Abstract
Plasmodium vivax and Plasmodium knowlesi depend on the Duffy-Binding Protein DBL domain (RII-PvDBP or RII-PkDBP) engaging Duffy Antigen/Receptor for Chemokines on red blood cells during invasion. Inhibition of this key interaction provides an excellent opportunity for parasite control. There are competing models for whether Plasmodium ligands engage receptors as monomers or dimers, resolution of which has profound implications for parasite biology and control. We report crystallographic, solution and functional studies of RII-PvDBP, showing dimerization is required for and driven by receptor engagement. This work provides a unifying framework for prior studies and accounts for the action of naturally-acquired blocking-antibodies and the mechanism of immune evasion. We show dimerization is conserved in DBL-domain receptor-engagement, and propose receptor-mediated ligand-dimerization drives receptor affinity and specificity. Since dimerization is prevalent in signaling, our studies raise the possibility that induced dimerization activates pathways for invasion.
Reticulocyte invasion by Plasmodium vivax and Plasmodium knowlesi requires binding of the Duffy-Binding Protein (PvDBP or PkDBP) to the Red Blood Cell (RBC) Duffy Antigen/Receptor for Chemokines (DARC)1–3. PvDBP is a leading vaccine candidate for P. vivax malaria because the absence of PvDBP-DARC interaction in Duffy-null individuals confers protection against P. vivax infection3. Understanding the structure, function and mechanism of this important receptor-ligand interaction may inform strategies for improved control. PvDBP contains a single 302 amino acid cysteine-rich Duffy Binding-Like (DBL) domain within its extracellular N-terminus called region II (RII-PvDBP)4. This region contains twelve conserved cysteines and is sufficient for binding to DARC5,6.
PvDBP is a member of the Erythrocyte Binding-Like (EBL) protein superfamily. Members of this protein family contain one or two extracellular cysteine-rich DBL domains (region II), a second extracellular cysteine-rich domain (region VI), a type I transmembrane domain, and a short cytoplasmic domain (Supplemental Fig. 1a)4. EBL proteins are trafficked to secretory microneme organelles of the blood-stage merozoite form of Plasmodium parasites for host cell invasion7,8. Unlike the single EBL protein found in P. vivax, P. falciparum has four EBL proteins – PfEBA-175, PfEBA-140, PfEBA-181 and PfEBL-19–13, which allows the P. falciparum merozoite multiple invasion pathways6,10,14. P. vivax only contains the single PvDBP in its genome suggesting there are no alternate invasion pathways15.
Plasmodium DBL domains mediate diverse receptor-ligand interactions critical for invasion, cytoadherence, sequestration and the pathogenesis of malaria12. Plasmodium parasites have adapted the DBL fold to recognize a variety of chemically and functionally diverse host receptors. DBL domains of erythrocyte invasion proteins, such as PvDBP, mediate RBC invasion via high-affinity interactions with distinct host cell receptors. Hypervariant Plasmodium falciparum Erythrocyte Membrane Protein-1 (PfEMP1) mediates cytoadherence and placental sequestration by binding several different receptors via DBL domains. Despite the widespread and critical nature of DBL-receptor interactions in Plasmodium pathogenesis, the molecular details of receptor recognition have yet to be fully characterized. The structural basis and mechanism for receptor recognition of PvDBP would serve as a good model for other DBL interactions.
PvDBP must bind its RBC receptor, DARC, via RII-PvDBP to initiate reticulocyte invasion. DARC is a homodimeric G-protein coupled receptor (GPCR)16 whose N-terminal 60 amino acids (DARC1–60) are sufficient for inhibiting PvDBP mediated RBC rosetting17,18. DARC1–60 contains two tyrosines (Tyr30 and Tyr41) which are post-translationally sulfated. Sulfation of DARC Tyr41 results in a 1000 fold increase in inhibition of RBC binding demonstrated by a change in Ki from low micromolar to low nanomolar. Thus, sulfation is a critical binding determinant for PvDBP.
The adaptive immune response plays a critical role in parasite control. Residents in endemic areas where P. vivax is prevalent have naturally acquired antibodies to PvDBP19–21. Mapping epitopes of naturally-acquired blocking-antibodies that prevent PvDBP binding to RBCs identified linear epitopes within RII-PvDBP22. The parasite evades the immune response through extensive sequence polymorphisms several of which are found in RII-PvDBP20,23. Although it is clear that RII-PvDBP is a critical target for the host immune response and for immune evasion by the parasite, the molecular basis for protection and immune evasion are unclear.
Here, we present the crystal structure of the clinically relevant RII-PvDBP, show that RII-PvDBP dimerization is required for receptor binding, and demonstrate that receptor-binding drives RII-PvDBP dimerization in solution. The structure reveals a putative DARC binding site and putative sulfotyrosine binding pocket formed by a RII-PvDBP dimer. The DARC binding site and sulfotyrosine binding pocket are distinct from residues previously thought to bind sulfotyrosine24. The dimer interface and critical DARC binding residues required for RBC binding are targeted by the immune response, and are structurally and functionally conserved. These results elucidate the molecular mechanism of DBL-receptor interactions and identify new targets for small molecule and vaccine therapeutics.
Results
Overall architecture of the RII-PvDBP homodimer
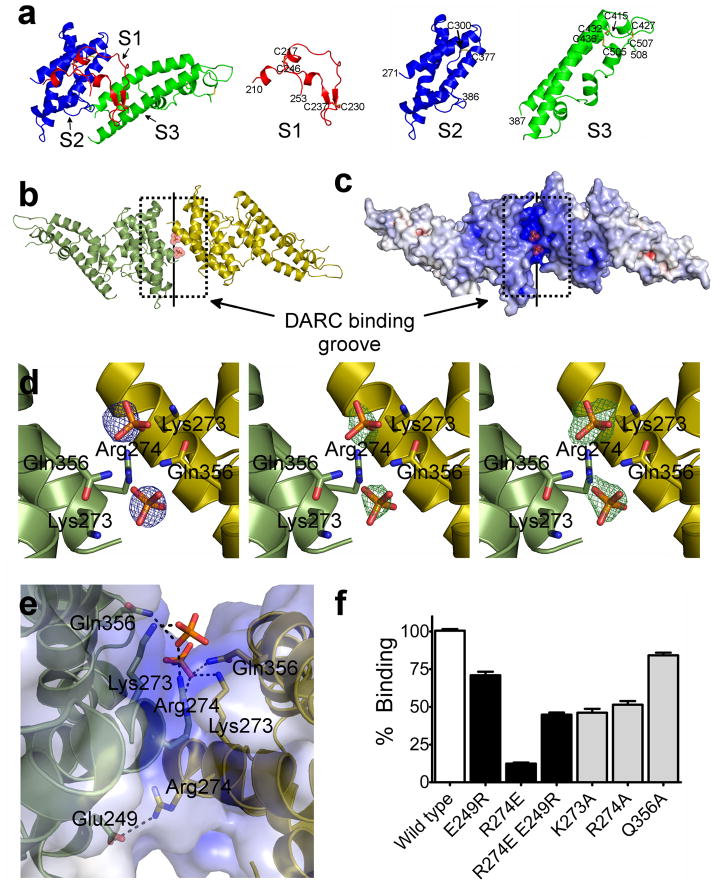
The single DBL domain from PvDBP (RII-PvDBP) is sufficient for binding to DARC5,6. We have determined a 1.95 Å crystal structure of RII-PvDBP (Fig. 1, Supplementary Fig. 2 and Table 1). RII-PvDBP is an elongated boomerang shaped molecule, composed primarily of α helices, with an antiparallel β hairpin near the N-terminus. RII-PvDBP is similar in sequence and structure to the DBL domains of RII-PfEBA-175 and RII-PkDBP, with RMSDs of 1.53, 1.84 and 1.06 Å, respectively (Supplementary Fig. 3). The RII-PvDBP DBL domain is composed of three subdomains that are stabilized by intra-subdomain disulfide bridges (Fig. 1a, Supplementary Fig. 1b). Subdomain 1 (S1) is composed of residues Asn211–Leu253 and has two intra-subdomain disulfides, Cys217–Cys246 and Cys230–Cys237. Subdomain 2 (S2) is composed of residues Tyr271–Glu386 and has one intra-subdomain disulfide, Cys300–Cys377. Subdomain 3 (S3) is composed of residues Pro387–Ser508 and has three intra-subdomain disulfides, Cys415–Cys432, Cys427–Cys507, and Cys436–Cys505 (Fig. 1a). All cysteines are involved in disulfide bridges, with a conserved disulfide bridging pattern seen in the DBL domains of P. falciparum EBA-175 (RII-PfEBA-175) and P. knowlesi DBP (RII-PkDBP) (Supplementary Fig. 1b).
Figure 1.
RII-PvDBP is composed of three subdomains, and a sulfotyrosine pocket within a DARC binding groove is formed by the RII-PvDBP dimer. (a) RII-PvDBP separated into the three subdomains. Subdomain 1 (S1 - red) contains the β-hairpin, subdomain 2 (S2 - blue) is a four helix bundle and subdomain 3 (S3 -green) forms a second helical bundle. (b) The RII-PvDBP dimer in ribbon representation, rotated by 180° along x. Monomers are in green and yellow. The DARC binding groove is outlined by a dashed box and the dimer interface is indicated by a solid line. (c) Electrostatic mapping of the RII-PvDBP dimer, rotated by 180° along x. The DARC binding groove is positively charged. (d) Density which clearly identifies selenate or phosphate at the RII-PvDBP dimer interface is shown. Left - Selenium anomalous difference map (blue mesh) from crystals grown in the presence of selenate, contoured at 4σ. Middle - the difference map (green mesh), contoured at 2.5σ, of crystals grown in phosphate, prior to addition of phosphates. Right - the omit map (green mesh), contoured at 2.5σ, of the final refined structure with the phosphates omitted. Phosphates are drawn in stick and colored red and yellow. Side chains of residues involved in interactions are shown in stick. (e) The putative sulfotyrosine binding pocket and the Arg274-Glu249 salt bridge shown. Phosphates are drawn in stick and colored red and yellow. Side chains of residues involved in interactions are shown in stick and contacts are depicted by dashed black lines. (f) Percentage of cells expressing point mutants of RII-PvDBP that bind RBCs relative to wildtype, shown with standard error. A paired two-tailed student t-test indicated that all mutants compared to wildtype have a p-value<0.0001. White bar – wildtype. Black bars – dimer mutants and rescue. Grey bars – sulfotyrosine binding mutants.
Table 1.
Data collection and refinement statistics
| 1.95 Å Native | 2.2 Å Selenate SAD | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 59.50, 66.97, 91.83 | 59.47, 66.85, 91.97 |
| α, β, γ (°) | 90, 108.055, 90 | 90, 108.103, 90 |
| Resolution (Å) | 20.0–1.95 (2.05–1.95) | 20.0–2.2 (2.30–2.20) |
| Rmerge | 0.069 (0.620) | 0.099 (0.582) |
| I / σI | 13.32 (1.95) | 13.24 (3.73) |
| Completeness (%) | 99.4 (99.5) | 99.8 (99.8) |
| Redundancy | 3.7 (3.7) | 5.3 (5.3) |
| Refinement | ||
| Resolution (Å) | 20.0–1.95 Å | |
| No. Reflections | 49,917 | |
| Rwork/Rfree | 19.50 / 24.11 | |
| No. Atoms | ||
| Protein | 9,217 | |
| Ligand Organic | 110 | |
| Ligand Inorganic | 20 | |
| Water | 364 | |
| B-factors | ||
| Protein | 44.42 | |
| Ligand Organic | 46.33 | |
| Ligand Inorganic | 76.98 | |
| Water | 41.71 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.009 | |
| Bond angles (°) | 1.066 | |
Values in parentheses are for highest-resolution shell.
Data were collected on a single crystal for each dataset
Two RII-PvDBP monomers are related by non-crystallographic symmetry, generating a dimer in the asymmetric unit (Fig. 1b,c). The dimeric architecture is created by homodimer contacts across subdomain 2. As shown below, the dimeric organization is required for functional receptor binding. A cluster of hydrophobic homodimer contacts are made by Phe267 and Leu270 of chain A with Ile277, Tyr278, Val282, and Tyr363 of chain B. A salt bridge across the two monomers at the dimer interface is created by Arg274 and Glu249.
The dimer creates a putative sulfotyrosine binding pocket
DARC sulfation results in a change in Ki of inhibition of RBC binding from micromolar to low nanomolar, indicating sulfation is critical for receptor recognition and binding18. To identify putative sulfotyrosine binding pockets we cocrystallized RII-PvDBP with sodium selenate (Table 1). Selenate is structurally similar to sulfate so we reasoned selenate should bind to and identify residues that form the sulfotyrosine binding pocket. Selenate was also selected as selenium-anomalous data could be collected to unambiguously identify the selenate binding sites.
We observed clear density at five sites in the anomalous maps for selenate at a sigma cutoff of greater than six. Of these five, two are located outside the minimal binding domain of RII-PvDBP, and are likely not functionally relevant. The remaining three sites are at the dimer interface. In the native structure, two of these sites are occupied by phosphates. Since phosphate, selenate and sulfate are stereochemically similar we propose these phosphate binding sites form a putative sulfotyrosine binding pocket that binds sulfated DARC (Fig. 1d). Lys273 and Gln356 of both chains and Arg274 of one chain create the pocket by directly contact the selenates (Fig. 1d). The putative sulfotyrosine binding pocket is generated by the RII-PvDBP dimer and located in a positively charged groove at the dimer interface. We propose this region is the DARC binding groove that engages the highly acidic DARC1–60 (Fig. 1c).
Our structural studies suggest an intact sulfotyrosine binding pocket is critical for DARC-dependent binding of RBC. We tested this model by mutating residues in the putative sulfotyrosine binding pocket to alanine and assayed the ability of the mutants to bind RBCs in a rosette binding assay. Mutations of any residue in the putative sulfotyrosine binding pocket resulted in a significant decrease in RBC rosetting (Fig. 1e, 1f and Supplementary Fig. 4). All decreases were significant compared to wildtype to a p-value<0.0001. These results are specific as mutation of residues coordinating a third phosphate located away from the binding groove has no effect on rosetting (Supplementary Fig. 4). These studies suggest Lys273, Arg274 and Gln356 coordinate a DARC sulfotyrosine residue. Other regions of DARC would bind to the DARC binding groove surrounding the putative sulfotyrosine binding pocket at the dimer interface.
RBC binding is abolished and rescued by dimer mutations
Our structural studies show PvDBP must dimerize to create the DARC binding groove and to bind RBCs. Arg274 and Glu249 form a salt bridge across two monomers at the dimer interface (Fig. 1e). This salt bridge is on the opposite face of the putative sulfotyrosine binding pocket and should not play a role in the DARC interaction. We found RBC rosetting was abolished by a RII-PvDBP R274E mutation and partially restored with a compensatory double mutation, RII-PvDBP R274E E249R (Fig. 1f and Supplementary Fig. 4). These data demonstrate that disruption and restoration of the RII-PvDBP dimer interface with a salt bridge switch mutation abolishes and rescues RBC rosetting, and that the dimer architecture is required for RBC binding.
Interestingly, RII-PvDBP R274E E249R does not fully recapitulate wildtype function (Fig. 1f) because the asymmetric RII-PvDBP dimer contains two Arg274 residues in either the Arg274-Glu249 salt bridge or the DARC binding groove on opposite sides of the RII-PvDBP homodimer (Fig. 1e). This suggests the complete loss of function in RII-PvDBP R274E is due to disruption of the wildtype dimer preventing formation of the DARC binding site, and the ~50% restoration of function in RII-PvDBP R274E E249R is the result of a dimeric assembly with an incomplete putative sulfotyrosine binding pocket. This is supported by RII-PvDBP R274A, which has ~50% functional activity due to the loss of Arg273 in the putative sulfotyrosine binding pocket (Fig. 1f).
DARC binding drives RII-PvDBP dimerization
The studies above demonstrate that both an intact sulfotyrosine binding pocket and a correct dimer interface are required for receptor recognition leading to RBC attachment. We next wanted to establish the mechanism of receptor engagement and the effect of receptor binding on dimerization. We collected small angle X-ray scattering (SAXS) data at several concentrations of RII-PvDBP in the presence and absence of unsulfated DARC1–60. SAXS allows for the determination of molecular envelopes in solution and is an excellent tool to examine changes of oligomeric state during RII-PvDBP receptor recognition.
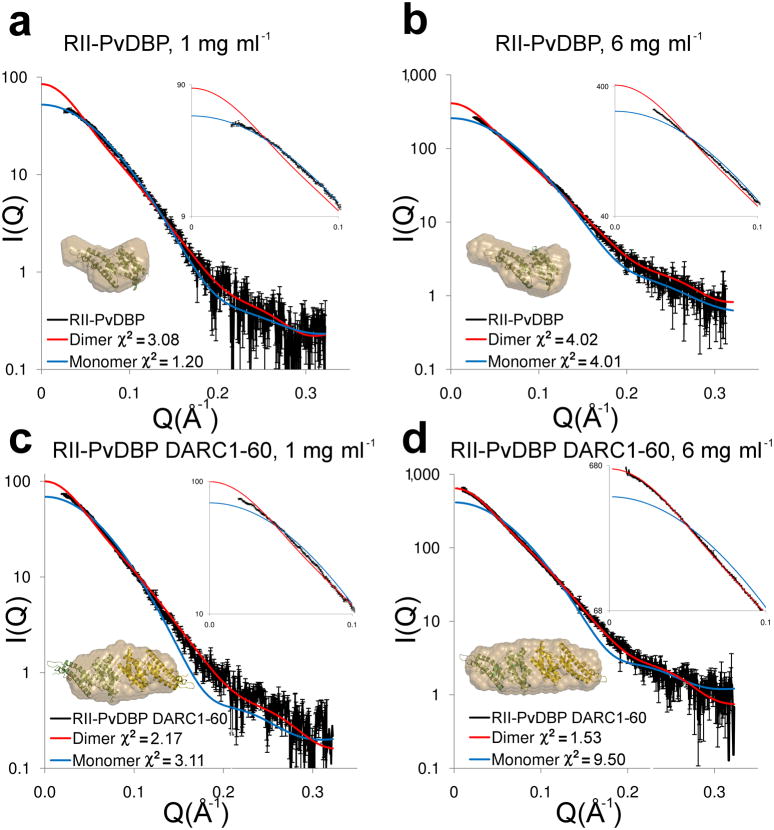
SAXS data for RII-PvDBP alone gave an excellent fit to predicted scattering of the crystal structure of monomeric RII-PvDBP with a χ2 of 1.2 at 1 mg ml−1, and a reasonable fit at 3 mg ml−1 with a χ2 of 3.89 (Fig. 2a, Supplementary Fig. 5a). These χ2 values were substantially lower than χ2 values for fits to the dimer at each concentration indicating RII-PvDBP is monomeric at these concentrations. Ab initio modeling returned molecular envelopes that closely match the monomeric RII-PvDBP structure (Fig. 2a,b, inserts, Supplementary Fig 5). At 6 mg ml−1 the experimental data for RII-PvDBP did not fit perfectly to calculated scattering from either the monomer or dimer structures suggesting a mixture of both species (monomer χ2 of 4.01, dimer χ2 of 4.02 - Fig. 2b). Thus, RII-PvDBP alone is predominantly monomeric in solution at lower concentrations.
Figure 2.
DARC binding drives dimerization of RII-PvDBP. Experimental (black) and theoretical SAXS plots for the monomer (blue) and dimer (red) at different concentrations. An expanded plot of the low-angle data (0 < Q < 0.1) that clearly delineates oligomeric state is shown in the top right insert. Ab initio reconstructions are overlayed on structures (bottom left insert) with monomers colored in green and yellow and molecular envelopes in sand. (a) RII-PvDBP at 1 mg ml−1. (b) RII-PvDBP at 6 mg ml−1. (c) RII-PvDBP–DARC1–60 at 1 mg ml−1. (d) RII-PvDBP–DARC1–60 at 6 mg ml−1.
Strikingly, addition of DARC1–60 drives dimerization of RII-PvDBP. SAXS data collected on an equimolar complex of RII-PvDBP and DARC1–60 at 1, 3 and 6 mg ml−1 gave excellent fits to the RII-PvDBP dimer crystal structure with χ2 of 2.17, 2.57 and 1.53, respectively (Fig. 2c,d, Supplementary Fig. 5b). Molecular envelopes derived from ab initio modeling also revealed excellent fits to the dimeric RII-PvDBP structure (Fig. 2c,d, Supplementary Fig. 5b). These data demonstrate that in solution DARC1–60 induces dimerization of RII-PvDBP leading to the dimeric architecture observed in the crystal structure. This is especially obvious when comparing the SAXS fits at 1 mg ml−1 (Fig. 2a,c) where RII-PvDBP alone clearly fits to a monomer (monomer fit χ2=1.2) while the RII-PvDBP–DARC complex is clearly dimeric (dimer fit χ2=2.2). The dimeric architecture in the crystal structure is induced by phosphates or selenates binding in the putative sulfotyrosine binding pocket mimicking DARC, while dimerization is induced in the solution studies by the addition of DARC1–60.
To unambiguously determine the mass of the receptor bound complex, we performed sedimentation equilibrium analytical ultracentrifugation experiments. The molecular weight of the RII-PvDBP–DARC1–60 complex was 90±4 kDa (Supplementary Fig. 6a). This is consistent with a theoretical mass of 88.4 kDa for a complex of two RII-PvDBP and two DARC1–60 molecules. We used isothermal titration calorimetry (ITC) to confirm our stoichiometry determination, as this technique directly measures stoichiometry as opposed to inferring stoichiometry from mass. Consistent with our SAXS and AUC measurements, ITC indicates the complex has a stoichiometry of 1:1 ratio of RII-PvDBP and DARC1–60 (Supplementary Fig. 6b). Together, these results demonstrate the receptor bound complex contains two molecules of RII-PvDBP bound to two molecules of DARC1–60.
It is clear that the concentrations of full receptor and ligand required to induce dimerization and binding in vivo will be much lower than the range studied here. This is because the experiments performed above were on the isolated soluble RII-PvDBP domain and a small fragment of DARC (60 residues).
Membrane anchoring of both PvDBP and DARC in vivo in addition to the dimeric nature of DARC16 will increase the propensity to dimerize even further. Lastly, the addition of a sulfate group on DARC1–60 is expected to further increase affinity for PvDBP and drive dimerization.
Functional regions of RII-PvDBP are under selective pressure
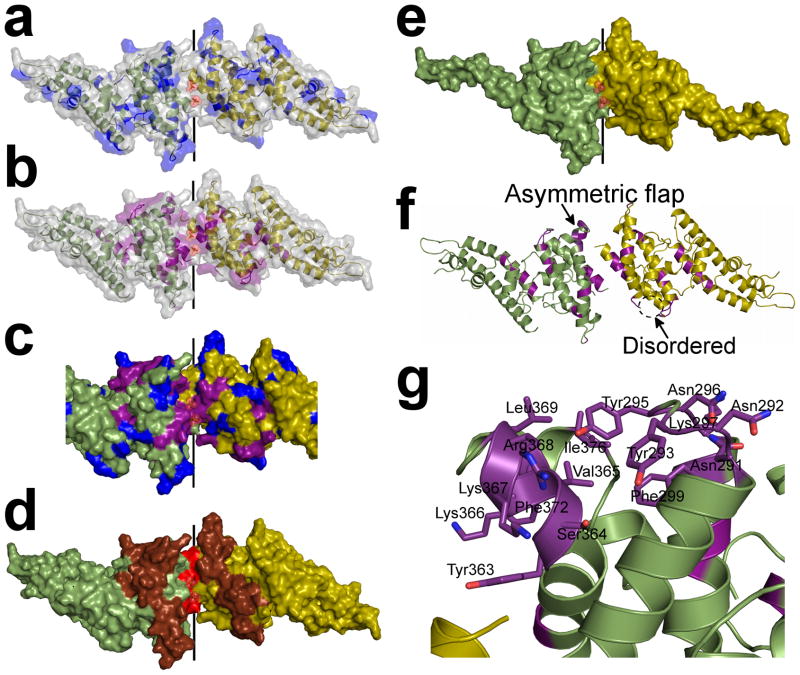
To visualize the effects of selective pressure on RII-PvDBP and test our receptor induced dimerization model, we mapped onto the structure known PvDBP polymorphisms (Fig. 3a,c and Supplementary Fig. 7) and amino acid substitutions which affect RBC rosetting (Fig. 3b,c and Supplementary Fig. 7)20,23,25,26. As the DARC binding groove and RII-PvDBP dimer interface are functionally critical, receptor induced dimerization predicts functional regions will be underrepresented by polymorphic residues and overrepresented by residues essential for DARC recognition.
Figure 3.
The sulfotyrosine pocket, DARC binding groove and dimer interface are under selective pressure and are targeted by blocking-antibodies. Monomers are in green and yellow. (a) Polymorphic residues20,23 (blue) are excluded from the dimer interface but evenly distributed over the remaining RII-PvDBP surface. (b) Amino acid substitutions which abrogate RBC rosetting25,26 (purple) map to the DARC binding groove and dimer interface. (c) Overlay of polymorphic residues (blue) and critical receptor binding residues (purple) on the dimer. The DARC binding groove at the dimer interface is composed of essential residues and devoid of polymorphisms. (d) Epitopes recognized by blocking-antibodies22 (red – most significant, brown – significant) map to the functional regions of RII-PvDBP which include the dimer interface and DARC binding groove. (e) The minimal binding domain of RII-PvDBP (residues 256–426)27 contains the full dimer interface and DARC binding groove. (f) A global view of the dimer which shows the asymmetric flap is disordered in chain A. Essential residues are colored in purple. (g) A detailed view of the asymmetric flap shows this region contains several essential residues suggesting a second potential DARC binding site.
As expected, regions spanning the DARC binding groove and dimer interface are devoid of documented polymorphic residues and heavily represented in essential residues (Fig. 3a,b,c and Supplementary Fig. 7). Polymorphisms are spread across non-functional regions suggesting the parasite generates variations in non-functional residues spread throughout the DBL domain for immune evasion.
Blocking-antibodies target functional regions of RII-PvDBP
The humoral immune response plays an important role during the pathogenesis of malaria. Receptor-induced dimerization predicts epitopes recognized by blocking-antibodies against PvDBP will map to the DARC binding groove or dimer interface. We mapped onto the structure epitopes targeted by blocking-antibodies from sera of individuals with naturally acquired immunity to P. vivax (Fig. 3d and Supplementary Fig. 7)22. Five epitopes that correlated significantly (p<0.05 22) with a block in PvDBP RBC rosetting are located in subdomain 2 of RII-PvDBP.
Mapping these five epitopes reveals the DARC binding groove and dimer interface are composed of the most significant epitope and surrounded by the four remaining epitopes (Fig. 3d and Supplementary Fig. 7). These results suggest protective antibodies target functional regions in RII-PvDBP leading to disruption of dimerization and/or prevention of receptor binding. Mapping blocking-antibody epitopes onto PvDBP shows the importance of the DARC binding groove and PvDBP dimerization in vivo and identifies structural regions targeted by the adaptive immune system in response to P. vivax.
The minimal binding domain and asymmetric flap of RII-PvDBP
The minimal region of RII-PvDBP required to bind RBCs extends from residues 256–426, spanning cysteines 4–827. This includes 170 residues of the 314 residue RII-PvDBP construct. Strikingly, this minimal binding domain retains all dimerization determinants and the putative sulfotyrosine binding pocket, further indicating that the functional elements defined in this study are critical for DARC recognition (Fig. 3e and Supplementary Fig. 7).
In the RII-PvDBP structure, residues 291–299 and 365–377 surround the DARC binding groove (Fig. 3f,g). In chain A, but not chain B, residues 365–377 are disordered indicating flexibility (Fig. 3f,g). Therefore, we termed the region spanning residues 291–299 and 365–377 the asymmetric flap. These residues are not part of the putative sulfotyrosine binding pocket but are required for DARC binding. Thus, we propose these residues form a second DARC contact. DARC1–60 binding may stabilize the asymmetric flap to form a tight complex.
VAR2CSA shares the dimer interface and receptor binding site
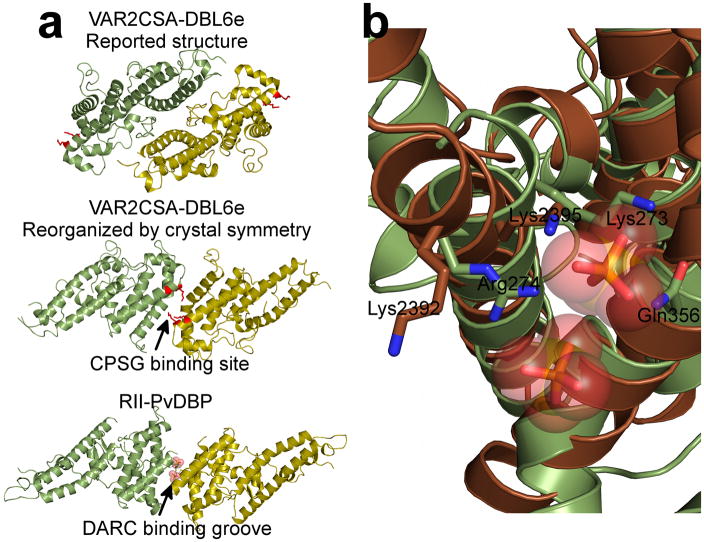
To test the generality of our receptor-mediated ligand-dimerization model we examined receptor recognition by other DBL domains. The VAR2CSA variant of PfEMP1 is the primary virulence determinant for placental malaria, and is associated with an estimated 75,000 to 200,000 infant deaths each year28. VAR2CSA is expressed on the surface of infected erythrocytes, and binds specifically to chondroitin sulfate proteoglycans (CSPG) in the intervillous space of the placenta29. VAR2CSA contains six DBL domains, which can individually bind CSPG30. A recent study has reported the structure of VAR2CSA domain DBL6ε31, the sixth DBL domain of VAR2CSA. DBL6ε crystallized with two copies in the asymmetric unit, but was proposed to function as monomer analogous to PkDBP.
We examined the crystal symmetry for the DBL6ε structure and found an unreported crystal packing contact is identical to RII-PvDBP’s dimer interface (Fig. 4a). Mutagenesis of several basic patches in DBL6ε identified two lysines, 2392 and 2395, as essential for CSPG binding31. Striking, these lysines align to RII-PvDBP’s putative sulfotyrosine binding pocket (Fig. 4b), with DBL6ε Lys2392 occupying the location of RII-PvDBP Arg274 and DBL6ε Lys2395 occupying the location of RII-PvDBP Lys273 (Fig. 4b). Lys2395 is also close to an acidic residue on the opposing subunit at the dimer interface analogous to the Arg274-Glu249 salt bridge. Thus, DBL6ε may dimerize and bind its receptor in a similar manner as RII-PvDBP. Further examination of the oligomeric state of DBL6ε is warranted and could explain the increase in receptor specificity and affinity of full length VAR2CSA over individual domains30.
Figure 4.
RII-PvDBP’s dimer interface and receptor binding site are conserved in VAR2CSA DBL6ε. (a) Examination of the crystal packing interfaces for VAR2CSA DBL6ε revealed a dimeric organization identical to the RII-PvDBP dimer. Critical VAR2CSA DBL6ε binding residues are shown in red and map to the putative sulfotyrosine binding pocket indicating that both the receptor binding pocket and dimer interface are conserved in these two DBL domains. Monomers are colored green/yellow in both cases. Top panel – reported asymmetric unit for DBL6ε, Middle panel – reorganized dimer based solely on crystal symmetry, Bottom panel – RII-PvDBP dimer. (b) Overlay of RII-PvDBP (green) and DBL6ε (brown) reveals critical binding residues for each protein superpose well (Lys273 and Arg274 from RII-PvDBP, and Lys2392 and Lys2395 from DBL6ε).
Discussion
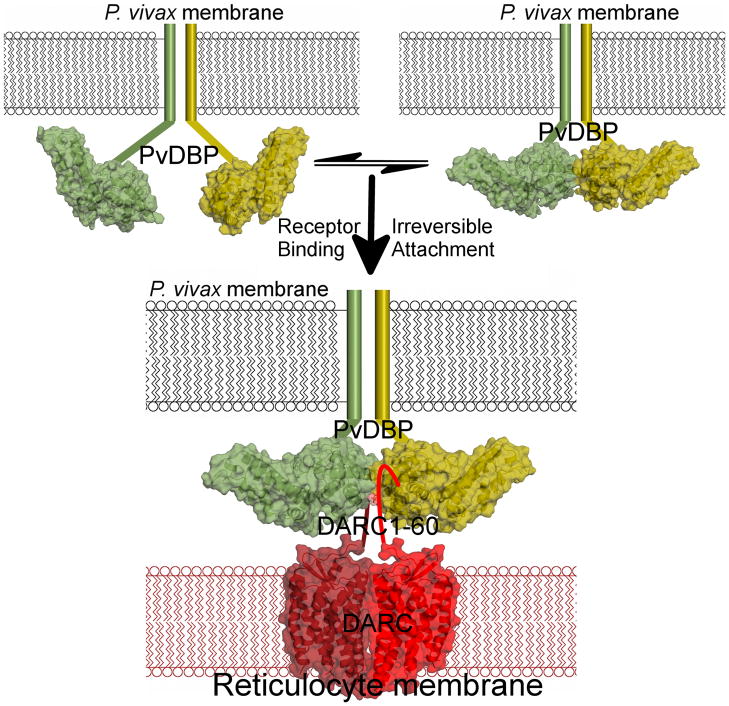
In an effort to elucidate the molecular basis and mechanism of receptor recognition by PvDBP, we performed a structural, functional and mechanistic study of RII-PvDBP’s interaction with the N-terminus of DARC. We show RII-PvDBP’s receptor binding site is formed by a RII-PvDBP homodimer and disruption and restoration of dimerization disrupts and restores RBC binding. We demonstrate dimerization is critical for and driven by receptor binding leading to the formation of a complex composed of two PvDBP and two DARC. We show recognition of DARC occurs concomitantly with dimerization of PvDBP, placing the N-termini of DARC in the DARC binding groove created upon dimerization. Thus, receptor recognition by PvDBP is through receptor-mediated ligand-dimerization (Fig. 5). This model is applicable to receptor recognition by DBL domains from PvDBP, PfEMP1 VAR2CSA and PfEBA-17532.
Figure 5.
PvDBP binds DARC via a model of receptor-mediated ligand-dimerization. PvDBP exists as an equilibrium of monomers and dimers that is shifted to dimerization upon receptor-binding. RII-PvDBP monomers are in green/yellow. The P. vivax membrane is in black and the reticulocyte membrane is in red. Flat lines represent portions of PvDBP not in the crystal structure. The DARC homodimer is represented by the crystal structure of a related GPCR, CXCR4’s, homodimeric membrane spanning region40, in dark/light red. DARC1–60 is shown as a flat line. Two PvDBP molecules bind two DARC molecules as indicated by our stoichiometry measurements.
This model is consistent with structural and functional studies of PfEBA-175 which proposed a dimeric RII-PfEBA-175 assembly upon receptor binding32. A subsequent crystal structure of RII-PkDBP from P. knowlesi DBP suggested this domain binds to DARC as a monomer as no dimeric contacts were observed in the crystal structure24, which is inconsistent with the data presented here. The RII-PkDBP structure likely is not dimeric as these crystals were grown in detergent (0.5% N-octyl β-d-glucopyranoside), which would disrupt biologically relevant oligomerization. Although the different models may be due to the different species, the structure of monomeric RII-PkDBP could represent the unbound monomer that dimerizes upon receptor binding.
Singh et al. further proposed a sulfotyrosine binding pocket24 although no direct interaction studies were presented. Furthermore, the monomeric model of binding and putative binding pocket cannot account for all DARC binding mutations reported in the literature25,26. The authors also proposed a “just in time release” strategy of immune evasion based on mapping of a subset of polymorphic residues to a face on the opposite side of their proposed DARC binding site24. However, polymorphisms are more widely dispersed than those examined by Singh et al.33 and cannot be accounted for by the monomeric model or “just in time release” strategy.
The receptor-mediated ligand-dimerization model proposed here clarifies the mutational, polymorphism and antibody data for PvDBP. This model shows mutations can be grouped into residues that affect the dimer interface, the putative sulfotyrosine binding pocket, the DARC binding groove and the asymmetric flap. Our data strongly suggest an alternate location for the sulfotyrosine binding pocket based on the identification of selenates and phosphates at the dimer interface coupled with functional studies.
Field isolates of RII-PvDBP show extensive sequence polymorphism20,23, which is thought to be an immune evasion mechanism. We have shown PvDBP immune evasion is mediated by generating polymorphisms in non-functional regions. Thus, immune evasion is achieved by changes at multiple regions rather than clustering polymorphisms on one face of the DBL domain as previously proposed24. This method of immune evasion is similar to that proposed for influenza virus hemagglutinin34. Strikingly, RII-PvDBP’s putative sulfotyrosine binding pocket, DARC binding groove and dimer interface are all targeted by the adaptive immune response and targeting these functional regions prevents RBC binding leading to a protective immunity. These studies identify critical regions of RII-PvDBP that could be exploited for therapeutic design. Targeting the interaction between RII-PvDBP and DARC is effective against P. vivax, as natural selection of a Duffy null phenotype has largely eliminated P. vivax in West Africa3.
Finally, PvDBP contains a region VI (RVI-PvDBP) with sequence similarity to region VI of PfEBA-175 (RVI-PfEBA-175, Supplementary Fig. 1). The crystal structure of RVI-PfEBA-175 showed that this domain dimerizes35, and the RVI-PvDBP domain is expected to increase the propensity of PvDBP to dimerize. Lastly, membrane anchoring of both PvDBP and the DARC homodimer, and sulfation of DARC are all likely to drive dimerization further supporting the receptor-mediated ligand-dimerization model.
Receptor-mediated dimerization would increase the specificity and affinity of the host-parasite interaction. Ligand oligomerization during receptor binding has also been proposed for the Toxoplasma gondii micronemal proteins MIC336 and MIC237. RBC invasion involves drastic changes within the merozoite which would require intracellular signaling. Dimerization is common in signaling pathways38 and it is possible that receptor-induced ligand-dimerization of RII-PvDBP transduces a signal for initiation of RBC invasion. Finally, this model has implications for other chemokine receptor-ligand interactions which are reliant on sulfotyrosine such as the HIV gp120–CCR5 interaction 39.
We show that blocking-antibodies prevent RBC binding by targeting epitopes at the dimer interface or in the DARC binding groove preventing dimerization and/or direct receptor binding. Our work provides structural details of RII-PvDBP regions relevant for vaccine design and predicts focusing the immune response to the RII-PvDBP dimer interface and receptor binding pocket epitopes could lead to the development of a potent neutralizing therapeutic. As receptor-mediated ligand-dimerization is a general mechanism by which EBL proteins engage receptors, disrupting EBL dimerization could be a viable strategy for therapeutic intervention for other Plasmodium species.
Finally, these data show that single DBL domains dimerize upon receptor binding, consistent with studies on the multi-DBL domain proteins PfEBA-17532 and PfEMP130. This model for receptor recognition is applicable to the large superfamily of EBL proteins. We propose a mechanism of receptor-mediated ligand-dimerization during the initiation of RBC invasion by Plasmodium parasites and upon receptor-binding during PfEMP1 mediated cytoadherence and sequestration. This model allows for the possibility that induced dimerization activates downstream pathways essential for invasion. Finally, receptor-mediated ligand-dimerization is expected to increase receptor specificity and affinity and may explain how DBL domains can recognize a wide variety of receptor chemistries.
Methods
Protein Expression, Purification and Complex Formation
A plasmid containing PvDBP DNA was a generous gift from Dr. Adams (University of South Florida). Amino acids 211–525 of PvDBP (RII-PvDBP) were expressed as inclusion bodies in E. coli. 200 mg of RII-PvDBP recovered from inclusion bodies with 6 M guanidinium hydrochloride were rapidly diluted into 2 liters of 400 mM L-Arginine, 10 mM ethylenediaminetetraacetic acid, 0.1 mM phenylmethanesulfonylfluoride, 2 mM reduced glutathione, 0.2 mM oxidized glutathione, and 200 mM tris(hydroxymethyl)aminomethane (pH 8.0). After stirring at 4 °C for 36 hours, soluble RII-PvDBP was concentrated in 350 ml Amicon concentrators and purified by size-exclusion chromatography.
Three cysteines (C4A, C51A and C54A) in DARC1–60 were mutated to alanine to prevent aberrant disulfide bridge formation that might complicate analyses of oligomerization. DARC1–60 was expressed in E. coli as an N-terminal His-tag with a Precission Protease cleavage site. Nickel-NTA chromatography followed by protease treatment and gel filtration resulted in a homogenous sample.
Prior to complex formation, RII-PvDBP and DARC were purified separately by size-exclusion chromatography to remove any trace aggregates in either sample. The RII-PvDBP/DARC complex was prepared by mixing purified RII-PvDBP and purified DARC1–60 at an equimolar ratio to a final concentration of 1 mg ml−1 in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.4 and 50 mM sodium chloride. This sample was then concentrated in an Amicon concentrator with a 3 kD molecular weight cutoff to the desired concentration for SAXS and AUC experiments. Protein concentrations were determined by absorbance measurements under denaturing conditions (6 M guanidinium hydrochloride, 10 mM dithiothreitol).
Crystallization and data collection
Native RII-PvDBP crystals were grown by hanging drop vapor diffusion by mixing 1 μl of protein solution at 15 mg ml−1 and 1 μl of reservoir solution containing 0.1 M ammonium phosphate and 23% (w/v) polyethylene glycol 3350. Selenate derivatized crystals were obtained by similar methods with a reservoir containing 0.1 M sodium selenate, 0.1 M ammonium chloride and 23% (w/v) polyethylene glycol 3350. Native and derivative crystals were cryoprotected by transfer to reservoir solutions supplemented with ethylene glycol and flash frozen in liquid nitrogen. Data were collected at beamline 4.2.2 at the Advanced Light Source, Lawrence Berkeley National Laboratory. Data collection statistics are shown in Table 1.
Structural studies
Selenium sites were identified in the derivative crystals using SHELX41. Heavy atom refinement and phasing was performed in SHARP42 leading to interpretable maps. Density modification of these starting phases in DM43 followed by autobuilding using ARP/wARP44 lead to a starting model with Rwork/Rfree of 22.4%/32.4%. Subsequent manual rebuilding in COOT45 and refinement in PHENIX46 against the native dataset lead to a final refined model with Rwork/Rfree of 18.64%/23.77% (Table 1). NCS restraints were initially imposed on the two copies of RII-PvDBP but were finally removed when it became evident that the two copies were not identical as observed in electron density maps. These low R-factors combined with the good Ramachandran plot statistics (allowed 91.5 %, additionally allowed 7.7 %, generously allowed 0.8 %, disallowed 0 %) calculated by Procheck indicated structure refinement was complete.
Functional studies
RII-PvDBP with a C-terminally fused GFP was cloned into plasmid pRE4 for surface expression in mammalian cells. Single amino acid mutations were introduced in RII-PvDBP using the Quickchange method (Stratagene). Fresh monolayers of HEK293T cells were cultured in 3.5 cm-diameter wells and transfected with 2 μg/ml DNA in PEI. The binding assay was performed 20 h after transfection. Human RBCs were added to each well in a 10% suspension, incubated at 37 °C for 1 h, and washed three times with PBS. Binding was quantified by counting rosettes observed over 10 fields of view at X200 magnification. Transfected HEK 293T cells with five or more attached RBCs were defined as positive rosettes, which is an accepted value for this assay25. In each experiment, three wells of HEK 293T cells were transfected for each mutation. Cell counting was performed using ImageJ (NIH) on randomized images. Three fields of view from ten independent transfections (final n=30) were counted for each sample (wildtype or mutant). Significance was tested by a paired two-tailed student t-test as the data were normally distributed and had large sample sizes (n=30).
Small Angle X-ray Scattering
SAXS experiments were performed at the SIBYLS beamline 12.3.1 at the ALS. 30 μL of purified RII-PvDBP or RII-PvDBP plus DARC1–60 were used for SAXS analyses. Data were acquired on a series of concentrations of 6, 3 and 1 mg ml−1. Samples, with corresponding matching buffers, were loaded into a 96-5 well plate (Nunc) and covered with protective film. Samples were automatically loaded into the cuvette with a Hamilton syringe robot47. SAXS data collection and processing were performed as described48. SAXS measurements were made at three X-ray exposures of either 0.1, 0.1 and 0.5 seconds for the 6 mg ml−1 samples or 0.5, 0.5, and 1 seconds for the lower concentrations at room temperature. Sample radiation damage was assessed by overlaying the two short exposures using PRIMUS49. SAXS-based ab initio modeling was performed with DAMMIF50. CRYSOL51 was used to derive theoretical scattering data from monomeric and dimeric RII-PvDBP, and to compare these data with the experimental results. SUPCOMB2052 was used to align SAXS reconstructions with the crystal structure.
Supplementary Material
Acknowledgments
We thank S. Beverley, D. Fremont, L. Joshua-Tor, D. Goldberg, D. Sibley, J. Vogel and E. Chen for constructive comments on the manuscript, J. Adams (University of South Florida) for providing RII-PvDBP DNA and D. Goldberg (Washington University) for additional reagents. We thank T. Lohman and R. Galletto for assistance with the AUC experiments, D. Fremont and T. Brett for assistance with the ITC experiments, J. Nix and the Molecular Biology Consortium Collaborative Access Team at the Advanced Light Source for assistance in crystallographic data collection, and G. Hura, K. Dyer and the Structurally Integrated Biology for Life Sciences Team at the Advanced Light Source for assistance in SAXS data collection funded by DOE IDAT grant contract number DE-AC02-05CH11231. This project is supported by grants from the National Institutes of Health (NIH) and the Edward Mallinckrodt, Jr. Foundation to N.H.T, and an NIH Training Grant and W. M. Keck Postdoctoral Fellowship to J.D.B.
Footnotes
Author Contributions J.D.B. performed functional assays, SAXS data analysis, AUC studies, ITC studies and structure analyses. J.A.Z. cloned, purified and crystallized RII-PvDBP. N.H.T. designed the study, analyzed SAXS data, collected, processed and refined X-ray data, and analyzed the structure. All authors were involved in writing the paper, discussed the results and commented on the manuscript. The authors declare no competing financial interests.
Accession codes The atomic coordinates and structure factors for the RII-PvDBP structure have been deposited in the protein data bank with accession number 3RRC.
References
- 1.Mason SJ, Miller LH, Shiroishi T, Dvorak JA, McGinniss MH. The Duffy Blood Group Determinants: Their Role in the Susceptibility of Human and Animal Erythrocytes to Plasmodium knowlesi Malaria. British Journal of Haematology. 1977;36:327–335. doi: 10.1111/j.1365-2141.1977.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 2.Miller L, Mason S, Clyde D, McGinniss M. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 3.Miller LH, Mason SJ, Dvorak JA. Erythrocytes receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 4.Adams JH, et al. A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci U S A. 1992;89:7085–9. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chitnis C, Miller L. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–4. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 7.Adams JH, et al. The duffy receptor family of plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell. 1990;63:141–153. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- 8.Haynes JD, et al. Receptor-like specificity of a Plasmodium knowlesi malarial protein that binds to Duffy antigen ligands on erythrocytes. The Journal of Experimental Medicine. 1988;167:1873–1881. doi: 10.1084/jem.167.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed MB, et al. Targeted disruption of an erythrocyte binding antigen in Plasmodium falciparum is associated with a switch toward a sialic acid-independent pathway of invasion. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7509–7514. doi: 10.1073/pnas.97.13.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaur D, Mayer DCG, Miller LH. Parasite ligand-host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. International Journal for Parasitology. 2004;34:1413–1429. doi: 10.1016/j.ijpara.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Narum DL, Fuhrmann SR, Luu T, Sim BKL. A novel Plasmodium falciparum erythrocyte binding protein-2 (EBP2/BAEBL) involved in erythrocyte receptor binding. Molecular and Biochemical Parasitology. 2002;119:159–168. doi: 10.1016/s0166-6851(01)00428-5. [DOI] [PubMed] [Google Scholar]
- 12.Adams JH, Blair PL, Kaneko O, Peterson DS. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 2001;17:297–9. doi: 10.1016/s1471-4922(01)01948-1. [DOI] [PubMed] [Google Scholar]
- 13.Camus D, Hadley T. A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science. 1985;230:553–556. doi: 10.1126/science.3901257. [DOI] [PubMed] [Google Scholar]
- 14.Lobo CA, Rodriguez M, Reid M, Lustigman S. Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl) Blood. 2003;101:4628–4631. doi: 10.1182/blood-2002-10-3076. [DOI] [PubMed] [Google Scholar]
- 15.Carlton JM, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–63. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakera A, Seeber RM, John AE, Eidne KA, Greaves DR. The Duffy Antigen/Receptor for Chemokines Exists in an Oligomeric Form in Living Cells and Functionally Antagonizes CCR5 Signaling through Hetero-Oligomerization. Molecular Pharmacology. 2008;73:1362–1370. doi: 10.1124/mol.107.040915. [DOI] [PubMed] [Google Scholar]
- 17.Chitnis C, Chaudhuri A, Horuk R, Pogo A, Miller L. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med. 1996;184:1531–1536. doi: 10.1084/jem.184.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choe H, et al. Sulphated tyrosines mediate association of chemokines and Plasmodium vivax Duffy binding protein with the Duffy antigen/receptor for chemokines (DARC) Molecular Microbiology. 2005;55:1413–1422. doi: 10.1111/j.1365-2958.2004.04478.x. [DOI] [PubMed] [Google Scholar]
- 19.Cole-Tobian Jennifer L, et al. Age Acquired Immunity to a Plasmodium vivax Invasion Ligand, the Duffy Binding Protein. The Journal of Infectious Diseases. 2002;186:531–539. doi: 10.1086/341776. [DOI] [PubMed] [Google Scholar]
- 20.Xainli J, Adams JH, King CL. The erythrocyte binding motif of plasmodium vivax duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol Biochem Parasitol. 2000;111:253–60. doi: 10.1016/s0166-6851(00)00315-7. [DOI] [PubMed] [Google Scholar]
- 21.Xainli J, et al. Age-dependent cellular immune responses to Plasmodium vivax Duffy binding protein in humans. J Immunol. 2002;169:3200–7. doi: 10.4049/jimmunol.169.6.3200. [DOI] [PubMed] [Google Scholar]
- 22.Chootong P, et al. Mapping Epitopes of the Plasmodium vivax Duffy Binding Protein with Naturally Acquired Inhibitory Antibodies. Infect Immun. 2010;78:1089–1095. doi: 10.1128/IAI.01036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gosi P, et al. Polymorphism patterns in Duffy-binding protein among Thai Plasmodium vivax isolates. Malar J. 2008;7:112. doi: 10.1186/1475-2875-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh SK, Hora R, Belrhali H, Chitnis CE, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–744. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- 25.VanBuskirk KM, Sevova E, Adams JH. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc Natl Acad Sci U S A. 2004;101:15754–9. doi: 10.1073/pnas.0405421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hans D, et al. Mapping binding residues in the Plasmodium vivax domain that binds Duffy antigen during red cell invasion. Mol Microbiol. 2005;55:1423–34. doi: 10.1111/j.1365-2958.2005.04484.x. [DOI] [PubMed] [Google Scholar]
- 27.Singh SK, et al. Definition of structural elements in Plasmodium vivax and P. knowlesi Duffy-binding domains necessary for erythrocyte invasion. Biochem J. 2003;374:193–8. doi: 10.1042/BJ20030622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steketee R, Nahlen B, Parise M, Menendez C. The burden of malaria in pregnancy in malaria-endemic areas. Am J Trop Med Hyg. 2001;64:28–35. doi: 10.4269/ajtmh.2001.64.28. [DOI] [PubMed] [Google Scholar]
- 29.Salanti A, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Molecular Microbiology. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava A, et al. Full-length extracellular region of the var2CSA variant of PfEMP1 is required for specific, high-affinity binding to CSA. Proc Natl Acad Sci U S A. 2010;107:4884–9. doi: 10.1073/pnas.1000951107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khunrae P, Philip JM, Bull DR, Higgins MK. Structural comparison of two CSPG-binding DBL domains from the VAR2CSA protein important in malaria during pregnancy. J Mol Biol. 2009;393:202–13. doi: 10.1016/j.jmb.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tolia NH, Enemark EJ, Sim BKL, Joshua-Tor L. Structural Basis for the EBA-175 Erythrocyte Invasion Pathway of the Malaria Parasite Plasmodium falciparum. Cell. 2005;122:183–193. doi: 10.1016/j.cell.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 33.McHenry AM, Adams JH. The crystal structure of P. knowlesi DBPalpha DBL domain and its implications for immune evasion. Trends Biochem Sci. 2006;31:487–91. doi: 10.1016/j.tibs.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson IA, Cox NJ. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol. 1990;8:737–71. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 35.Withers-Martinez C, et al. Malarial EBA-175 region VI crystallographic structure reveals a KIX-like binding interface. J Mol Biol. 2008;375:773–81. doi: 10.1016/j.jmb.2007.10.071. [DOI] [PubMed] [Google Scholar]
- 36.Cerede O, Dubremetz JF, Bout D, Lebrun M. The Toxoplasma gondii protein MIC3 requires pro-peptide cleavage and dimerization to function as adhesin. EMBO J. 2002;21:2526–2536. doi: 10.1093/emboj/21.11.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jewett TJ, Sibley LD. The Toxoplasma Proteins MIC2 and M2AP Form a Hexameric Complex Necessary for Intracellular Survival. Journal of Biological Chemistry. 2004;279:9362–9369. doi: 10.1074/jbc.M312590200. [DOI] [PubMed] [Google Scholar]
- 38.Klemm JD, Schreiber SL, Crabtree GR. Dimerization as a regulatory mechanism in signal transduction. Annu Rev Immunol. 1998;16:569–92. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- 39.Farzan M, et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–76. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 40.Wu B, et al. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010;330:1066–71. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 42.de la Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Meth Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 43.Acta Crystallogr. D50. Daresbury, UK: 1994. CCP4: Collaborative Computational Project No. 4; p. 760. [Google Scholar]
- 44.Cohen SX, et al. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr D Biol Crystallogr. 2004;60:2222–9. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 47.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–12. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 49.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- 50.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svergun D, Barberato C, Koch MHJ. CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- 52.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. Journal of Applied Crystallography. 2001;34:33–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.