Summary
Cryptochromes are blue light receptors that mediate light regulation of gene expression in all major evolution lineages, but the molecular mechanism underlying cryptochrome signal transduction remains not fully understood [1, 2]. It has been reported that cryptochromes suppress activity of the multifunctional E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) to regulate gene expression in response to blue light [3, 4]. But how plant cryptochromes mediate light suppression of COP1 activity remains unclear. We report here that Arabidopsis CRY2 (cryptochrome 2) undergoes blue light-dependent interaction with the COP1-interacting protein SUPPRESSOR OF PHYTOCHROME A 1 (SPA1) [5, 6]. We demonstrate that SPA1 acts genetically downstream from CRY2 to mediate blue light suppression of the COP1-dependent proteolysis of the flowering-time regulator CONSTANS (CO) [7, 8]. We further show that blue light-dependent CRY2-SPA1 interaction stimulates CRY2-COP1 interaction. These results reveal for the first time a wavelength-specific mechanism by which a cryptochrome photoreceptor mediates light regulation of protein degradation to modulate developmental timing in Arabidopsis.
Results and Discussions
CRY2 Interacts with SPA1 in Response to Blue Light
In a previous study, we used the blue light-differentiated yeast two-hybrid screen to look for proteins that interact with CRY2 in a blue light-dependent manner [9]. Several clones we isolated in that screen correspond to SPA1. SPA1 is a kinase-like coiled-coil/WD-repeat protein that interacts and activates the E3 ubiquitin ligase activity of COP1 [6, 10, 11]. Given the known function of SPA1 in light signal transduction, we conducted a detailed investigation of the CRY2-SPA1 interaction. We first examined and confirmed the blue light-dependent interaction between CRY2 and SPA1 by using yeast two-hybrid assay with both the β-galactosidase (Figure 1A) and the histidine auxotrophy assays (Figure S1,available online). The CRY2-SPA1 interaction is dependent on not only the wavelength of light (Figure 1A) but also the photon density of light (Figure 1B). As expected, the blue light-dependent CRY2-SPA1 interaction requires the FAD chromophore of CRY2 because the CRY2D387A mutant impaired in FAD-binding failed to interact with SPA1 in response to the same blue-light treatment [9] (Figures 1A and 1B and Figure S1). SPA1 is one of the four SPA quartet genes (SPA1, SPA2, SPA3, and SPA4) that play partially redundant functions in Arabidopsis [12, 13]. We found that all SPA quartet proteins interact with CRY2, although SPA1 showed the most robust interaction with CRY2 in response to blue light (Figure S2). We focused on the analysis of CRY2-SPA1 interaction for the rest of this study.
Figure 1. CRY2 Undergoes Blue Light-Dependent Interaction with SPA1.
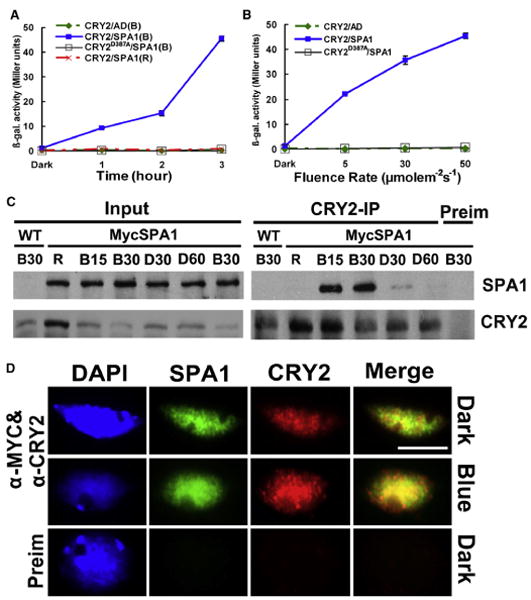
(A and B) Yeast two-hybrid experiments showing the wavelength- and fluence rate-dependent CRY2-SPA1 interaction. Yeast cells expressing the indicated proteins were irradiated with red light (R, 50 μmolem−2s−1) or blue light (B, 50 μmolem−2s−1) for the time indicated (A), or irradiated with blue light of the fluence rates indicated for 3 hr (B). β-galactosidase activities were assayed and the standard deviations (n = 3) are shown.
(C) Co-IP experiments showing the blue light-dependent CRY2-SPA1 complex formation in plant cells. Fourteen-day-old wild-type (WT) and transgenic plants expressing Myc-SPA1 were transferred to red light (R, 20 μmolem−2s−1) for 18 hr, then exposed to blue light (20 μmolem−2s−1) for 15 min (B15) or 30 min (B30). After blue light treatment, aliquots of plant samples were transferred to darkness for 30 min (D30) or 60 min (D60). Immunoblots of the total protein extracts (Input) and immunoprecipitation (IP) product of the CRY2 antibody or preimmune serum (Preim) were probed by the Myc antibody, stripped, and reprobed by the CRY2 antibody.
(D) A coimmunostaining experiment showing the blue light-induced colocalization (indicated by yellow spots) of CRY2 (red) or Myc-SPA1 (green). The images of the same cell from separate color channels were merged by the merge program of Photoshop and shown (Merge). The scale bar indicates 5 μm.
Also see Figure S1.
We next examined the blue light-dependent CRY2-SPA1 interaction in transgenic plants expressing the 35S∷MycSPA1 transgene by using a coimmunoprecipitation assay. Wild-type and MycSPA1-expressing plants grown in long-day (LD) photoperiods for two weeks were transferred (adapted) to red light; the red light-adapted plants were exposed to blue light for the coimmunoprecipitation assay. Some aliquots of the plant samples were transferred to darkness after blue light treatment to further access the light effect. Figure 1C shows that a similar amount of the MycSPA1 protein was detected in red light-adapted plants with or without blue light treatment (Figure 1C, Input). Little MycSPA1 was coprecipitated by the CRY2 antibody in red light-adapted controls (Figure 1C, CRY2-IP, R), suggesting the lack of CRY2-SPA1 interaction in response to red light. In contrast, MycSPA1 protein was coprecipitated with CRY2 in plants exposed to blue light for 15 or 30 min (Figure 1C, CRY2-IP, B15, B30), although the abundance of CRY2 decreased as the consequence of the blue light-dependent CRY2 degradation (Figure 1C, CRY2-IP, B15, B30) [14, 15]. In plants transferred to darkness after blue light treatment, the amount of MycSPA1 coprecipitated by CRY2 decreased markedly within 30 min, and it was almost undetectable after 60 min of dark treatment (Figure 1C, CRY2-IP, D30, D60). These results clearly demonstrate that blue light stimulates formation of the CRY2-SPA1 protein complex in plant cells. Consistent with the coimmunoprecipitation (co-IP) experiment, an immunostaining experiment shows that blue light enhances colocalization of the CRY2 and MycSPA1 proteins in the nuclear bodies (Figure 1D). Taken together, we concluded that CRY2 undergoes blue light-dependent physical interaction with SPA1.
SPA1 Is Required for the CRY2-Mediated Photoperiodic Regulation of Floral Initiation
To better understand the molecular nature of the blue light-dependent CRY2-SPA1 interaction, we analyzed the domain structures of CRY2 and SPA1 required for their interaction. CRY2 has two domains, the N-terminal photolyase homologous region (PHR) domain and the C-terminal cryptochrome C-terminal extension (CCE) domain [2, 16] (Figure 2A). PHR is the evolutionarily conserved chromophore-binding domain; CCE is an effector domain that interacts with COP1 [17–21]. SPA1 is composed of three domains: the N-terminal kinase-like domain, the central coiled-coil domain, and the C-terminal WD-repeat domain (Figure 2A). It has been demonstrated that the kinase-like domain is a regulatory domain for SPA1 [22], whereas the coiled-coil domain and the WD-repeat domain interact with COP1 and the COP1 substrates, respectively [10, 23]. A detailed yeast two-hybrid analysis demonstrates that the PHR domain of CRY2 (Figures 2B and 2C) and the kinase-like domain of SPA1 (Figures 2D and 2E) are necessary and sufficient for the wavelength-specific and fluence rate-dependent CRY2-SPA1 interaction. For example, the SPA1 N-terminal fragment containing the kinase-like domain alone showed highest activity interacting with CRY2, whereas the SPA1 C-terminal fragment of 509 residues lacking the kinase-like domain, referred to as CT509, failed to interact with CRY2 (Figures 2D and 2E). Consistent with this result, CT509 also showed no interaction with CRY2 in transgenic Arabidopsis plants expressing the Myc-tagged CT509 (Figure 2F and Figure S4). Interestingly, transgenic plants expressing CT509 in the spa1 mutant background showed delayed flowering in continuous light (LL) or in LD photoperiods (Figures 2G and 2H, p < 0.01) but showed similar flowering time as the spa3 parent in short-day (SD) photoperiods (Figures 2G and 2H). This observation appears to suggest that CT509 may affect flowering time in LD but not SD. Because CT509 interacts strongly with COP1 [10] and that the other three SPA proteins also interact with COP1, CRY2, and each other [13]; the CT509 fragment lacking the CRY2-interacting domain may act as a competitive inhibitor of COP1 or other SPA quartet proteins to affect their activity. This interpretation would be consistent with a hypothesis that SPA1 is involved in the CRY2-dependent photoperiodic control of floral initiation.
Figure 2. The Structures of the CRY2 and SPA1 Involved in Protein-Protein Interaction or Photoperiodic Flowering.
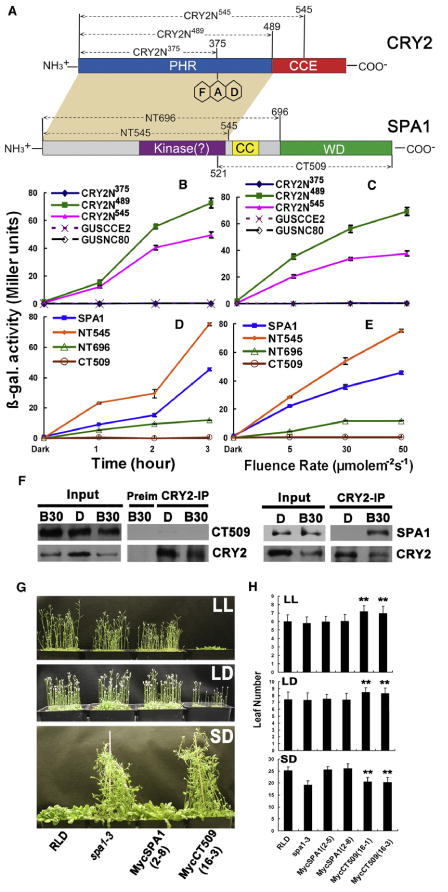
(A) Diagrams depicting the linear structures of CRY2 and SPA1: NT545: SPA1 N-terminal 545 residues; NT696: SPA1 N-terminal 696 residues; CT509, SPA1 C-terminal 509 residues. CRY2N545, CRY2N489, and CRY2N375 are the CRY2 N-terminal fragments containing residue 1 to the residues numbered by the superscripts. GUSCCE2 and GUSNC80 are the fusion proteins of GUS fused to CRTY2 C-terminal fragment of residues 486–612 and 486–565, respectively [16].
(B–E) β-galactosidase activities in the yeast two-hybrid assays showing interactions of different domains of CRY2 (as the BD-fusion bait proteins) interacting with SPA1 (B and C) or interactions of different domains of SPA1 (as the AD-fusion prey proteins) interacting with CRY2 (D and E). Yeast cells expressing indicated baits and preys combinations were irradiated by blue light (50 μmolem−2s−1) for the time indicated (B and D) or irradiated with blue light of different fluence rates (5–50 μmolem−2s−1) for 3 hr (C and E). Standard deviations (n = 3) are shown.
(F) Co-IP experiments showing the lack of interaction between CRY2 and the CT509 fragment of SPA1. Transgenic plants expressing Myc-tagged CT509 (CT509) or the Myc-tagged SPA1 (SPA1) were grown in LD photoperiods, transferred to dark for 18 hr, and exposed to blue light (20 μmolem−2s−1) for 30 min. Immunoblots of the protein extracts (Input) and the IP product of the CRY2 antibody (CRY2-IP) or preimmune serum (Preim) were probed by the Myc antibody (SPA1), stripped, and reprobed by the CRY2 antibody (CRY2).
(G) The transgenic lines expressing the Myc-tagged CT509 fragment of SPA1 showing delayed flowering in LD but accelerated flowering in SD photoperiods. The wild-type (RLD), spa1-3 (RLD accession), two independent lines expressing the 35S∷MycSPA transgene in spa1-3 mutant background (MycSPA1, the line 2-8), or the 35∷MycSPACT509 transgene in the spa1-3 mutant background (MycCT509, the line 16-3) were grown in LL, LD photoperiods (16 hr light/8 hr dark), or SD (8 hr light/16 hr dark) for 23 days (G) or until all plants flowered (H). Numbers of the rosette leaves at the time of emerging of floral buds, and the standard deviations (n > 20) are shown (**p < 0.01).
Also see Figure S2.
To test whether light-dependent CRY2-SPA1 interaction is required for the CRY2 signal transduction in photoperiodic sensing, we analyzed the genetic interaction between CRY2 and SPA1 (Figure 3 and Figure S3). Figure 3 shows that under LD photoperiods, the cry2 mutant, but not the spa1 mutant, flowered significantly later than wild-type plants, whereas the cry2spa1 double mutant flowered at about the same time as the spa1 monogenic mutant (Figures 3A and 3B). Because both cry2 and spa1 are recessive mutations [5, 24], these results demonstrate that the spa1 mutant is epistatic to cry2 and that SPA1 acts downstream of CRY2 to mediate the CRY2 regulation of photoperiodic flowering. Additional genetic analyses of other spa mutants demonstrate that spa1 is necessary and sufficient to suppress the late-flowering phenotype of the cry2 mutation (Figure S2). This result suggests that, among the SPA quartet genes, SPA1 is the primary regulator required for the CRY2-dependent control of flowering time, although the possible involvement of other SPA proteins cannot be excluded. Interestingly, despite the genetic, physical, and functional interactions between CRY2 and SPA1 and the role of SPA1 in protein ubiquitination, SPA1 is not directly involved in the blue light regulation of CRY2 expression, because neither the spa1 or spa1234 quadruple mutations affected the blue light-dependent CRY2 degradation [25](B. Liu and C. Lin, unpublished data).
Figure 3. SPA1 Is Required for the CRY2-Dependent Blue Light Suppression of CO Degradation and the CRY2-Dependent Promotion of Flowering.
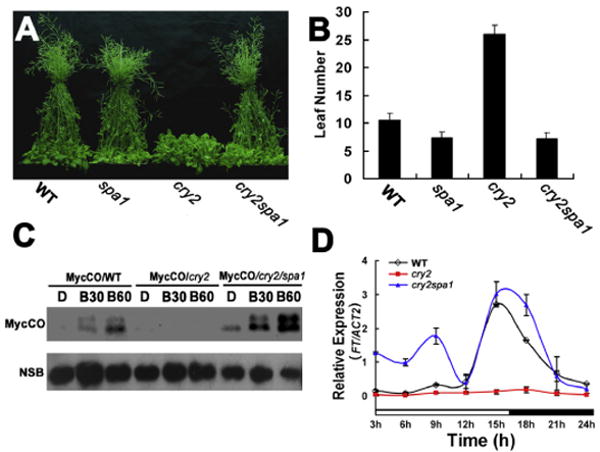
(A and B) The wild-type (WT: F1 hoterozygate of RLD × Col), spa1 (RLD), cry2 (Col), and cry2/spa1 (F3 homozygate of RLD × Col) plants grown in LD photoperiods (16 hr light/8 hr dark) for 20 days (A) or until all plants flowered (B). The number of rosette leaves at flowering and the standard deviations (n > 20) are shown (B).
(C) Immunoblot showing that the blue light-dependent suppression of CO degradation was impaired in the cry2 mutant but restored in the cry2spa1 double-mutant plants. Fourteen-day-old transgenic plants expressing the 35∷MycCO transgene in the wild-type (MycCO/WT), the cry2 mutant (MycCO/cry2), or the cry2spa1 double mutant backgrounds (Myc-CO/cry2spa1) were adapted in the darkness for 48 hr, and then exposed to blue light (30 μmolem−2s−1) for 30 min or 60 min. The immunoblots were probed with Myc antibody (MycCO). A nonspecificity band was included for the loading control (NSB).
(D) A quantitative PCR showing the reduced FT expression in the cry2 mutant but restored in the spa1cry2 double mutant (see also Figure S3).
It has been previously reported that COP1 interacts with the floral regulator CO to cause CO ubiquitination and degradation [26, 27]; SPA1 activates COP1 to promote the COP1-dependent CO degradation [10, 28], whereas cryptochromes mediate blue-light suppression of the COP1 activity and CO degradation [29–31]. We reasoned that CRY2 may interact with SPA1 to suppress COP1 activity and CO degradation in response to blue light. To test this possibility, we prepared transgenic plants expressing the 35S∷Myc-CO transgene in the wild-type, cry2, and cry2spa1 mutants and compared blue-light effects on the abundance of the Myc-CO protein in the lines that express the similar level of the Myc-CO protein under SD photoperiods (Figure S3). In this experiment, transgenic lines expressing the 35S∷Myc-CO transgene in different genetic backgrounds were grown in SD photoperiods, adapted in the darkness for 2 days, transferred to blue light for 30 or 60 min, and the level of the Myc-CO protein were analyzed by immunoblot. Figure 3C shows that the level of Myc-CO protein increased significantly in response to blue light in the wild-type plants. Because the expression of Myc-CO is under the control of the constitutive 35S promoter, the increased Myc-CO protein level is most likely due to the blue light suppression of CO degradation [31]. No such increase of the Myc-CO protein level in response to blue light was observed in the cry2 mutant background, which is consistent with the hypothesis that CRY2 mediates blue light suppression of CO degradation [31] Importantly, the blue light suppression of Myc-CO degradation was restored in the cry2spa1 double mutant (Figure 3C). As expected, the mRNA expression of the CO target gene, FT, also showed a significant reduction in the cry2 mutant, but it was restored to the wild-type level in the spa1cry2 double mutant (Figure 3D). These results clearly demonstrate that SPA1 acts as a signaling molecule to mediate CRY2-dependent control of CO protein stability, FT transcription, and floral initiation in response to blue light.
The CRY1-SPA1 Interaction Enhances the CRY2-COP1 Interaction in Response to Blue Light
We next investigated how CRY2-SPA1 interaction in response to blue light may affect the CRY2 function. Given that CRY2 undergoes blue light-dependent interaction with SPA1 (Figures 1 and 2), SPA1 mediates CRY2 regulation of flowering time (Figure 3), and SPA1 and CRY2 physically interact with COP1 to activate or suppress the COP1 activity, respectively [10, 11, 20, 21, 23, 26–28], we reasoned that blue light may affect interactions of these three mutually interactive proteins (CRY2, SPA1, and COP1). We tested this possibility by using the yeast three-hybrid assay (Figures 4A and 4B). In this experiment, the protein-protein interaction between one pair of proteins (bait and prey) is tested in the absence or presence of the third protein (bait-mate) in response to blue light. Expression of the bait-mate protein was controlled by the methionine-suppressible Met25 promoter in yeast cells (Figure S4) [32]. We found that CRY2 (expressed as the bait-mate) did not affect the COP1-SPA1 interaction in yeast cells grown in the dark or under blue light (Figure 4A). In contrast, expression of SPA1 (as the bait-mate) significantly enhanced the CRY2-COP1 interaction in response to blue light (Figure 4B). In the absence of SPA1, blue light showed no apparent effect on the CRY2-COP1 interaction in yeast cells. In the presence of SPA1, the CRY2-COP1 interaction increased more than 5-fold when the total fluence of blue light increased by less than 2-fold (Figure 4B). On the basis of this result, we hypothesized that the blue light-dependent CRY2-SPA1 interaction enhances the CRY2-COP1 interaction to suppress the COP1 activity.
Figure 4. The CRY2-SPA1 Interaction Enhances the CRY2-COP1 Interaction in Response to Blue Light.
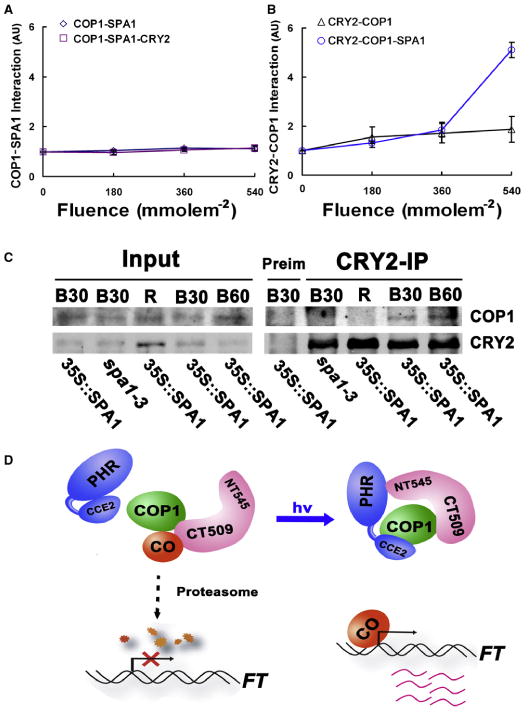
(A and B) Yeast three-hybrid experiments showing the SPA1-dependent enhancement of the CRY2-COP1 interaction in response to blue light. Yeast cells expressing the indicated proteins (bait-prey-bait mate) were grown in the dark (0 μmolem−2s−1) or blue light (50 μmolem−2s−1) for up to 3 hr before the β-galactosidase assay, with the total fluence of blue light (50 μmolem−2s−1 × treatment time in seconds) indicated underneath. The relative bait-prey interaction was presented as an arbitrary unit (AU), which is calculated by the formula AU = [miller units (light)]/[miller unit (dark)]; the AU of dark-treated samples is set to 1. The standard deviations (n = 3) are shown.
(C) Co-IP experiments showing the SPA1-dependent CRY2-COP1 interaction in response to blue light. The wild-type, spa1-3 mutant and transgenic plants expressing the 35∷MycSPA1 transgene in the spa1-3 background were grown in LD photoperiods for two weeks, transferred to dark for 18 hr, and then exposed to red light (R, 20 μmolem−2s−1) for 60 min, or to blue light (20 μmolem−2s−1) for 30 min (B30) or 60 min (B60). Immunoblots of the total protein extracts (Input) and IP product prepared by the CRY2 antibody (CRY2-IP) or preimmune serum (Pre-im) were probed by the COP1 antibody (COP1), stripped, and reprobed by the CRY2 antibody (CRY2). Arrows indicate COP1 or CRY2.
(D) A hypothetic model depicting CRY2-mediated blue light suppression of the COP1 activity. According to this model, SPA1 interacts with COP1 to activate COP1 activity for the ubiquitination and degradation of CO in the absence of light. In response to blue light, photoexcited CRY2 interacts with SPA1 to stimulate the CRY2-COP1 interaction, resulting in suppression of the COP1 activity, decreased degradation of CO, and increased FT transcription (see also Figure S4).
To further test this possibility, we examined how SPA1 and blue light affect formation of the CRY2-COP1 complex in plants by using a coimmunoprecipitation assay. In this experiment, the spa1 mutant plants and transgenic plants expressing MycSPA1 in the spa1 background were grown in a LD photoperiod for 2 weeks, transferred to darkness for 18 hr, and then exposed different wavelengths of light for 30 or 60 min. Figure 4C shows that, as expected, a similar amount of COP1 was detected in different samples, whereas the level of CRY2 decreased in plants exposed to blue light (Figure 4C, Input). Little COP1 protein was coprecipitated with CRY2 by the CRY2 antibody in plants treated with red light (Figure 4C, CRY2-IP, R). However, COP1 was clearly coprecipitated with CRY2 in the MycSPA1-overexpressing plants treated with blue light for 30 or 60 min (Figure 4C, CRY2-IP, B30, and B60; and Figure S4). In contrast, little COP1 was coprecipitated with CRY2 in the spa1 mutant plants treated with blue light (Figure 4C, CRY2-IP, spa1-3, and B30; and Figure S4). Importantly, the actual increase of the CRY2-COP1 interaction in response to blue light is likely to be more robust than what we observed in this experiment because the abundance of CRY2 decreases as a consequence of the blue light-induced CRY2 degradation [33](Figure 4C, Input). These results demonstrated, for the first time, that a cryptochrome could undergo blue light-dependent interaction with COP1. The fact that the blue light dependence of cryptochrome-COP1 interaction was not observed in previous reports for CRY1 [20, 21] but was observed in our study for CRY2 suggests a different mode of action of the two cryptochromes. Indeed, we and others recently found that Arabidopsis CRY1 interacts with SPA1 in response to blue light, but the structure-function relationship and the mode of action of the CRY1-SPA1-COP1 complex seem different from that of the CRY2-SPA1-COP1 complex [34, 35].
On the basis of previous discoveries and the findings presented in this study that (1) CRY2 undergoes blue light-dependent interaction with SPA1 (Figure 1), (2) overexpression of the SPA1 effector domain incapable of interacting with CRY2 suppresses photoperiodic sensitivity in transgenic plants (Figure 2), (3) SPA1 is required for the blue light-dependent CRY2 suppression of CO degradation (Figure 3), and (4) CRY2-SPA1 interaction enhances CRY2-COP1 interaction in response to blue light (Figures 4A–4C), we propose a working hypothesis to explain how CRY2 mediates blue light suppression of the COP1 activity to affect flowering time. According to this model, in the absence of a light signal, SPA1 interacts with COP1 to activate COP1-dependent ubiquitination and degradation of transcription regulators, such as CO, resulting in suppression of FT transcription and floral initiation. In response to blue light, the photoexcited CRY2 interacts with SPA1, which enhances CRY2-COP1 interaction to suppress the COP1 activity and CO degradation; this series of events leads to floral initiation in response to photoperiodic signals. Exactly how CRY2 suppress the COP1 activity remains to be further investigated.
Experimental Procedures
Supplementary Material
Acknowledgments
The authors thank Xingwang Deng, Hongquan Yang, and Haiyang Wang for kindly providing experimental materials used in this report. Works in authors' laboratories are supported in part by National Institute of Health (GM56265 to C.L.), the 985 Higher Education Enhancement grant (to Hunan University), University of California Los Angeles (UCLA) faculty research grants and Sol Leshin Ben Gurion University-UCLA Academic Cooperation programs.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at doi:10.1016/j.cub.2011.03.048.
References
- 1.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 2.Yu X, Liu H, Klejnot J, Lin C. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2010. The cryptochrome blue-light receptors. 10.1199/tab.0135, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 4.Yi C, Deng XW. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 2005;15:618–625. doi: 10.1016/j.tcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 6.Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- 7.Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 8.Searle I, Coupland G. Induction of flowering by seasonal changes in photoperiod. EMBO J. 2004;23:1217–1222. doi: 10.1038/sj.emboj.7600117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 10.Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003;17:2642–2647. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003;423:995–999. doi: 10.1038/nature01696. [DOI] [PubMed] [Google Scholar]
- 12.Laubinger S, Fittinghoff K, Hoecker U. The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell. 2004;16:2293–2306. doi: 10.1105/tpc.104.024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell. 2008;20:2307–2323. doi: 10.1105/tpc.107.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin C. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell. 2007;19:3146–3156. doi: 10.1105/tpc.107.053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C. Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci USA. 2007;104:7289–7294. doi: 10.1073/pnas.0701912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin C, Shalitin D. Cryptochrome structure and signal transduction. Annu Rev Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- 18.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. Structure of the photolyaselike domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- 21.Yang HQ, Tang RH, Cashmore AR. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell. 2001;13:2573–2587. doi: 10.1105/tpc.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Wang H. The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J. 2006;47:564–576. doi: 10.1111/j.1365-313X.2006.02811.x. [DOI] [PubMed] [Google Scholar]
- 23.Hoecker U, Quail PH. The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem. 2001;276:38173–38178. doi: 10.1074/jbc.M103140200. [DOI] [PubMed] [Google Scholar]
- 24.Hoecker U, Xu Y, Quail PH. SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature. 2002;417:763–767. doi: 10.1038/nature00815. [DOI] [PubMed] [Google Scholar]
- 26.Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008;27:1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell. 2008;20:292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development. 2006;133:3213–3222. doi: 10.1242/dev.02481. [DOI] [PubMed] [Google Scholar]
- 29.Osterlund MT, Deng XW. Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 1998;16:201–208. doi: 10.1046/j.1365-313x.1998.00290.x. [DOI] [PubMed] [Google Scholar]
- 30.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 31.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 32.Tirode F, Malaguti C, Romero F, Attar R, Camonis J, Egly JM. A conditionally expressed third partner stabilizes or prevents the formation of a transcriptional activator in a three-hybrid system. J Biol Chem. 1997;272:22995–22999. doi: 10.1074/jbc.272.37.22995. [DOI] [PubMed] [Google Scholar]
- 33.Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C. Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell. 2009;21:118–130. doi: 10.1105/tpc.108.061663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011 doi: 10.1101/gad.2025011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lian HL, He SH, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011 doi: 10.1101/gad.2025111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


