Abstract
NF-κB activation is essential for RANKL-induced osteoclast formation. IL-4 is known to inhibit the RANKL-induced osteoclast differentiation, while at the same time promote macrophage fusion to form multinucleated giant cells (MNG). Several groups have proposed that IL-4 inhibition of osteoclastogenesis is mediated by suppressing the RANKL-induced activation of NF-κB. However, we found that IL-4 did not block proximal, canonical NF-κB signaling. Instead, we found that IL-4 inhibited alternative NF-κB signaling and induced p105/50 expression. Interestingly, in nfκb1−/− bone marrow macrophages (BMM), the formation of both multinucleated osteoclast and MNG induced by RANKL or IL-4 respectively was impaired. This suggests that NF-κB signaling also plays an important role in IL-4-induced macrophage fusion. Indeed, we found that the RANKL-induced and IL-4-induced macrophage fusion were both inhibited by the NF-κB inhibitors IKK2 inhibitor, and NEMO inhibitory peptide. Furthermore, overexpression of p50, p65, p52 and RelB individually in nfκb1−/− or nfκb1+/+ BMM enhanced both giant osteoclast and MNG formation. Interestingly, knockdown of nfκb2 in wild type BMM dramatically enhanced both osteoclast and MNG formation. In addition, both RANKL- and IL-4-induced macrophage fusion were impaired in NIK−/− BMM. These results suggest IL-4 influences NF-κB pathways by increasing p105/p50 and suppressing RANKL-induced p52 translocation, and that NF-κB pathways participate in both RANKL- and IL-4- induced giant cell formation.
Introduction
Macrophages reside in all tissues and undergo a homotypic fusion process under specific conditions. There are two major types of macrophage-derived multinucleated giant cells, osteoclasts in bone and multinucleated giant cells (MNG) found in chronic inflammatory reactions(1). The osteoclast resorbs mineralized matrix while the MNG engulfs large foreign bodies. Macrophage fusion is mediated through protein-protein interaction. Several proteins important for macrophage fusion have been identified, including macrophage fusion receptor (MFR), CD47, CD44, E cadherin, CD36 and DC-STAMP(2–8); however, the molecular mechanism controlling macrophage fusion and the pathways which determine whether macrophages will form MNG or osteoclasts remain poorly understood.
RANKL signals macrophages to differentiate into osteoclasts and to undergo fusion forming large, mutlinucleated cells expressing high levels of the enzyme tartrate-resistant acid phosphatase (TRAP)(9). This process is dependent on induction of NFATc1 and NF-κB. RANKL can activate NF-κB by either the canonical or alternative pathways(10). The canonical pathway is mediated by the NEMO-dependent activation of IKK2 leading to the phosphorylation and degradation of the classic inhibitors of κB (IκB α,ε). Degradation of IκB leads to the translocation of the classic NF-κB dimer consisting of p50 and p65/RelA. The p50 subunit is derived from the nfkb1 gene that also encodes p105. The activation of this pathway is very rapid and is independent of new protein synthesis. The alternative pathway is mediated by the NIK-dependent activation of IKK1 that leads to the phosphorylation and partial degradation of the p100 precursor (derived from the nfkb2 gene) to generate the p52 subunit. p100 also acts as an IκB, and thus its degradation allows the generation of p52 and the release of NF-κB dimers including p52/RelB(11). Mice lacking expression of nfkb1 or nfkb2 have modest defects in osteoclastogenesis while the double deficient mice have severe osteoporosis caused by the absence of osteoclasts(12).
In contrast to RANKL, IL-4 signals macrophages to differentiate to the alternative type (13,14) and to undergo fusion forming multinucleated giant cells (MNG) expressing high levels of arignase I and chitinases (15). This IL-4 induced macrophage differentiation and MNG formation is dependent on STAT6 (15,16). The IL-4-induced expression of the fusion-related molecules E-cadherin and DC-STAMP were also STAT6 dependent. These fusion molecules are important for the formation of both osteoclasts and MNG (15,17). In the presence of both RANKL and IL-4, the IL-4-induced differentiation was dominant. IL-4 suppressed the RANKL-induced osteoclast differentiation and promoted MNG formation in a STAT6 dependent manner (15,16).
The effect of IL-4 on the activation of NF-κB in this setting is unclear. There are several studies reporting that IL-4 inhibited osteoclast differentiation through inhibition of the RANKL-induced activation of the classical NF-κB pathway(18–20). However, in our report, we did not observe a direct effect of IL-4 on RANK signaling (16). Others have shown that NF-κB is essential for macrophage fusion induced by culture at high cell density(21) and for the expression of fusion-related genes such as DC-STAMP (22).
Therefore, in this study, we carefully investigated the role of NF-κB in IL-4-induced MNG formation and whether IL-4 regulates RANKL-induced NF-κB activation. We found that NF-κB signaling is also required for IL-4- induced MNG formation. IL-4 treatment did not inhibit the RANKL-induced activation of the NF-κB canonical pathway but inhibited the activation of the alternative pathway. In addition, IL-4 induced the expression of p105/p50 and stimulated the accumulation of p50 in the nucleus in a NIK-dependent manner. Overexpression of NF-κB dimer proteins enhanced macrophage fusion while knockdown of p100 enhanced macrophage fusion. These results indicate that NF-κB pathways regulate macrophage fusion induced by IL-4 treatment, in addition to RANKL-induced osteoclast formation, although the specific pathways are different.
Material and Methods
Cell culture
Nonadherent bone marrow mononuclear cells were isolated from femurs and tibias of 4 to 8 week old female wild type C57BL/6 (Taconic Laboratories) mice, nfκb1-deficient mice on the C57BL/6 background (provided by Dr. Ranjan Sen, National Institute of Aging, NIH, Baltimore, MD), or NIK-deficient mice on the C57BL/6/129 background (developed by Dr. Robert Schreiber (23) and provided by Dr. Barry Sleckman, Washington University at St. Louis, MO). Cells were cultured overnight in α-MEM media (Biowhittaker) to deplete adherent stromal cells. These nonadherent bone marrow mononuclear cells were cultured for 3 days in α-MEM media supplemented with penicillin, streptomycin, glutamine (Biowhittaker), 10% fetal bovine serum and 20 ng/mL recombinant mouse M-CSF (R&D systems, Minneapolis, MN) to generate bone-marrow derived macrophages (hereafter also called BMM). Mature osteoclasts were obtained after 5–6 days culture in the presence of 20 ng/mL M-CSF and 150 ng/mL RANKL (provided by Dr. Mehrdad Tondravi, National Cancer Institute). BMM were also treated with 20 ng/mL recombinant mouse M-CSF and 10 ng/mL IL-4 (R&D systems) to generate MNG. In the RANKL plus IL-4 group, BMM were treated with 20 ng/mL recombinant mouse M-CSF, 150 ng/mL RANKL and 10 ng/mL IL-4 for various time points.
TRAP and crystal violet staining
Osteocalsts were identified by high levels of TRAP staining. TRAP staining was performed as previously described (24). MNG were identified using crystal violet staining. Cells were fixed and stained with 1% crystal violet in 95% ethanol, washed with water, and air dried as described (15).
Immunoblotting
Total cell lysate preparation and immunoblotting were performed as described previously (16). Briefly, Cell lysates were fractionated on 4–20% or 7.5% SDS PAGE gels, and electrophoretically transferred to PVDF membranes (Millipore). Membranes were blocked and probed with rabbit anti-p105/50 (Abcam, Cambridge, MA), rabbit anti p100/52 (Cell Signaling Technology, Danvers, MA), rabbit anti-p65 (RelA), -RelB, -STAT6, -Lamin A, anti-IκBα, and mouse anti-Actin —all from from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The ratio of p100/p52 was calculated by densitometric analysis using Quantity One software (Bio-rad).
Cell fractionation
Cells were lysed in lysis buffer A (10 mM HEPES, pH 7.9; 10 mM KCL; 10 mM EDTA, 1 mM DTT, 0.4% IGEPAL, 1μg/ml pepstatin A, leupeptin, aprotinin). The supernatant was collected by centrifugation and considered the cytoplasmic fraction. The precipitate, was solubilized in nuclear lysis buffer B (20 mM HEPES, pH 7.9; 400 mM NaCl; 1 mM EDTA; 10% Glycerol, 1 mM DTT, 1μg/ml pepstatin A, leupeptin, aprotinin.), centrifuged, and the nuclear fraction was collected as supernatant lysates.
EMSA
An NF-κB consensus oligonucleotide derived from the TNF promoter κB3 site(25) was labeled with biotin using the 3′ end DNA labeling kit (Pierce). The consensus probe sequence is AACAGGGGGCTTTCCCTCCT. EMSA assay was performed using a chemiluminescient EMSA kit (Pierce). Briefly, 2 μg of nuclear lysate was incubated with the probe in reaction buffer (1× binding buffer, 2.5% glycerol, 5 mM MgCl2, 50 ng/μl polydI-dC, 0.05% NP-40) for 30 minutes. Reactants were loaded onto a 6% native polyacrylamide gel. The bands were transfered onto positively charged nylon membrane (Roche). The DNA was crosslinked by UV crosslinker. The membrane was incubated and exposed according to the kit instruction.
Real time PCR
Total RNA was extracted from cultured cells using the RNeasy kit (Invitrogen). First strand of cDNA was transcribed from 2 μg of RNA with Superscript RT kit (Invitrogen). Quantitation of the target gene relative to that of β-actin was determined using the SYBR green dye method and was detected on an ABI7700/SDS platform (Applied Biosystems). The primers used were the following:
p100/52: 5′ GCCCGAGCGTTGCTGGACTA 3′
5′ CGCCGAGGTACTGAGCGTGAT 3′ 184bp
RelB: 5′ ACACCGGGTACACCCACATAGC 3′
5′ TGGATGCCCAGGTTGTTAAAGC 3′ 114bp
DC-STAMP: 5′ CGCTGTGGACTATCTGCTGT 3′
5′ CACTGAGACGTGGTTTAGGAAT 3′ 186bp
E-cadherin: 5′ TCCAACAGGGACAAAGAAACAA 3′
5′ TGACACGGCATGAGAATAGAGG 3′ 171bp
CD44: 5′ CCCAGCCAGTGACAGGTTC 3′
Antisense 5′ CGGCAGGTTACATTCAAATCG 3′ 193bp
Lentivirus infection and gene transfection
p50, p65, p52, RelB and p100 were generated by PCR using cDNA plasmids containing p50, p65 ( provided by Dr. Janet Stavnezer from University of Massachusetts(26)), NF-κB2 and RelB (open Biosystems) as templates. All constructs were cloned into the PLVX-puro lentiviral vector (developed by Clonetech, provided by Dr. Ricardo Feldman from University of Maryland). p50 and p65 were also cloned into the pcDNA3.1 myc-HisB vector (Invitrogen). The primers for p52 (5′-ataactcgagatggacaattgctacgatccag, 3′-ataagggccctca tcgctgcagtgtgtcaagggc) were designed to introduce a XhoI site at the 5′ end, a termination codon after the amino acid 442, and ApaI site at 3′ end(27). PLVX-puro, PsPax2 and PMD.2G (developed by Dr. Didier Trono, EIA, provided by Dr. Ricardo Feldman from University of Maryland) were cotransfected to 293T cells by using Fugene6 reagent (Roche). For the shRNA experiment, p100/52-PLKO.1 lentivirus vector (Sigma-Aldrich) and PsPax2 and PMD.2G were cotransfected to 293T cells. After 24 hours, transfection medium was replaced with fresh medium. Sodium pyruvate (1 mM) and HEPES (25 mM) were added into the fresh medium. Viral supernatant was collected at 48 hours and 72 hours after infection; the supernatant was filtered through a 0.45μm filter and incubated with primary BMM for 72 hours. Then the infection medium was replaced by induction medium composed of 150 ng/ml RANKL alone, 10 ng/ml IL-4 alone, or RANKL plus IL-4 respectively. The cells were kept in culture for 5 days and harvested for staining, real time RT-PCR, cell fractionation and immunoblot. p50, p65 pcDNA3.1 myc-HisB vectors were transfected to 293T cells using Fugene6 reagent. After 24 hours, total cell lysate was harvested for EMSA assay as previous described(28).
NF-κB inhibitor assay
BMM were treated with various concentrations of in solution™ IKK2 inhibitor (Calbiochem, SC-514), IKKγ NEMO binding domain inhibitory peptide, or control peptide (Imgenex, IMG-2000) for 5 days. Cells were stained for TRAP and crystal violet respectively.
Microscopy
Samples were fixed and pictured at room temperature on a Nikon eclipse E400 microscope using a 4× (plan APO) or 10×(plan fluor) objective lens (Nikon corporation). Digital images were captured using a CCD camera and processed using CoolSNAP version 1.2 software (Roper Scientific Inc.). Three random fields (40×) per well were counted. The average area per giant cell and the total area of giant cells were measured as described (15) using ImagePro plus software (Media cybernetics). The average size of giant cells was obtained by dividing the total giant cell size by the total giant cell number. The results are shown as the mean± SD of triplicate fields.
Statistical analysis
Results were analyzed by using Microsoft Excel (Microsoft). P values were caculated using a paired student t test. Statistical significance was determined when a P value less than 0.05
Results
IL-4 did not block activation of the NF-κB canonical pathway by RANKL
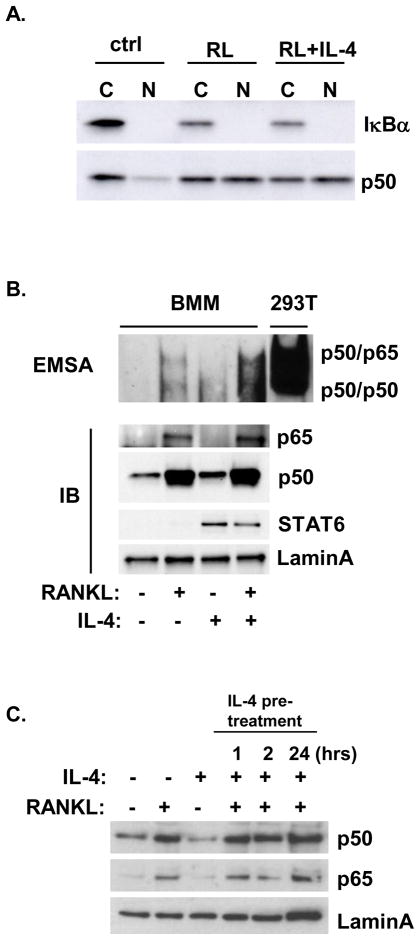
Several groups reported that IL-4 was capable of inhibiting the rapid RANKL-induced activation of NF-κB, however we did not observe any effect of IL-4 on RANK signaling (16,19,29). To examine whether IL-4 influenced the RANKL-induced canonical NF-κB pathway, BMM was treated with cytokine combinations for 30 minutes (Figure 1). As expected, we found that RANKL induced the degradation of IκBα and the nuclear translocation of p50 (Figure 1A); however, the presence of IL-4 had no effect on these rapid RANKL-induced responses. EMSA assay was also performed to analyze NF-κB DNA-binding activity. The sequence of the TNF κB3 site was used as the NF-κB probe, which has been shown to interact with high affinity with both p50-p65 heterodimers and p50 homodimers(25). Total lysate of 293T cells co-transfected with myc-tagged p50 and p65 cDNA was run simultaneously with BMM nuclear lysate to identify the NF-κB dimers (Supplemental Figure 1). RANKL induced NF-κB dimer formation in primary BMM after 30 minutes; concurrent addition of IL-4 did not inhibit the RANKL-induced NF-κB dimer formation (Figure 1B). Treatment of primary BMM with RANKL rapidly induced the translocation of p50 and p65 to the nucleus; IL-4 did not inhibit the RANKL-induced translocation of either p50 or p65. IL-4 treatment induced STAT6 translocation to the nucleus. We also performed IL-4 pretreatment experiments. We found that pretreatment of BMM with IL-4 for 1, 2, or 24 hours did not suppress the translocation of p50 or p65 induced by RANKL (Figure 1C). These results indicate that IL-4 does not block the rapid activation of the canonical NF-κB pathway by RANKL.
Figure 1. Effect of RANKL and IL-4 on the rapid activation of NF-κB.
Primary BMM were cultured in the presence or absence of RANKL (150 ng/ml) or IL-4 (10 ng/ml) as indicated for 30 minutes. Cytoplasmic (C) and nuclear (N) lysates were prepared as described in the Methods section. A. Lysates were analyzed by western blotting with anti-IκBα or anti-p50 as indicated. B. Top panel. Nuclear lysates were incubated with the biotin-labeled κB oligo and analyzed by EMSA assay as described in the Methods section. Total lysates of 293T cells transfected with p50 and p65 were run simultaneously as reference. Bottom panel, Nuclear lysates were analyzed by western blotting using anti-p65, anti-p50, anti-STAT6, or anti-lamin A antibodies. C. BMM were pretreated with 10 ng/ml IL-4 for various times and then were treated with RANKL (150 ng/ml) or IL-4 (10 ng/ml) as indicated for 30 minutes. Nuclear lysates were harvested and analyzed using anti-p50, anti-p65, or anti-lamin A antibodies.
IL-4 upregulated p105/50 expression after 48 hours
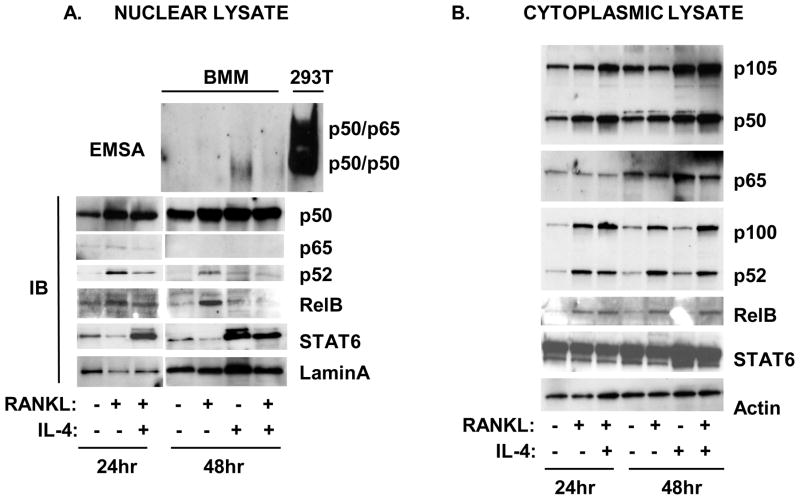
We also examined longer time periods of stimulation. After longer time periods (48 hours), the RANKL -induced NF-κB binding was barely detectable (Figure 2A). Surprisingly, IL-4 treatment in the presence or absence of RANKL induced a single NF-κB band that comigrated with the p50 homodimer position in the control 293T cell lysates (Figure 2A). RANKL-induced the accumulation of p50 in the nucleus at these longer times of stimulation (24 and 48 hours), however there was very little p65 remaining in the nuclear fraction. IL-4 enhanced the expression of p105/50 in the cytoplasm in the presence or absence of RANKL and increased the amount of p50 in the nucleus after 48 hours (Figure 2A,B). These results indicate that IL-4 upregulates p105/50 expression and enhances the accumulation of p50 in the nucleus.
Figure 2. Effect of RANKL and IL-4 on the slow activation of NF-κB.
Primary BMM were cultured in the presence or absence of RANKL (150 ng/ml) or IL-4 (10 ng/ml) for various times as indicated. Nuclear and cytoplasmic lysates were prepared as described in the Methods section. A. Top panel. Nuclear lysates were incubated with the biotin-labeled κB oligo and analyzed by EMSA assay as described in the Methods section. Total lysates of 293T cells transfected with p50 and p65 were run simultaneously as reference. Bottom panel, Nuclear lysates were analyzed by western blotting using anti-p50, anti-p65, anti-p52, anti-RelB, anti-STAT6, or anti-lamin A antibodies. B. Cytoplasmic lysates were analyzed by western blot using anti-p105/p50, anti-p65, anti-p100/p52, anti-RelB, anti-STAT6, or anti-actin antibodies.
IL-4 inhibited activation of the NF-κB alternative pathway by RANKL
We next examined the effect of IL-4 on the alternative NF-κB pathway. We found that IL-4 reduced the RANKL-induced nuclear translocation of NF-κB p52 and RelB (Figure 2A). We also observed changes in the cytoplasmic fraction (Figure 2B). RANKL increased p100/52 and RelB expression in the cytoplasm after 24 hours. In the presence of IL-4, the amount of cytoplasmic p100 and the p100/p52 ratio were altered. In the presence of RANKL, the ratio of p100/p52 was 0.7. However, in the presence of RANKL plus IL-4, the ratio was 1.7. These results suggest that IL-4 inhibits activation of the slower alternative NF-κB pathway by RANKL and alters the RANKL-induced processing of p100 to p52.
Nfκb1 is necessary for RANKL- and IL-4-induced macrophage fusion
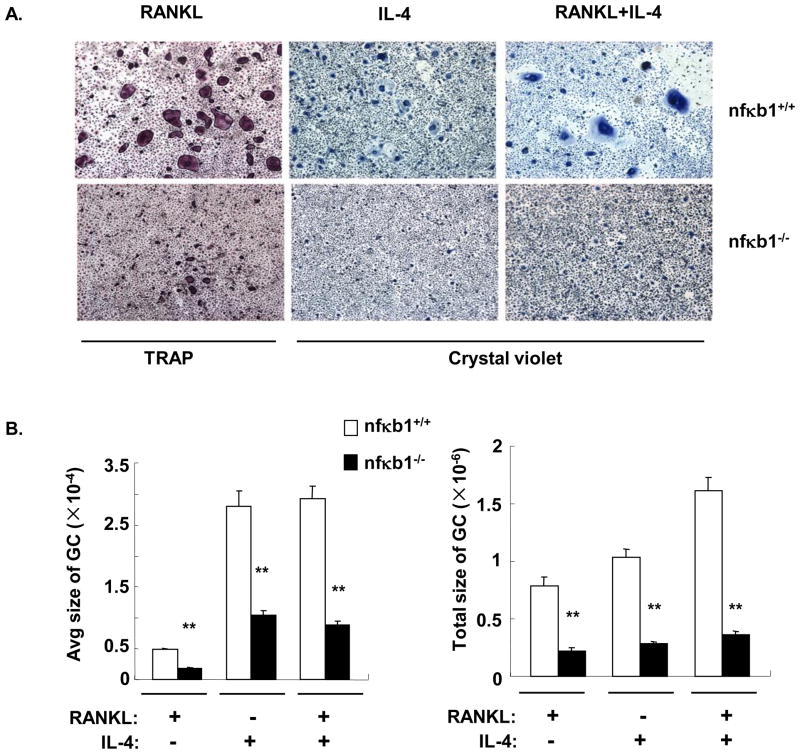
In order to determine the biological impact of the increase in p105/50, nfκb1−/− BMM, whick lack both p105 and p50, were treated with cytokines for 5 days to induce macrophage fusion. Interestingly, the formation of giant cells induced by RANKL (osteoclasts detected by TRAP staining), or by IL-4 or RANKL plus IL-4 (MNG detected by crystal violet staining) was impaired in nfκb1−/− BMM (Figure 3A). The average size of each multinucleated cell (nuclei>2) and the total area were significantly lower in the absence of nfκb1 (Figure 3B). This suggests that p105/50 may be important for macrophage fusion in general. While the RANKL-induced formation of TRAP+ osteoclasts was reduced in the absence of nfκb1, IL-4 was still able to suppress the TRAP+ osteoclasts formation in these cultures (Supplemental Figure 2A).
Figure 3. Both RANKL-induced osteoclast formation and IL-4-induced MNG formation were impaired in BMM lacking nf κb1.
A. BMM were prepared from nfκb1+/+ or nfκb1−/−mice and plated at 1×106 cells/ml for osteoclast differentiation and 2×106 cells/ml for IL-4-induced MNG differentiation. The cells were cultured in the presence or absence of RANKL or IL-4 as indicated for 5 days. TRAP staining was performed to detect osteoclasts and crystal violet staining was performed to detect MNG. Representative fields (40×) are shown for each group. B. The average area per giant cell (GC, nuclei>2) and the total area of giant cells were measured using ImagePro plus software as described in Materials and Methods. The results are shown as the mean± SD of triplicate fields. Unit: μm2. **, p<0.01.
We also examined the alternative pathway in BMM derived from nfκb1−/− mice. The RANKL-induced increase in p100/52 and RelB protein was still observed in the nfκb1−/− cells (Supplemental Figure 2B). Furthermore, RANKL was still able to induce the nuclear translocation of p52, p65, and RelB. IL-4 also suppressed the RANKL-induced nuclear translocation of p52 and RelB. Taken together these results indicate that modulation of p105/50 by IL-4 was not essential for its inhibitory effect on osteoclast differentiation and TRAP expression. The effect of RANKL and IL-4 on the NF-κB alternative pathway was not affected by the lack of p105/50.
NF-κB inhibitors suppress both osteoclast and MNG formation
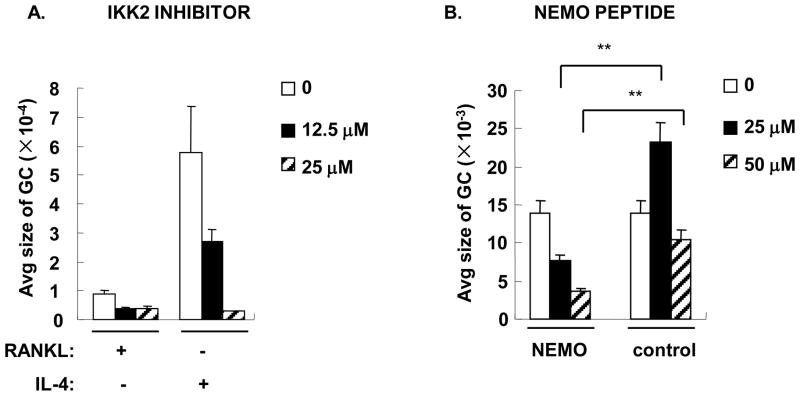
To confirm a role for NF-κB in macrophage fusion, we utilized several selective inhibitors of NF-κB (Supplemental Figure 3 and Figure 4). The In solution™ IKK2 inhibitor completely abrogated RANKL- and IL-4-induced macrophage fusion (Figure 4A). Similar results were obtained using PDTC (data not shown). NF-κB essential modulator (NEMO) forms an IκB kinase complex with IKK1 and IKK2 and is essential for both IκBα and p105 phosphorylation, degradation and NF-κB pathway activation(30). A cell permeable NEMO binding domain (NBD) inhibitory peptide blocked association of NEMO with the IKK complex and inhibited cytokine-induced NF-κB activation(31). To further identify the role of the IKK complex in IL-4 induced macrophage fusion, BMM were treated with IL-4 in the presence of NBD inhibitory peptide or control peptide. NEMO inhibitory peptide inhibited IL-4-induced MNG formation but control peptide did not (Supplemental Figure 3B and Figure 4B). These results suggest that a NEMO-dependent pathway is involved in IL-4-induced MNG formation.
Figure 4. IKK2 inhibitor and NEMO peptide suppressed both osteoclast and MNG formation.
BMM were plated and treated with cytokines as shown in Figure 3 in the presence or absence of various concentrations of in solution™ IKK2 inhibitor (A) or NEMO peptide or control peptide (B) for 5 days. TRAP or crystal violet staining was performed to identify giant cells (shown in supplemental data). The average size per giant cell (GC, μm2) was measured and statistical analysis was performed as in Figure 3. *, p<0.05, **, p<0.01.
Overexpression of NF-κB proteins enhanced both osteoclast and MNG formation
To confirm that NF-κB proteins participated in macrophage fusion, cDNA encoding p50, p65, p52, and RelB were transduced by lentiviral vector in nfκb1−/− or nfκb1+/+ BMM (Supplemental Figure 4). Overexpression of p50, p52, p65, or RelB in nfκb1−/− BMM restored macrophage fusion induced by RANKL or IL-4 (Supplemental Figure 4A). Overexpression of these proteins also enhanced IL-4-induced fusion in nfκb+/+ BMM (Supplemental Figure 4B). Overexpression of these proteins was confirmed by western blotting analysis (Supplemental Figure 4C).
NF-κB2 (p100/52) knockdown altered RANKL-induced osteoclast and IL-4- induced MNG formation
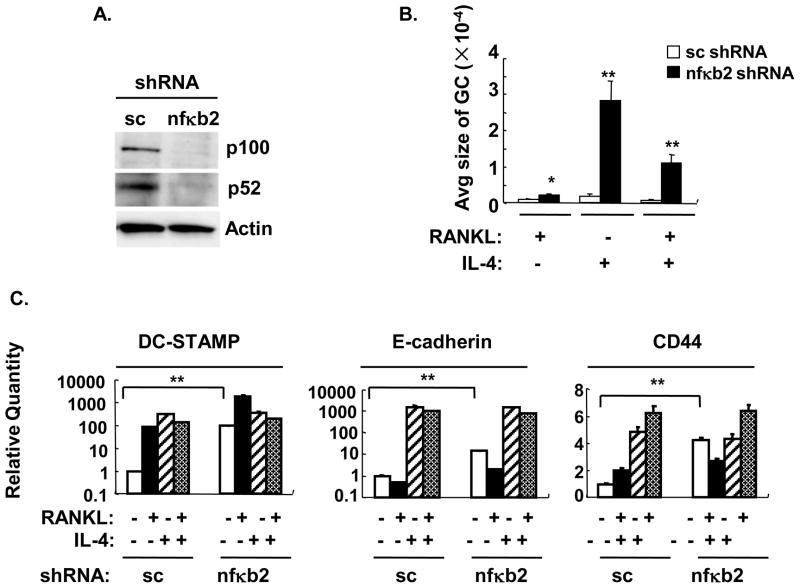
In order to examine the role of the alternative NF-κB pathway in RANKL- and IL-4-induced macrophage fusion, nfκb2 shRNA or scrambled control was transduced by lentivirus to wild type BMM (Figure 5 and Supplemental Figure 5). Transduction of BMM with nfκb2 shRNA resulted in substantial knock-down of p100 and p52 (Figure 5A). Nfκb2 knockdown enhanced RANKL-induced macrophage fusion which is in accordance with the study that nfκb2-deficient BMM showed enhanced sensitivity to RANKL (32). Surprisingly, nfκb2 knockdown also significantly enhanced the IL-4- and RANKL plus IL-4-induced macrophage fusion (Figure 5 and Supplemental Figure 5). These results suggest that p100/52 (Nfκb2) acts as an inhibitor of macrophage fusion. Since p52 transduction enhanced macrophage fusion, the inhibitory effect is likely mediated by p100 itself. Indeed, dimers of p100 have been identified as IκBδ (11). Interestingly, nfκb2 shRNA knockdown significantly enhanced the basal expression of CD44, DC-STAMP and E cadherin in wild type BMM (M-CSF treatment, Figure 5C) while the RANKL-induced or IL-4-induced expression was unchanged.
Figure 5. Nfκb2 knockdown by shRNA enhanced both RANKL-induced osteoclast and the IL-4-induced MNG formation.
Wild type BMM were transduced with lentivirus containing nfκb2 or scrambled (sc) shRNA and were cultured in cytokines for 5 days. A. Total cell lysates were analyzed by western blotting for p100/p52 or actin as indicated. B. TRAP or crystal violet staining was performed (shown in supplemental data). The average area per giant cell (GC) was measured (μm2) and statistical analysis was performed as shown in Figure 3. **, p<0.01. C. Fusion-related gene expression was measured by real time RT-PCR. The results are shown as the mean± SD of triplicate samples. *, p<0.05, **, p<0.01.
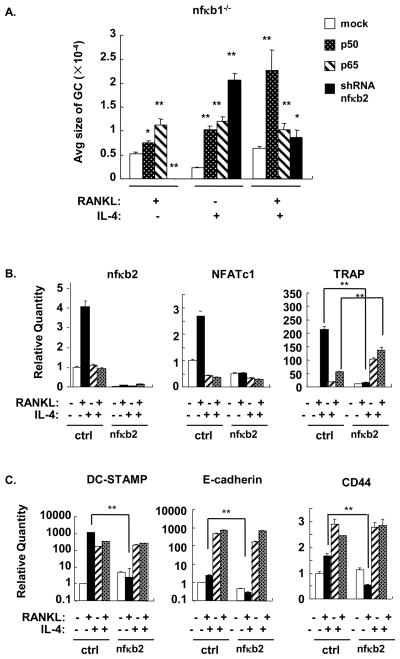
To determine whether BMM lacking both NF-κB1 (p105/50) and NF-κB2 (p100/52) would have a defect in macrophage fusion, nfκb2 shRNA was transduced by lentivirus to nfκb1-deficient BMM (Supplemental Figure 5B and Figure 6). The RANKL-induced formation of osteoclasts was completely abrogated by the knockdown of nfκb2 in the nfκb1−/−background while overexpression of p50 or p65 enhanced osteoclast formation. This result agrees with previous reports showing that NF-κB1/NF-κB2 double knock out mice have a defect in osteoclastogenesis and confirms the efficiency of nfκb2 knockdown(33). In contrast, knockdown of nfκb2 enhanced the IL-4-induced formation of MNG in the nfκb1−/−background in the absence and presence of RANKL (Supplemental Figure 5B and Figure 6A). However, the MNG observed with nfκb2 knockdown in the nfκb1−/− background were smaller than MNG formed in nfκb1+/+ BMM (Supplemental Figure 5) revealing a contribution for p105/p50 in IL-4-induced fusion in a cell devoid of p100/p52.
The mRNA level of osteoclastogenic and fusion related genes were examined by real time PCR (Figure 6B,C). Analysis of mRNA for nfκb2 demonstrated the efficiency of knockdown in these cells. In the absence of nfκb1, RANKL was still able to induce expression of NFATc1 and TRAP and the fusion related genes demonstrating redundancy among NF-κB family members for responsiveness to RANKL. As expected, nfκb2 knockdown completely abrogated the RANKL-induced expression of NFATc1 and TRAP and the fusion-related genes DC-STAMP, E-cadherin, and CD44 in nfκb1−/− BMM. The effect of nfκb2 knockdown on IL-4-induced gene expression was different. DC-STAMP, E-cadherin and CD44 were significantly upregulated in the presence of IL-4. Nfκb2 knockdown significantly enhanced TRAP expression in IL-4 and RANKL plus IL-4 treated BMM without affecting NFATc1 expression (Figure 6B). This is consistent with our previous findings that IL-4-induced TRAP expression is mediated by STAT6 and is unrelated to NFATc1(24,34). These results suggest that p100/p52 inhibits the expression of STAT6 dependent genes induced by IL-4.
Figure 6. Effect of nfκb2 shRNA knockdown in nf κb1−/− BMM on fusion- and osteoclast-related genes.
A. NFκb1−/− BMM were transduced with lentivirus encoding p50, p65, or nfκb2 shRNA. After transduction, cells were cultured in the indicated cytokines for 5 days. A. TRAP or crystal violet staining was performed to identify giant cells (shown in supplemental data). The average size per giant cell (GC, μM2) was measured and statistical analysis was performed as in Figure 3. *, p<0.05, **, p<0.01. B,C. Total RNA was prepared and real time RT-PCR was performed to measure osteoclast- (B) and fusion- (C) -related gene expression. The results are shown as the mean± SD of triplicate samples. **, p<0.01.
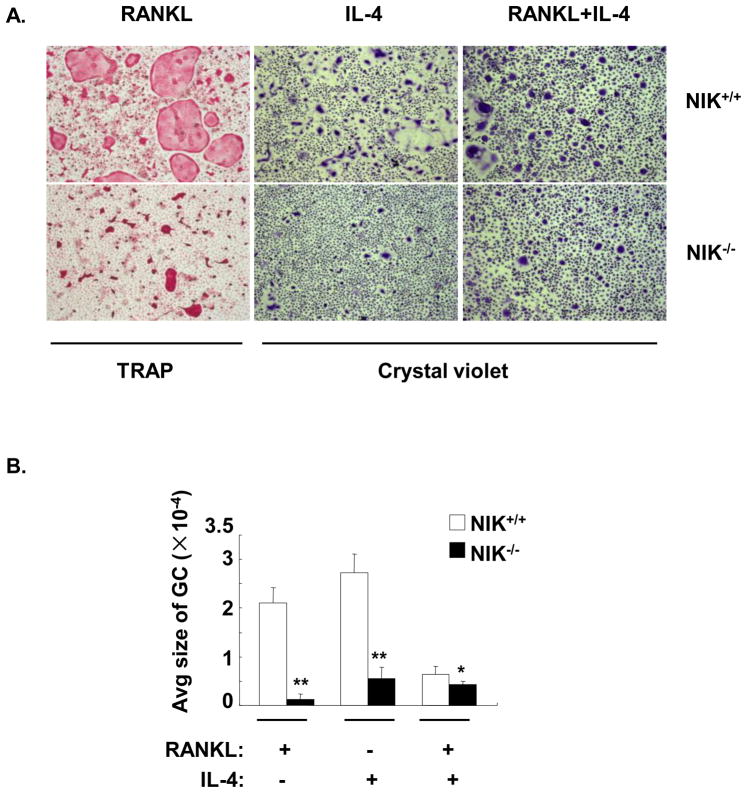
Both RANKL- and IL-4-induced macrophage fusion were abrogated in NIK deficient BMM
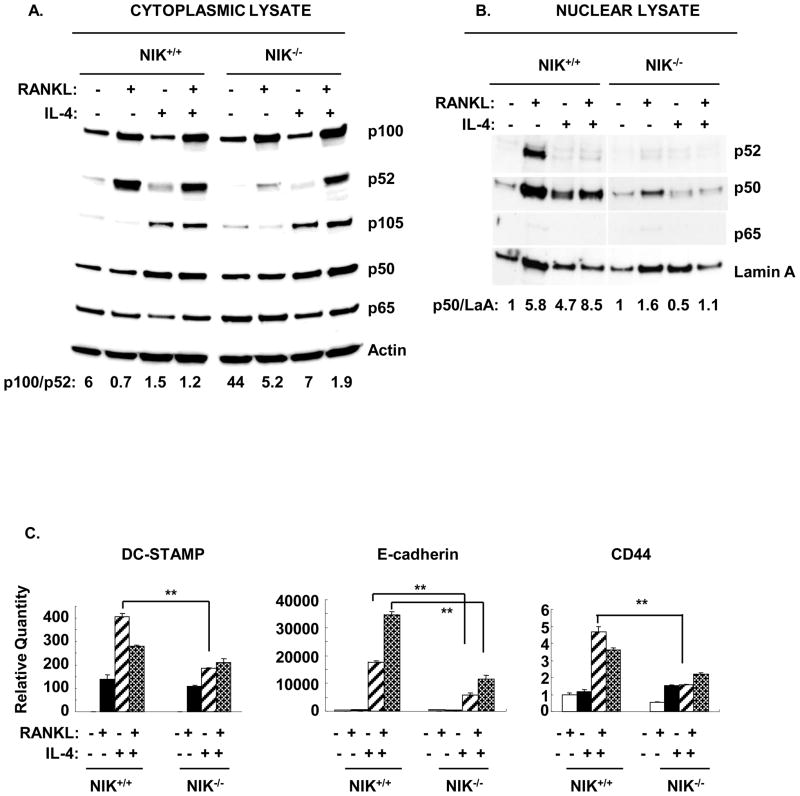
NF-κB inducing kinase (NIK) activates IKK1, which phosphorylates p100. After phosphorylation, p100 undergoes partial degradation to form p52(35). NIK−/− mice demonstrate reduced p100 processing and p52 generation(23). The nfκb2 shRNA knockdown strategy depletes both p100 and p52 expression and therefore cannot distinguish their respective roles in macrophage fusion. To clarify the roles of p100 and p52 in RANKL and IL-4 induced macrophage fusion, NIK−/− BMM were prepared. Both RANKL- and IL-4-induced fusion were significantly reduced in NIK−/− BMM as compared to controls (Figure 7A, B). Cytoplasmic and nuclear extracts of these cultures were evaluated by western blotting. In wild type BMM, RANKL induced the expression of p100 and enhanced the formation of p52 in the cytoplasm. The RANKL-stimulated increase in p52 in the cytoplasm was greatly reduced in the absence of NIK, as expected. The ratio of p100/p52 was significantly increased from 0.7 in wild type cells to 5.2 in NIK−/− BMM. In contrast, treatment of wild type BMM with IL-4 induced expression of p105 and p50 in the cytoplasm. This IL-4-induced response was intact in NIK−/− BMM. As shown in Figure 2, the combination of IL-4 with RANKL resulted in an increase in the p100/p52 ratio in wild type cells. However, in the absence of NIK, IL-4 plus RANKL still induced the production of p52; in this case the amount of p100 was in excess of p52 (p100/p52 ratio 1.9) (Figure 8A).
Figure 7. Both RANKL- and IL-4-induced macrophage fusion were impaired in NIK−/− BMM.
NIK+/+ and NIK−/− BMM were treated with cytokines as indicated for 5 days. A. TRAP or crystal violet staining was performed and representative digital images are shown (40×). B. The average area per GC (μm2) was measured and statistical analysis was performed as shown in Figure 3. **, p<0.01.*, p<0.05. C. Total RNA was prepared and real time RT-PCR was performed to measure fusion-related gene expression in NIK+/+ and NIK−/− BMM. The results are shown as the mean± SD of triplicate samples. **, p<0.01.
Figure 8. IL-4-induced accumulation of p50 in the nucleus and expression of fusion-related genes is dependent on NIK.
BMM were treated as described in Figure 7. A. Cytoplasmic and nuclear lysates were analyzed by western blotting for NF-κB transcription factor subunit expression. The p100/p52 ratio in cytoplasmic lysate was calculated by densitometry using Quantity one software. The p50/Lamin A ratio in nuclear lysate was calculated by densitometry and all values were normalized to the M-CSF only group in NIK+/+ and NIK−/− respectively.
In wild type BMM, RANKL stimulated the nuclear translocation of both p52 and p50 (Figure 8B). The levels of p65 in the nucleus were almost negligible. IL-4 alone induced the accumulation of p50 in the nucleus. Addition of IL-4 inhibited the RANKL-induced translocation of p52. In NIK−/− BMM, the pattern of nuclear translocation was quite different. In the absence of NIK, RANKL did not induce translocation of p52 and the amount of p50 in the nucleus was reduced. Furthermore, the IL-4-induced accumulation of p50 in the nucleus was greatly reduced in the absence of NIK even though IL-4 induced expression of p105 and p50 in the cytoplasmic fraction. The IL-4-induced expression of the fusion related genes DC-STAMP, CD44 and E-cadherin were reduced in NIK−/− BMM as compared to wild type (Figure 8C). Taken together these results demonstrate the important role for NIK in both RANKL- and IL-4-induced macrophage fusion. They also support the model that excess p100 in the cytoplasm acts as an inhibitor of NF-κB dimers and suppresses macrophage fusion. In support of this model, we found that overexpression of p100 in BMM suppressed RANKL- and IL-4-induced macrophage fusion (Supplemental Figure 5A).
Discussion
IL-4 is known to induce macrophage fusion to form MNG by a STAT6-dependent process requiring protein-protein interaction(36). While directly driving MNG formation, IL-4 suppressed the RANKL-induced formation of osteoclasts, cells that also form by macrophage fusion. Several studies showed that inhibition of IκBα degradation and prevention of p65 translocation were responsible for the IL-4 inhibitory effect on osteoclast formation(18–20,37). However, the regulation of NF-κB signaling by IL-4 and the potential consequences of this regulation are highly controversial. Some studies showed that IL-4 suppressed NF-κB activity and inhibited pro-inflammatory cytokine production(38,39). The inhibition of IκBα degradation and suppression of p65 translocation were proposed as the inhibitory mechanism by which IL-4 modulates cellular responses to stress(40). Other mechanisms of regulation were also proposed. For example, IL-4 inhibited TNF-α stimulated E selectin expression through STAT6 competition for an NF-κB DNA binding site in the promoter(41). IL-4/STAT6 also can suppress NF-κB activity through competing for coactivators such as CBP (CREB binding protein)(42). However, there are also reports showing that activation of the heavy chain germline epsilon promoter by IL-4 required both STAT6 and NF-κB binding(43,44). STAT6 and NF-κB were found to directly associate and mediate the synergistic activation of IL-4 induced transcription(45). In B cells, the ability of IL-4-stimulated STAT6 to bind to DNA required NF-κB activity(46). Our previous study showed that NF-κB activation plays an important role in the IL-4-induced protection from apoptosis(47). In addition, another report showed that NF-κB activation is required for CD44-mediated macrophage fusion (21). Taken together, these studies argue that IL-4 activates signaling pathways, including STAT6, that can cross-talk with constitutive or inducible NF-κB pathways. However, the characterization of this cross-talk has remained elusive.
Using a variety of pharmacologic, knockout, knockdown, and overexpression approaches, we investigated the contribution of the NF-κB pathways to IL-4 and RANKL-induced macrophage fusion. While focused on macrophage fusion and cross-talk with RANKL, these studies have implications for the mechanisms by which IL-4 cooperates with other NF-κB-activating receptors including TLR4, B-cell antigen receptor, T-cell receptor and CD40. We found that IL-4 treatment and pretreatment did not inhibit the rapid RANKL-induced activation of the NF-κB canonical pathway. However, we found that IL-4 inhibited the RANKL-induced p52 and RelB translocation which suggests that IL-4 inhibits the RANKL-activated alternative pathway (Figure 9). We do not know the mechanism by which IL-4 suppresses p52/RelB translocation, but it may be due, in part, to an alteration in processing of p100. We found a moderate increase in the p100/p52 ratio in cells cultured in RANKL plus IL-4 as compared to cells cultured in RANKL alone. Since p100 can act as an IκB (IκBδ) and retain p52/RelB and p50/RelA dimers in the cytoplasm (11), an increase in the relative amount of p100 may prevent NF-κB dimer translocation.
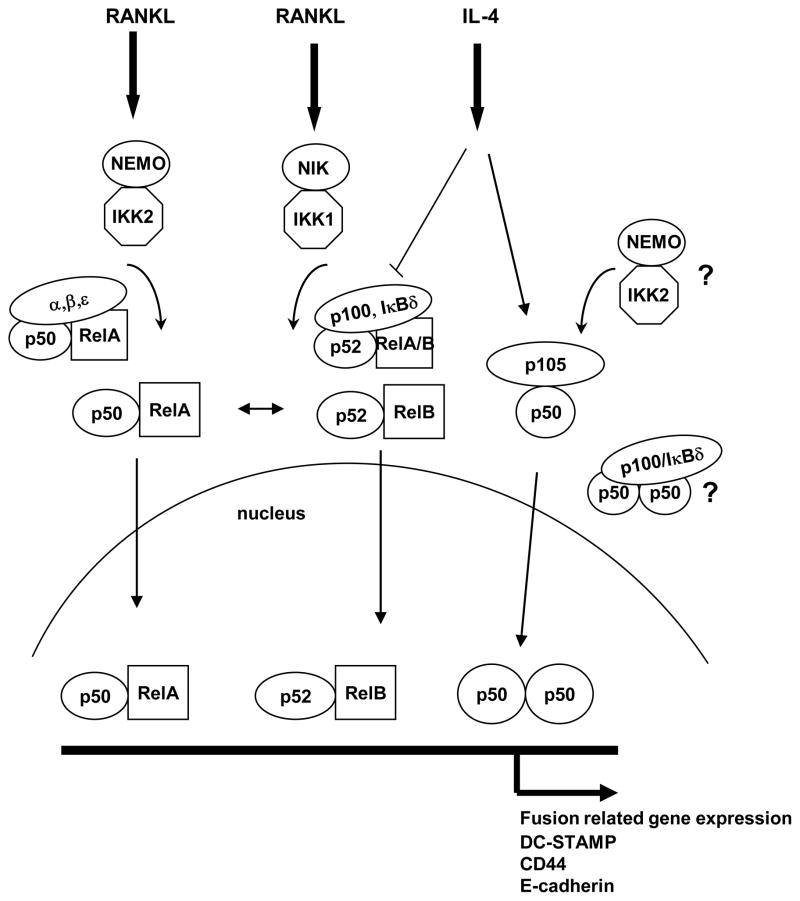
Figure 9. Model for signaling cross-talk.
RANKL rapidly activates the classical (canonical) NF-κB pathway consisting of activation of the classic IKK complex (NEMO-IKK2), degradation of IκBα, and release of p50/p65(RelA) dimers for translocation to the nucleus. IL-4 did not influence this pathway. RANKL also more slowly activates the alternative NF-κB pathway consisting of activation of the alternative NIK-dependent IKK complex (IKK1-IKK1), partial cotranslational proteolysis of p100, generation of p52, and release of p52/RelB(orRelA) dimers for translocation to the nucleus. Intact p100 can function as an IκB (IκBδ) and retains p50/p65 and p52/RelB(A) dimers in the cytoplasm (11). IL-4 altered the relative ratio of p100 to p52 in the cytoplasm, and prevented the RANKL-induced nuclear translocation of p52 and RelB. IL-4 treatment alone did not induce the rapid translocation of p50 or p65, and did not induce the slow translocation of p52 or RelB. However, IL-4 treatment did increase the amount of cytoplasmic p105 and p50 after 48 hours and increased the amount of p50 in the nucleus. Based on EMSA data, IL-4 favored the formation of p50/p50 homodimers. The accumulation of p50 in the nucleus induced by IL-4 was dependent on NIK, while the increase in cytoplasmic p105/p50 was not. IL-4-induced macrophage fusion, known to be STAT6 dependent, was also dependent on NEMO-IKK2 and NIK, and was inhibited by p100. The mechanism(s) for this dependence is unknown, but could be due to processing of p105 by NEMO-IKK2 to p50 and retention of p50/p50 in the cytoplasm by p100.
IL-4 did not induce rapid NF-κB translocation by itself. IL-4 enhanced p105/50 expression and p50/50 homodimer formation after 48 hours treatment. The nfκb1 gene, that encodes p105 and p50, was required for IL-4-induced macrophage fusion while ectopic overexpression of p50 in nfκb1−/− BMM significantly enhanced IL-4-induced MNG formation. Interestingly, NIK, which regulates the inducible proteolysis of p100, was required for IL-4-induced MNG formation, the accumulation of p50 in the nucleus, and induction of fusion related genes in response to IL-4. However, NIK was not required for the induction of cytoplasmic p105/p50 by IL-4. These results suggest that IL-4-induced upregulation of p105 and accumulation of p50 in the nucleus play a critical role in MNG formation and that NIK participates in the IL-4 driven accumulation of nuclear p50 (Figure 9). The mechanism by which NIK regulates this process is unknown. It is possible the inability to process p100 leads to excess cytoplasmic p100 which could act to sequester p50/p50 homodimers (Figure 9). The importance of p105/p50 in these IL-4-driven responses are consistent with several other studies showing a crucial role for p50 in the orchestration of alternative activated macrophage-driven inflammatory reactions and inhibition of the classical activation(48,49).
The NF-κB inhibitor IKK2 inhibitor suppressed both osteoclast and MNG formation induced by RANKL or IL-4 treatment respectively. The NEMO inhibitory peptide also inhibited IL-4 induced MNG formation. The classical IKK complex, composed of IKK1, IKK2 and the regulatory subunit NEMO triggers IκBα degradation and also regulates p105 proteolysis to form p50(50,51). However, only IKK1 but not IKK2 triggers p100 phosphorylation and proteolysis to form p52(52,53). The finding that the IKK2 inhibitor and NEMO-peptide blocked IL-4-induced fusion further suggests that the classic IKK complex plays an important role in RANKL- and IL-4-induced macrophage fusion. Since IL-4 did not induce translocation of p65 at any time points studied, the mechanism by which this complex controls IL-4-induced events is unclear. It is possible that constitutive activity of this complex is required to generate the p50 needed for IL-4-induced fusion.
Overexpression of p52 or RelB and nfκb2 knockdown enhanced RANKL- and IL-4-induced macrophage fusion, while overexpression of p100 reduced IL-4-induced macrophage fusion. Since p52 was shown to enhance macrophage fusion, these results suggest that p100 plays a negative role in MNG formation. NIK deficient BMM have an increased p100/p52 ratio and reduced fusion, suggesting that NIK acts to generate p52 in an inducible fashion, as observed during RANKL signaling, and to decrease the amount of p100. Several studies have also shown that p100 is a negative regulator of RANKL-induced osteoclast formation (32,54,55). These results indicate that the alternative pathway also participates in RANKL- and IL-4-induced macrophage fusion. However, IL-4 alone did not induce p52 and RelB translocation and inhibited the RANKL-induced nuclear translocation of p52 and RelB. These results suggest that p52/RelB are not necessary for IL-4-induced MNG formation.
Knockdown of nfκb2 in nfκb1-deficient BMM completely blocked osteoclast differentiation. On the other hand, nfκb2 knockdown in nfκb1-deficient BMM enhanced IL-4-induced MNG formation. This suggests that IL-4-induced MNG formation may not require both NF-κB1 and NF-κB2. However, IKK2 and NEMO inhibitory peptide completely blocked MNG formation. There are two possible explanations for these somewhat paradoxical findings. One is that NF-κB transcription factor dimers are not absolutely required for IL-4 induced macrophage fusion, and act only to modulate the activity of STAT6. Another possibility is that other NF-κB transcription factor dimers may participate in this process in the absence of NF-κB1 and NF-κB2. Overexpression of either p65 or RelB enhanced IL-4-induced MNG formation. In addition, Rel proteins can form homodimeric and heterodimeric complexes in the absence of NF-κB1 and NF-κB2. For example, p65 can form a relatively stable homodimer complex and is rapidly translocated to the nucleus after stimulation(56). p65 and c-Rel also can form heterodimer complexes and induce the expression of NF-κB responsive genes(57). Further investigation will be required to determine the particular NF-κB pathways and which transcription factor dimers are involved in the IL-4-induced fusion process.
Knockdown of nfκb2 in wild type BMM significantly enhanced the basal expression of CD44, DC-STAMP and E cadherin. This may partially explain why nfκb2 knockdown enhanced both RANKL-induced osteoclast and IL-4-induced MNG formation. The fusion associated genes would already be upregulated and the fusion machinery would be ready for rapid response to fusion stimuli. Compared with responses observed in nfκb2 knockdown in nfκb1-deficient BMM, the basal levels of CD44, DC-STAMP and E cadherin were significantly higher in nfκb2 knockdown of wild type BMM. This further suggests that p105/50 plays a critical role in the expression of these fusion related genes. Expression of these downstream genes induced by IL-4 does not require RANKL- activated NF-κB translocation. It’s still not clear if these fusion-related genes are regulated directly by NF-κB binding to their promoters.
Our previous study indicated that IL-4 modestly enhanced TRAP expression in BMM through STAT6 binding to the TRAP promoter (24). Interestingly, we found that nfκb2 knockdown significantly enhanced IL-4-induced TRAP expression. One possibility for this result is that NF-κB molecules coordinate and enhance STAT6 function in the absence of p100. Another possibility is that p100 has a direct inhibitory effect on STAT6 function. The possible regulation of STAT6 by p100 will require further investigation. The physiological function of TRAP expression in the IL-4-induced MNG is still unknown.
The mechanism of macrophage fusion shares several characteristics with the process of phagocytosis(58,59). Raveh et al. showed that IL-4 and IFN-γ alone or together enhanced mannose receptor mediated phagocytosis (60). However, fusion and phagocytosis can be uncoupled based on their differential susceptibility to Rac1 inhibition. In this model, inhibition of fusion did not inhibit phagocytosis (61). In another study, increasing the stimulus of phagocytosis did not inhibit cell fusion (62). Varin et al. indicated that nonopsonic phagocytic uptake of N meningitids and several other microbial particles were markedly reduced in IL-4 treated peritoneal macrophages although IL-4 induced macrophage fusion (63). Papadimitriou et al. showed that plastic film-induced multinucleate giant cells (>7 nuclei) lost phagocytic ability(64). These studies show that fusion and phagocytosis are different processes. Phagocytosis may not be enhanced through cell fusion. It will be interesting to further investigate whether macrophage phagocytosis will be affected by manipulating NF-κB signaling.
Implanted biomaterials, some infected parasites, or products of infected parasites are subject to a foreign body reaction (65,66). During that process, implants or parasites are surrounded by macrophages that fuse to form mulinuclated giant cells, as well as a collagenous and avascular fibrotic capsule and finally form granuloma (67,68). This damaging effect can lead to reduced functionality, failure of implants, or organ fibrosis (69–71). This foreign body reaction is highly dependent on IL-4 and IL-13 (67,72). Our new findings indicate that NF-κB signaling plays an essential role in both RANKL- and IL-4-induced macrophage fusion. Since NF-κB inhibitors can block both osteoclast and MNG formation, it may be possible to develop NF-κB inhibitors as therapeutic agents to prevent Th2 type giant cell and granuloma formation. This concept provides a new explanation for the finding that glucocorticoids, potent NF-κB inhibitors (73), effectively reduced both Schistosoma mansoni egg-induced granuloma formation, maturation (74) and strongly inhibited granuloma formation induced by implanted biomaterial (65).
Supplementary Material
Acknowledgments
We’d like to acknowledge Dr. Ranjan Sen for providing nfκb1−/− mice. We’d like to acknowledge Dr. Robert Schreiber and Dr. Barry Sleckman for providing NIK−/− mice. We also would like to acknowledge Dr. Janet Stavnezer, Dr. Ricardo Feldman, and Dr. Didier Trono for generously supplying plasmid cDNA for these studies, and Ms. Mary Kaileh and and Mr. Ryan Irwin for excellent technical assistance.
This work was supported by PHS grants AI059775 and AI038985 (ADK).
Reference List
- 1.Anderson JM. Multinucleated giant cells. Curr Opin Hematol. 2000;7:40–47. doi: 10.1097/00062752-200001000-00008. [DOI] [PubMed] [Google Scholar]
- 2.Saginario C, Sterling H, Beckers C, Kobayashi R, Solimena M, Ullu E, Vignery A. MFR, a putative receptor mediating the fusion of macrophages. Mol Cell Biol. 1998;18:6213–6223. doi: 10.1128/mcb.18.11.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han X, Sterling H, Chen Y, Saginario C, Brown EJ, Frazier WA, Lindberg FP, Vignery A. CD47, a ligand for the macrophage fusion receptor, participates in macrophage multinucleation. J Biol Chem. 2000;275:37984–37992. doi: 10.1074/jbc.M002334200. [DOI] [PubMed] [Google Scholar]
- 4.Yagi M, Miyamoto T, Sawatani Y, Iwamoto K, Hosogane N, Fujita N, Morita K, Ninomiya K, Suzuki T, Miyamoto K, Oike Y, Takeya M, Toyama Y, Suda T. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J Exp Med. 2005;202:345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sterling H, Saginario C, Vignery A. CD44 occupancy prevents macrophage multinucleation. J Cell Biol. 1998;143:837–847. doi: 10.1083/jcb.143.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helming L, Winter J, Gordon S. The scavenger receptor CD36 plays a role in cytokine-induced macrophage fusion. J Cell Sci. 2009;122:453–459. doi: 10.1242/jcs.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helming L, Gordon S. Molecular mediators of macrophage fusion. Trends Cell Biol. 2009;19:514–522. doi: 10.1016/j.tcb.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Van den Bossche J, Bogaert P, van HJ, Guerin CJ, Berx G, Movahedi K, Van den Bergh R, Pereira-Fernandes A, Geuns JM, Pircher H, Dorny P, Grooten J, De BP, Van Ginderachter JA. Alternatively activated macrophages engage in homotypic and heterotypic interactions through IL-4 and polyamine-induced E-cadherin/catenin complexes. Blood. 2009;114:4664–4674. doi: 10.1182/blood-2009-05-221598. [DOI] [PubMed] [Google Scholar]
- 9.Walsh NC, Cahill M, Carninci P, Kawai J, Okazaki Y, Hayashizaki Y, Hume DA, Cassady AI. Multiple tissue-specific promoters control expression of the murine tartrate-resistant acid phosphatase gene. Gene. 2003;307:111–123. doi: 10.1016/s0378-1119(03)00449-9. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 11.Basak S, Kim H, Kearns JD, Tergaonkar V, O’Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iotsova V, Caamano J, Loy J, Yang Y, Lewin A, Bravo R. Osteopetrosis in mice lacking NF-kappaB1 and NF-kappaB2. Nat Med. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 13.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 14.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Moreno JL, Mikhailenko I, Tondravi MM, Keegan AD. IL-4 promotes the formation of multinucleated giant cells from macrophage precursors by a STAT6-dependent, homotypic mechanism: contribution of E-cadherin. J Leukoc Biol. 2007;82:1542–1553. doi: 10.1189/jlb.0107058. [DOI] [PubMed] [Google Scholar]
- 16.Moreno JL, Kaczmarek M, Keegan AD, Tondravi M. IL-4 suppresses osteoclast development and mature osteoclast function by a STAT6-dependent mechanism: irreversible inhibition of the differentiation program activated by RANKL. Blood. 2003;102:1078–1086. doi: 10.1182/blood-2002-11-3437. [DOI] [PubMed] [Google Scholar]
- 17.Yagi M, Ninomiya K, Fujita N, Suzuki T, Iwasaki R, Morita K, Hosogane N, Matsuo K, Toyama Y, Suda T, Miyamoto T. Induction of DC-STAMP by alternative activation and downstream signaling mechanisms. J Bone Miner Res. 2007;22:992–1001. doi: 10.1359/jbmr.070401. [DOI] [PubMed] [Google Scholar]
- 18.Mangashetti LS, Khapli SM, Wani MR. IL-4 inhibits bone-resorbing activity of mature osteoclasts by affecting NF-kappa B and Ca2+ signaling. J Immunol. 2005;175:917–925. doi: 10.4049/jimmunol.175.2.917. [DOI] [PubMed] [Google Scholar]
- 19.Abu-Amer Y. IL-4 abrogates osteoclastogenesis through STAT6-dependent inhibition of NF-kappaB. J Clin Invest. 2001;107:1375–1385. doi: 10.1172/JCI10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei S, Wang MW, Teitelbaum SL, Ross FP. Interleukin-4 reversibly inhibits osteoclastogenesis via inhibition of NF-kappa B and mitogen-activated protein kinase signaling. J Biol Chem. 2002;277:6622–6630. doi: 10.1074/jbc.M104957200. [DOI] [PubMed] [Google Scholar]
- 21.Cui W, Ke JZ, Zhang Q, Ke HZ, Chalouni C, Vignery A. The intracellular domain of CD44 promotes the fusion of macrophages. Blood. 2006;107:796–805. doi: 10.1182/blood-2005-05-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yagi M, Ninomiya K, Fujita N, Suzuki T, Iwasaki R, Morita K, Hosogane N, Matsuo K, Toyama Y, Suda T, Miyamoto T. Induction of DC-STAMP by alternative activation and downstream signaling mechanisms. J Bone Miner Res. 2007;22:992–1001. doi: 10.1359/jbmr.070401. [DOI] [PubMed] [Google Scholar]
- 23.Yin L, Wu L, Wesche H, Arthur CD, White JM, Goeddel DV, Schreiber RD. Defective lymphotoxin-beta receptor-induced NF-kappaB transcriptional activity in NIK-deficient mice. Science. 2001;291:2162–2165. doi: 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- 24.Yu M, Moreno JL, Stains JP, Keegan AD. Complex regulation of tartrate-resistant acid phosphatase (TRAP) expression by interleukin 4 (IL-4): IL-4 indirectly suppresses receptor activator of NF-kappaB ligand (RANKL)-mediated TRAP expression but modestly induces its expression directly. J Biol Chem. 2009;284:32968–32979. doi: 10.1074/jbc.M109.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohuslav J, V, Kravchenko V, Parry GC, Erlich JH, Gerondakis S, Mackman N, Ulevitch RJ. Regulation of an essential innate immune response by the p50 subunit of NF-kappaB. J Clin Invest. 1998;102:1645–1652. doi: 10.1172/JCI3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen CH, Stavnezer J. Interaction of stat6 and NF-kappaB: direct association and synergistic activation of interleukin-4-induced transcription. Mol Cell Biol. 1998;18:3395–3404. doi: 10.1128/mcb.18.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Zhang B, Yang L, Ding J, Ding HF. Constitutive production of NF-kappaB2 p52 is not tumorigenic but predisposes mice to inflammatory autoimmune disease by repressing Bim expression. J Biol Chem. 2008;283:10698–10706. doi: 10.1074/jbc.M800806200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan JJ, McReynolds LJ, Keegan A, Wang LH, Garfein E, Rothman P, Nelms K, Paul WE. Growth and gene expression are predominantly controlled by distinct regions of the human IL-4 receptor. Immunity. 1996;4:123–132. doi: 10.1016/s1074-7613(00)80677-9. [DOI] [PubMed] [Google Scholar]
- 29.Wei S, Wang MW, Teitelbaum SL, Ross FP. Interleukin-4 reversibly inhibits osteoclastogenesis via inhibition of NF-kappa B and mitogen-activated protein kinase signaling. J Biol Chem. 2002;277:6622–6630. doi: 10.1074/jbc.M104957200. [DOI] [PubMed] [Google Scholar]
- 30.Heissmeyer V, Krappmann D, Hatada EN, Scheidereit C. Shared pathways of IkappaB kinase-induced SCF(betaTrCP)-mediated ubiquitination and degradation for the NF-kappaB precursor p105 and IkappaBalpha. Mol Cell Biol. 2001;21:1024–1035. doi: 10.1128/MCB.21.4.1024-1035.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.May MJ, D’Acquisto F, Madge LA, Glockner J, Pober JS, Ghosh S. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science. 2000;289:1550–1554. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- 32.Novack DV, Yin L, Hagen-Stapleton A, Schreiber RD, Goeddel DV, Ross FP, Teitelbaum SL. The IkappaB function of NF-kappaB2 p100 controls stimulated osteoclastogenesis. J Exp Med. 2003;198:771–781. doi: 10.1084/jem.20030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iotsova V, Caamano J, Loy J, Yang Y, Lewin A, Bravo R. Osteopetrosis in mice lacking NF-kappaB1 and NF-kappaB2. Nat Med. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 34.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 35.Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 36.McInnes A, Rennick DM. Interleukin 4 induces cultured monocytes/macrophages to form giant multinucleated cells. J Exp Med. 1988;167:598–611. doi: 10.1084/jem.167.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirayama T, Dai S, Abbas S, Yamanaka Y, Abu-Amer Y. Inhibition of inflammatory bone erosion by constitutively active STAT-6 through blockade of JNK and NF-kappaB activation. Arthritis Rheum. 2005;52:2719–2729. doi: 10.1002/art.21286. [DOI] [PubMed] [Google Scholar]
- 38.Clarke CJ, Taylor-Fishwick DA, Hales A, Chernajovsky Y, Sugamura K, Feldmann M, Foxwell BM. Interleukin-4 inhibits kappa light chain expression and NF kappa B activation but not I kappa B alpha degradation in 70Z/3 murine pre-B cells. Eur J Immunol. 1995;25:2961–2966. doi: 10.1002/eji.1830251037. [DOI] [PubMed] [Google Scholar]
- 39.Donnelly RP, Crofford LJ, Freeman SL, Buras J, Remmers E, Wilder RL, Fenton MJ. Tissue-specific regulation of IL-6 production by IL-4. Differential effects of IL-4 on nuclear factor-kappa B activity in monocytes and fibroblasts. J Immunol. 1993;151:5603–5612. [PubMed] [Google Scholar]
- 40.Nelson G, Wilde GJ, Spiller DG, Kennedy SM, Ray DW, Sullivan E, Unitt JF, White MR. NF-kappaB signalling is inhibited by glucocorticoid receptor and STAT6 via distinct mechanisms. J Cell Sci. 2003;116:2495–2503. doi: 10.1242/jcs.00461. [DOI] [PubMed] [Google Scholar]
- 41.Bennett BL, Cruz R, Lacson RG, Manning AM. Interleukin-4 suppression of tumor necrosis factor alpha-stimulated E-selectin gene transcription is mediated by STAT6 antagonism of NF-kappaB. J Biol Chem. 1997;272:10212–10219. doi: 10.1074/jbc.272.15.10212. [DOI] [PubMed] [Google Scholar]
- 42.Ohmori Y, Hamilton TA. Interleukin-4/STAT6 represses STAT1 and NF-kappa B-dependent transcription through distinct mechanisms. J Biol Chem. 2000;275:38095–38103. doi: 10.1074/jbc.M006227200. [DOI] [PubMed] [Google Scholar]
- 43.Stutz AM, Woisetschlager M. Functional synergism of STAT6 with either NF-kappa B or PU.1 to mediate IL-4-induced activation of IgE germline gene transcription. J Immunol. 1999;163:4383–4391. [PubMed] [Google Scholar]
- 44.Delphin S, Stavnezer J. Characterization of an interleukin 4 (IL-4) responsive region in the immunoglobulin heavy chain germline epsilon promoter: regulation by NF-IL-4, a C/EBP family member and NF-kappa B/p50. J Exp Med. 1995;181:181–192. doi: 10.1084/jem.181.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen CH, Stavnezer J. Interaction of stat6 and NF-kappaB: direct association and synergistic activation of interleukin-4-induced transcription. Mol Cell Biol. 1998;18:3395–3404. doi: 10.1128/mcb.18.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thieu VT, Nguyen ET, McCarthy BP, Bruns HA, Kapur R, Chang CH, Kaplan MH. IL-4-stimulated NF-kappaB activity is required for Stat6 DNA binding. J Leukoc Biol. 2007;82:370–379. doi: 10.1189/jlb.1106707. [DOI] [PubMed] [Google Scholar]
- 47.Zamorano J, Mora AL, Boothby M, Keegan AD. NF-kappa B activation plays an important role in the IL-4-induced protection from apoptosis. Int Immunol. 2001;13:1479–1487. doi: 10.1093/intimm/13.12.1479. [DOI] [PubMed] [Google Scholar]
- 48.Porta C, Rimoldi M, Raes G, Brys L, Ghezzi P, Di LD, Dieli F, Ghisletti S, Natoli G, De BP, Mantovani A, Sica A. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proc Natl Acad Sci U S A. 2009;106:14978–14983. doi: 10.1073/pnas.0809784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saccani A, Schioppa T, Porta C, Biswas SK, Nebuloni M, Vago L, Bottazzi B, Colombo MP, Mantovani A, Sica A. p50 nuclear factor-kappaB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 2006;66:11432–11440. doi: 10.1158/0008-5472.CAN-06-1867. [DOI] [PubMed] [Google Scholar]
- 50.Salmeron A, Janzen J, Soneji Y, Bump N, Kamens J, Allen H, Ley SC. Direct phosphorylation of NF-kappaB1 p105 by the IkappaB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J Biol Chem. 2001;276:22215–22222. doi: 10.1074/jbc.M101754200. [DOI] [PubMed] [Google Scholar]
- 51.Lang V, Janzen J, Fischer GZ, Soneji Y, Beinke S, Salmeron A, Allen H, Hay RT, Ben-Neriah Y, Ley SC. betaTrCP-mediated proteolysis of NF-kappaB1 p105 requires phosphorylation of p105 serines 927 and 932. Mol Cell Biol. 2003;23:402–413. doi: 10.1128/MCB.23.1.402-413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 53.Xiao G, Cvijic ME, Fong A, Harhaj EW, Uhlik MT, Waterfield M, Sun SC. Retroviral oncoprotein Tax induces processing of NF-kappaB2/p100 in T cells: evidence for the involvement of IKKalpha. EMBO J. 2001;20:6805–6815. doi: 10.1093/emboj/20.23.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao Z, Xing L, Boyce BF. NF-kappaB p100 limits TNF-induced bone resorption in mice by a TRAF3-dependent mechanism. J Clin Invest. 2009;119:3024–3034. doi: 10.1172/JCI38716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soysa NS, Alles N, Weih D, Lovas A, Mian AH, Shimokawa H, Yasuda H, Weih F, Jimi E, Ohya K, Aoki K. The pivotal role of the alternative NF-kappaB pathway in maintenance of basal bone homeostasis and osteoclastogenesis. J Bone Miner Res. 2010;25:809–818. doi: 10.1359/jbmr.091030. [DOI] [PubMed] [Google Scholar]
- 56.Ganchi PA, Sun SC, Greene WC, Ballard DW. A novel NF-kappa B complex containing p65 homodimers: implications for transcriptional control at the level of subunit dimerization. Mol Cell Biol. 1993;13:7826–7835. doi: 10.1128/mcb.13.12.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parry GC, Mackman N. A set of inducible genes expressed by activated human monocytic and endothelial cells contain kappa B-like sites that specifically bind c-Rel-p65 heterodimers. J Biol Chem. 1994;269:20823–20825. [PubMed] [Google Scholar]
- 58.McNally AK, DeFife KM, Anderson JM. Interleukin-4-induced macrophage fusion is prevented by inhibitors of mannose receptor activity. Am J Pathol. 1996;149:975–985. [PMC free article] [PubMed] [Google Scholar]
- 59.McNally AK, Anderson JM. Multinucleated giant cell formation exhibits features of phagocytosis with participation of the endoplasmic reticulum. Exp Mol Pathol. 2005;79:126–135. doi: 10.1016/j.yexmp.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 60.Raveh D, Kruskal BA, Farland J, Ezekowitz RA. Th1 and Th2 cytokines cooperate to stimulate mannose-receptor-mediated phagocytosis. J Leukoc Biol. 1998;64:108–113. [PubMed] [Google Scholar]
- 61.Jay SM, Skokos E, Laiwalla F, Krady MM, Kyriakides TR. Foreign body giant cell formation is preceded by lamellipodia formation and can be attenuated by inhibition of Rac1 activation. Am J Pathol. 2007;171:632–640. doi: 10.2353/ajpath.2007.061213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jay SM, Skokos EA, Zeng J, Knox K, Kyriakides TR. Macrophage fusion leading to foreign body giant cell formation persists under phagocytic stimulation by microspheres in vitro and in vivo in mouse models. J Biomed Mater Res A. 2010;93:189–199. doi: 10.1002/jbm.a.32513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Varin A, Mukhopadhyay S, Herbein G, Gordon S. Alternative activation of macrophages by IL-4 impairs phagocytosis of pathogens but potentiates microbial-induced signalling and cytokine secretion. Blood. 2010;115:353–362. doi: 10.1182/blood-2009-08-236711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Papadimitriou JM, Robertson TA, Walters MN. An analysis of the Phagocytic potential of multinucleate foreign body giant cells. Am J Pathol. 1975;78:343–358. [PMC free article] [PubMed] [Google Scholar]
- 65.Morais JM, Papadimitrakopoulos F, Burgess DJ. Biomaterials/tissue interactions: possible solutions to overcome foreign body response. AAPS J. 2010;12:188–196. doi: 10.1208/s12248-010-9175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kellermeyer RW, Warren KS. The role of chemical mediators in the inflammatory response induced by foreign bodies: comparison with the schistosome egg granuloma. J Exp Med. 1970;131:21–39. doi: 10.1084/jem.131.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Higgins DM, Basaraba RJ, Hohnbaum AC, Lee EJ, Grainger DW, Gonzalez-Juarrero M. Localized immunosuppressive environment in the foreign body response to implanted biomaterials. Am J Pathol. 2009;175:161–170. doi: 10.2353/ajpath.2009.080962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luttikhuizen DT, Harmsen MC, van Luyn MJ. Cellular and molecular dynamics in the foreign body reaction. Tissue Eng. 2006;12:1955–1970. doi: 10.1089/ten.2006.12.1955. [DOI] [PubMed] [Google Scholar]
- 69.Zhao Q, Topham N, Anderson JM, Hiltner A, Lodoen G, Payet CR. Foreign-body giant cells and polyurethane biostability: in vivo correlation of cell adhesion and surface cracking. J Biomed Mater Res. 1991;25:177–183. doi: 10.1002/jbm.820250205. [DOI] [PubMed] [Google Scholar]
- 70.Boynton EL, Henry M, Morton J, Waddell JP. The inflammatory response to particulate wear debris in total hip arthroplasty. Can J Surg. 1995;38:507–515. [PubMed] [Google Scholar]
- 71.Andrade ZA. Schistosomiasis and liver fibrosis. Parasite Immunol. 2009;31:656–663. doi: 10.1111/j.1365-3024.2009.01157.x. [DOI] [PubMed] [Google Scholar]
- 72.Jankovic D, Kullberg MC, Noben-Trauth N, Caspar P, Ward JM, Cheever AW, Paul WE, Sher A. Schistosome-infected IL-4 receptor knockout (KO) mice, in contrast to IL-4 KO mice, fail to develop granulomatous pathology while maintaining the same lymphokine expression profile. J Immunol. 1999;163:337–342. [PubMed] [Google Scholar]
- 73.Auphan N, Didonato JA, Rosette C, Helmberg A, Karin M. Immunosuppression by glucocorticoids: inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 74.Pyrrho AS, Ramos JA, Neto RM, Silva CS, Lenzi HL, Takiya CM, Gattass CR. Dexamethasone, a drug for attenuation of Schistosoma mansoni infection morbidity. Antimicrob Agents Chemother. 2002;46:3490–3498. doi: 10.1128/AAC.46.11.3490-3498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.