATOH8
With the contribution of our sequencing data, we were able to perform multiple sequence alignments of ATOH8 at genomic sequence level and protein level. Genomic sequence alignment confirms that vertebrate ATOH8 consists of three exons, consistent with the annotation from GenBank. Additionally, we found that an alternative third exon may exist in mammals, which has been verified by EST (AL831857.1) from the amygdala of the human brain. Whether this isoform is expressed in other tissues and how it functions is currently unknown. Interestingly, a potential DNA loop may form around exon3b of primate ATOH8, which may provide a novel regulation for alternative splicing. Once the loop forms, the access to splicing exon3b may be blocked. As a result, exon3a would have to be utilized during alternative splicing.
The protein alignment shows that in vertebrates the bHLH domain of ATOH8 is highly conserved, suggesting that the bHLH domain of ATOH8 is functionally important in different organisms. However, some variation exists. By sequence comparison, we have found that the frog and zebrafish ATOH8 do not have the proline-rich region, and that only mammals develop the serine-rich region. Such regions could be termed as simple sequence repeats (SSRs). They are evolutionary labile and often variable between species and could fine-tune the function of transcription factors [16]. A few studies have shown that SSRs are functional and could be associated with molecular or morphological divergence: one study shows that stretches of proline and glutamine, when fused to the DNA-binding domain of the GAL4 transcription factor, could activate gene transcription. The activity of transcription increases with repeat length [17]; another study shows that the length of glutamine/alanine-repeats in the gene Runx-2 is correlated with the degree of dorsoventral nose bend and midface length in dogs and other carnivores [18]. It would be very interesting to check whether similar structural differences found in ATOH8 among different species could lead to its functional varieties. We have discovered that the chicken ATOH8 lacks approximately 50 amino acids in the downstream of N-terminus and the serine-rich region compared with mammalian ATOH8. Correspondingly, the functional differences of ATOH8 in the mouse and chicken, according to previous studies are distinguishable. In an earlier study, ATOH8 is shown to promote neurogenesis in the mouse retina [4], while a recent report demonstrates that ATOH8 inhibits neuronal differentiation in the chicken retina [9]. According to the latter study, ATOH8 in the chicken retina functions as a transcriptional activator. In contrast, the mouse ATOH8 acts as a transcriptional repressor in the development of pancreas [7]. Although we cannot exclude other factors such as different cellular contexts coordinating the ATOH8 function, the structural differences of ATOH8 itself in these two species may contribute to the functional divergences. With regard to the extended C-terminus of the predicted opossum ATOH8, direct evidence such as from RT-PCR is required to validate its cDNA sequence.
Regulatory elements of ATOH8
After sequence analysis, we find that the sequences upstream of eutherian mammalian ATOH8 orthologues are mainly CpG islands without TATA box. The GC content of these islands is more or less commensurate with that of presently reported human and mouse CpG island promoters, approximately 67% and 64% respectively [19]. Instead, the TATA box is present upstream of ATOH8 orthologues of frog and zebrafish with ordinary GC content, while in chicken, the upstream sequence contains both CpG islands and TATA box. Both CpG islands and TATA box are characteristic of gene promoters [20]. In human genome, about half of the potential promoter regions for human genes are located in CpG islands and about 32% potential promoter regions contain a putative TATA box motif [21]. The transition from a TATA box-type promoter to a CpG island-type promoter reflects the variation of the regulation of ATOH8 gene expression. TATA box promoters have one single transcription initiation site. Any change in the functional region of the promoter would lead to significant phenotypic alterations. In contrast, CpG island-type promoters usually have multiple transcription start sites (TSS). Which TSS is adopted depends on the cellular status, allowing the fine-tuning of gene expression responding to minor changes in cellular environment [22], [23]. In this way, the CpG island-type promoter of ATOH8 may be functionally more dynamic than the TATA-box promoter to regulate ATOH8 expression in a particular cellular environment, thus providing a more specific regulation of ATOH8 expression. Indeed, the mouse ATOH8 armed with CpG islands promoter, based on presently available studies, seems more widely expressed during embryonic development than its chicken and zebrafish orthologues. The mouse ATOH8 transcripts are present in multiple tissues including the developing brain, retina, cortical plates, kidney, pancreas, heart, liver, lung, stomach, intestine and spleen [4], [6], [7]. In comparison, zebrafish ATOH8 is mainly expressed in the developing retina and skeletal muscle during embryonic development [8]. In chicken, the expression of ATOH8 is concentrated in the myotome, eyes and podocytes (data not shown). It seems that ATOH8 expression extends to more organs during evolution. This expansion, if it is true, indicates that ATOH8 may have multiplied its functions in various organogeneses through evolution, which truly requires the flexibility of the CpG islands promoter to regulate its expression in multiple tissues. For the upstream region of the opossum ATOH8, both CpG islands and TATA elements are absent. This may be due to the unique character of the opossum genome. Analysis at genomic level indicates that the opossum has a lower GC content (37.7%) and lower density of CpG islands (0.9%) than other amniotes (40.9%–41.8% GC content and 1.7–2.2% CpG islands density) [24].
Experimentally, we identify that both 944 bp and 442 bp fragments upstream of the start codon of human ATOH8 are effective in driving the expression of their downstream sequences in the chicken embryo, revealing that the 442 bp fragment at least contains effective if not optimal components as the promoter of hATOH8. It also tells us that the CpG islands type promoter of human ATOH8 can function in the chicken cells. It will be desirable to know if the TATA promoter of ATOH8 can work in the mammals as well.
Remote regulation of gene expression in human chromosome 2
Comparative genomics have shown that human and chimpanzee genome sequences are nearly identical with only 1.23% nucleotide divergences [25]. The focus of study thus turns to differences rather than similarities of the genome between human and chimpanzee. The most striking difference is the reduction of the chromosome from n = 24 in the chimpanzee to n = 23 in the human, caused by the fusion of two medium-sized acrocentric chromosomes in the chimpanzee that give rise to the long and short arms of the large submetacentric human chromosome 2, the human “fusion chromosome 2” [26], [27], [28], [29]. This difference between human and chimpanzee at chromosome level may be one of the reasons causing the divergence of the two species.
In the interphase nucleus, chromosomes with corresponding proteins occupy discrete territories. Recently, interchromosomal and intrachromosomal interactions during interphase have been shown to regulate gene transcription in the mammalian nucleus [30]. To quote a few, the expression of olfactory receptor genes is regulated by the association of their promoters with loci from another chromosome [31]; interaction of different loci within one single chromosome is implicated in the DNA recombination to attain antigen specificity of B and T cells [32], [33]. It is proposed that the distant interchromosal and intrachromosomal interactions render remote regulatory elements and corresponding proteins to approach closely their target genes, thus coordinating the expression of these genes [34]. From our chromosome painting data, we demonstrated that human chromosome 2 containing the ATOH8 gene in its short arm is located in a restricted territory in the nuclei of interphase cells, while in chimpanzee, orangutan, rhesus monkey and marmoset the two chromosomes which are respectively orthologous to the long and short arms of human fusion chromosome 2 are within separate chromatin territories in interphase nuclei. Based on the hypothesis mentioned above, the differences of gene components in the chromosome(s) and protein components in corresponding chromosome territory(ies) between human and non-human primates, resulting from the fusion of two chromosomes, may differentially regulate the expression of genes, including ATOH8, in the relevant chromosome(s).
In summary, we analyzed the phylogeny of ATOH8 gene, and revealed the diversification of this gene across different species, especially for the vertebrate orthologues. Such diversification may significantly and delicately contribute to the various functions of ATOH8 in different cellular contexts.
Methods
Resequencing of ATOH8 gene loci and cDNA of several species
Based on known sequence information of ATOH8 gene loci of several species, we designed primers to perform genomic PCR to fill sequence gaps of ATOH8 gene loci for the chimpanzee, rhesus monkey, dolphin, horse, dog and cat. Genomic DNA was extracted from blood with Quick-gDNA mini prep kit (Zymo Research, Germany). PCRs were performed with Hot-start PCR kit (Qiagen). PCR products were directly sequenced or cloned into pDrive vector (Qiagen) for subsequent sequencing. ‘Primer walking’ strategy was adopted to sequence through the whole length of the PCR products. To complete missing sequence of the chicken ATOH8 gene locus, Southern blot was conducted following previously described protocols [35] from chicken BAC clone 13M24 (provided by Prof. Dr. MA Grownen, Wageningen Agricultural University, the Netherlands). The BAC DNA was digested with KpnI or/and EcoRI and separated on 0.8% agarose gel. The digoxigenin (DIG) labelled probe was synthesized with PCR using the conserved exon 1 of human or frog ATOH8 as the template. Based on results from Southern blot, a 3.4kb fragment was selected and cloned into pDrive vector (Qiagen) for sequencing, which proved to contain the upstream region and the first exon of chicken ATOH8. For RT-PCR, the chicken and frog total mRNA was extracted from chicken and frog embryos with Trizol (Sigma). Primers were designed and RT-PCR was performed with one-step RT-PCR kit (Qiagen).
Bioinformatic sequence analysis
Searches for sequences homologous to NET/ATOH8 in invertebrates were performed in GenBank using tBLASTn (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?). Multiple gene loci sequence comparisons of ATOH8 from different species were performed with the mVista program provided by VISTA (http://genome.lbl.gov/vista/index.shtml). Length of CpG islands of upstream region of ATOH8 was evaluated by CpG islands searcher (http://cpgislands.usc.edu/).
Construction of ATOH8 gene tree
In this study, two phylogenetic data sets have been analyzed. First, an ATOH8 cDNA sequence alignment of 18 vertebrate species along with the ATOH8 orthologues from Ciona intestinalis and Drosphila melanogaster was carried out using the translation alignment method and the software MAFFT version 6 program [36] (E-INS-I and L-INSI-algorithms) implemented as plugin in the Geneious software [37]. Second, a protein sequence alignment was constructed of species from several evolutionarily very different lineages bearing the ATOH8/NET-like bHLH motif from examined metazoan species and the ancestral protein Amq1 from sponge as outgroup with MAFFT. Reliably aligned blocks of the very divergent protein sequences were extracted from the alignment using the software gBlocks version 0.91 [38].
An appropriate nucleic and amino acid substitution model was determined using jModeltest version 0.1.1 [39], [40] and ProtTest version 2.4 [41]. MrBayes version 3.1.2 [42], RAxML version 7.2.7 [43] and PhyML [44] for the protein data set were employed to estimate phylogenetic trees. Bayesian analyses with MrBayes were conducted by computing 10×106 Markov Chain Monte Carlo generations in four parallel runs, each with four chains. Trees were sampled every 100th generation. Convergence was determined by comparing the split frequencies of the runs and the first 1000 trees were excluded for the nucleotide and protein alignment. Using RAxML, the most likely tree was determined by conducting 10 slow ML searches on 10 randomized stepwise addition parsimony trees. Furthermore, a total of 10000 rapid bootstrap replicates have been computed with RAxML. These were mapped onto the MrBayes tree. For PhyML, branch length and topology optimization were used and 1000 bootstrap replicates were performed. For both alignments the Bayesian trees were shown in this study and branches with posterior probabilities below 0.75 were collapsed to polytomies to display only trees with strong phylogenetic signal.
Construction of ATOH8 promoter- EGFP reporter system
Human BAC clone (RZPDB737E072124D) containing hATOH8 gene locus was purchased from Germany Genome Resource Center and cultured in 200ml LB-medium with 100 µg/ml chloramphenicol at 30°C for 48 hours. The plasmid DNA was isolated using Large Construct Kit (Qiagen) in accordance with the manufacturer's instructions. The DNA was digested with KpnI and separated on 0.8% agarose gel. A band (8.5 kb) was isolated from the gel and cloned into KpnI pre-linearised pDrive vector. The ends of the expected insert in the plasmid have been sequenced for confirmation of the presence of human ATOH8 gene locus. The cloned fragment (8.533 kb) covers the region from 8.3 kb upstream to 217 bp downstream of the hATOH8 start codon. A 944 bp and a 442 bp fragment upstream of hATOH8 start codon derived from the 8.5 kb fragment were pieced together with hATOH8 coding sequence, and cloned into pIRES2-EGFP (Clontech) vector by replacing the CMV promoter respectively. The control plasmid was constructed by replacement of the CMV promoter with an adaptor (adaptor sequence: Forward: 5′ CTAGCTAAGC TTGCCCGCGC CATGAAGCAC ATCCCGGTCC 3′; and Reverse: 5′ TCGAGGACCG GGATGTGCTT CATGGCGCGG GCAAGCTTAG 3′). The constructed plasmids were sequenced for verification (Eurofins, Germany).
In ovo electroporation and imaging of embryos
Fertilized chicken eggs were incubated to HH stage 17–18. The embryo was exposed by opening a window in the egg shell. 3–6µg/µl of DNA plasmids was electroporated into somites of the chicken embryo according to procedures described [45]. After 12–16 hours of re-incubation, the embryos were photographed under an epifluorescence microscope (MZFLIII, Leica) attached with a digital camera (DC300F, Leica).
In situ hybridization
A 1.3 kb human ATOH8 cDNA fragment cloned into pDrive was used as template to synthesize digoxigenin-labelled RNA probe. The plasmid was linearised with XbaI and subsequent in vitro transcription was performed with T7 RNA polymerase for the synthesis of antisense probe. BamHI was used to linearize the plasmid to synthesize sense probe as control with SP6 RNA polymerase. Fertilized chicken eggs obtained from a local breeder were incubated at 38°C, 80% humidity. Embryos were staged as described [46], sacrificed and fixed in 4% PFA/PBT. Whole mount in situ hybridization was performed as described previously [47].
FISH and chromosome painting
To perform the comparative FISH on ATOH8 gene location in chromosomes of human and non-human primates, the FISH probes were prepared from the human BAC clone (RP11-439L14) containing whole hATOH8 gene locus. The human BAC clone DNA which contains hATOH8 gene was isolated using a Large Construct Kit (Qiagen). The probe labelling was performed via PCR with random primers. The FISH processes was according to the protocols as described previously [48]. Briefly, the peripheral lymphocytes from human (Homo sapiens, HSA), chimpanzee (Pan troglodytes, PTR), orangutan (Pongo pygmaeus, PPY), rhesus macaque (Macaca mulatta, MMU) and lymphoblastoid cells from the common marmoset (Callithrix jacchus, CJA) were cultured. The preparation of chromosomes of human and non-human primates was carried out on the pre-treated glass slides. The human whole-chromosome painting probe for human chromosome 2 [48] was used to unequivocally assign hybridization signals to human chromosome 2 and the orthologous chromosomes in non-human primates, either for the territories in interphase cells or for the packaged chromosomes in metaphase cells. The new orthologous numbering is used for chimpanzee chromosomes in accordance to McConkey's suggestion [48]. The slides were counterstained with DAPI (4′, 6-diamidino-2-phenylindole; 0.14 µg/ml) and mounted in Vectashield (Vector Laboratories). Results were evaluated using a Zeiss Axiophot epifluorescence microscope equipped with single-bandpass filters for excitation of red, green, and blue (Chroma Technologies, Brattleboro, VT). During exposures, only excitation filters were changed allowing for pixel-shift-free image recording. Images of high magnification and resolution were obtained using a black-and-white CCD camera (Photometrics Kodak KAF 1400; Kodak, Tucson, AZ) connected to the Axiophot.
Supporting Information
Ensembl species tree. The species tree represents the mostly accepted phylogeny of analyzed species and is provided by Ensembl. Species used for analysis of ATOH8 phylogeny is marked with red.
(TIF)
The adapted phylogenetic tree of ATOH8 gene constructed by Ensembl. The tree was generated using TreeBeST pipeline. The tree is based on an ATOH8 sequence alignment of ATOH8 orthologues detected in the Ensembl genome database.
(TIF)
The distribution of CpG islands in the upstream region of ATOH8. The length of CpG islands in the ATOH8 upstream region of 17 vertebrate species is presented. In opossum, frog and zebrafish, there are no CpG islands in the upstream region of ATOH8.
(TIF)
The location of loop donor (LD) in chromosomes of primates.
(DOC)
Accession numbers of ATOH8/NET cDNA and protein orthologues.
(DOC)
Sequences of NET orthologues used for phylogenetic analysis.
(XLS)
Acknowledgments
We thank Dr. RP Crooijmans and Prof. Dr. MA Grownen (Wageningen Agricultural University, the Netherlands) for providing us a chicken BAC clone, BAC13M24, Germany Genome Resource Center (ImaGenes) for providing us a human BAC clone (RZPDB737E072124D) and Dr. S Wiemann (German Cancer Research Center) for providing us a human ATOH8 EST clone (DKFZp761E2117). We thank Ms. U. Pein, Ms. E. Gimbel, Ms. S. Glaser and Ms. U Baur for their technical assistance. We also thank all the researchers who submitted their data about ATOH8 into the bioinformatic databases in UCSC (http://genome.ucsc.edu), Ensembl (http://www.ensembl.org), Vista (http://genome.lbl.gov/vista/index.shtml), ChickEST (http://www.chick.manchester.ac.uk/) and NCBI (http://www.ncbi.nlm.nih.gov), which served as a resource and reference for annotation to this study.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The project was supported by an EU's Sixth Framework Program grant to Prof. Dr. B. Brand-Saberi (MYORES, 511978), by the DFG (GRK 1104) and the DAAD D/08/01771. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang Y, Chen K, Yao Q, Zheng X, Yang Z. Phylogenetic analysis of zebrafish basic helix-loop-helix transcription factors. J Mol Evol. 2009;68:629–640. doi: 10.1007/s00239-009-9232-7. [DOI] [PubMed] [Google Scholar]
- 2.Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, et al. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ledent V, Paquet O, Vervoort M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002;3:RESEARCH0030. doi: 10.1186/gb-2002-3-6-research0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inoue C, Bae SK, Takatsuka K, Inoue T, Bessho Y, et al. Math6, a bHLH gene expressed in the developing nervous system, regulates neuronal versus glial differentiation. Genes Cells. 2001;6:977–986. doi: 10.1046/j.1365-2443.2001.00476.x. [DOI] [PubMed] [Google Scholar]
- 5.Brentrup D, Lerch H, Jackle H, Noll M. Regulation of Drosophila wing vein patterning: net encodes a bHLH protein repressing rhomboid and is repressed by rhomboid-dependent Egfr signalling. Development. 2000;127:4729–4741. doi: 10.1242/dev.127.21.4729. [DOI] [PubMed] [Google Scholar]
- 6.Ross MD, Martinka S, Mukherjee A, Sedor JR, Vinson C, et al. Math6 expression during kidney development and altered expression in a mouse model of glomerulosclerosis. Dev Dyn. 2006;235:3102–3109. doi: 10.1002/dvdy.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynn FC, Sanchez L, Gomis R, German MS, Gasa R. Identification of the bHLH factor Math6 as a novel component of the embryonic pancreas transcriptional network. PLoS One. 2008;3:e2430. doi: 10.1371/journal.pone.0002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao J, Zhou J, Liu Q, Lu D, Wang L, et al. Atoh8, a bHLH transcription factor, is required for the development of retina and skeletal muscle in zebrafish. PLoS One. 2010;5:e10945. doi: 10.1371/journal.pone.0010945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubo F, Nakagawa S. Cath6, a bHLH atonal family proneural gene, negatively regulates neuronal differentiation in the retina. Dev Dyn. 2010;239:2492–2500. doi: 10.1002/dvdy.22381. [DOI] [PubMed] [Google Scholar]
- 10.Thomas N, Kenrick M, Giesler T, Kiser G, Tinkler H, et al. Characterization and gene expression profiling of a stable cell line expressing a cell cycle GFP sensor. Cell Cycle. 2005;4:191–195. doi: 10.4161/cc.4.1.1405. [DOI] [PubMed] [Google Scholar]
- 11.Ducray F, Idbaih A, de Reynies A, Bieche I, Thillet J, et al. Anaplastic oligodendrogliomas with 1p19q codeletion have a proneural gene expression profile. Mol Cancer. 2008;7:41. doi: 10.1186/1476-4598-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freire P, Vilela M, Deus H, Kim YW, Koul D, et al. Exploratory analysis of the copy number alterations in glioblastoma multiforme. PLoS One. 2008;3:e4076. doi: 10.1371/journal.pone.0004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, et al. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 15.Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, et al. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 18.Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci U S A. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki Y, Tsunoda T, Sese J, Taira H, Mizushima-Sugano J, et al. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 2001;11:677–684. doi: 10.1101/gr.164001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 23.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 24.Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe H, Hattori M. [Chimpanzee genome sequencing and comparative analysis with the human genome]. Tanpakushitsu Kakusan Koso. 2006;51:178–187. [PubMed] [Google Scholar]
- 26.Dutrillaux B. Chromosomal evolution in primates: tentative phylogeny from Microcebus murinus (Prosimian) to man. Hum Genet. 1979;48:251–314. doi: 10.1007/BF00272830. [DOI] [PubMed] [Google Scholar]
- 27.Yunis JJ, Prakash O. The origin of man: a chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- 28.JW IJ, Baldini A, Ward DC, Reeders ST, Wells RA. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci U S A. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.IJdo JW, Baldini A, Ward DC, Reeders ST, Wells RA. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci U S A. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunez E, Kwon YS, Hutt KR, Hu Q, Cardamone MD, et al. Nuclear receptor-enhanced transcription requires motor- and LSD1-dependent gene networking in interchromatin granules. Cell. 2008;132:996–1010. doi: 10.1016/j.cell.2008.01.051. [DOI] [PubMed] [Google Scholar]
- 31.Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Spicuglia S, Franchini DM, Ferrier P. Regulation of V(D)J recombination. Curr Opin Immunol. 2006;18:158–163. doi: 10.1016/j.coi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, et al. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat Immunol. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 34.Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132:929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Russell D. Molecular Cloning: a laboratory manual; In: Argentine J, editor. New York: Cold Spring Harbor Laboratory Press; 2001. pp. 6.47–46.49. [Google Scholar]
- 36.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drummond AJ AB, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A. Geneious v4.7, Geneious website. 2009. Available: http://www.geneious.com/. Accessed July 12, 2011.
- 38.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 39.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 40.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 41.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 42.Ronquist FaJPH. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 43.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 45.Scaal M, Gros J, Lesbros C, Marcelle C. In ovo electroporation of avian somites. Dev Dyn. 2004;229:643–650. doi: 10.1002/dvdy.10433. [DOI] [PubMed] [Google Scholar]
- 46.Hamburger V, Hamilton HL. A series of normal stage in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- 47.Nieto M, Patel K, Wilkinson DG. In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol. 1996;51:219–235. doi: 10.1016/s0091-679x(08)60630-5. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt J, Kirsch S, Rappold GA, Schempp W. Complex evolution of a Y-chromosomal double homeobox 4 (DUX4)-related gene family in hominoids. PLoS One. 2009;4:e5288. doi: 10.1371/journal.pone.0005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ensembl species tree. The species tree represents the mostly accepted phylogeny of analyzed species and is provided by Ensembl. Species used for analysis of ATOH8 phylogeny is marked with red.
(TIF)
The adapted phylogenetic tree of ATOH8 gene constructed by Ensembl. The tree was generated using TreeBeST pipeline. The tree is based on an ATOH8 sequence alignment of ATOH8 orthologues detected in the Ensembl genome database.
(TIF)
The distribution of CpG islands in the upstream region of ATOH8. The length of CpG islands in the ATOH8 upstream region of 17 vertebrate species is presented. In opossum, frog and zebrafish, there are no CpG islands in the upstream region of ATOH8.
(TIF)
The location of loop donor (LD) in chromosomes of primates.
(DOC)
Accession numbers of ATOH8/NET cDNA and protein orthologues.
(DOC)
Sequences of NET orthologues used for phylogenetic analysis.
(XLS)