Abstract
Eukaryote cells dramatically reorganize their long chromosomal DNAs to facilitate their physical segregation during mitosis. The internal organization of folded mitotic chromosomes remains a basic mystery of cell biology; its understanding would likely shed light on how chromosomes are separated from one another as well as into chromosome structure between cell divisions. We report biophysical experiments on single mitotic chromosomes from human cells, where we combine micromanipulation, nanonewton-scale force measurement and biochemical treatments to study chromosome connectivity and topology. Results are in accord with previous experiments on amphibian chromosomes, and support the “chromatin network” model of mitotic chromosome structure. Prospects for studies of chromosome-organizing proteins using siRNA expression knockdowns, as well as for differential studies of chromosomes with and without mutations associated with genetic diseases are also discussed.
1. Introduction
Eukaryote cells dramatically reorganize their genomes as they prepare for cell division (figure 1(a)). This reorganization appears in the light microscope as “condensation” of relatively dispersed interphase chromosomes which are not easily seen as individual chromosome fibers, into dense, rope-like mitotic chromatids. The structure of mitotic chromosomes is only partially understood, but it is thought that a key function of the mitotic condensation process is to facilitate accurate segregation of different chromosomes [1, 2, 3], a necessary condition for normal cell division and proliferation.
Figure 1.
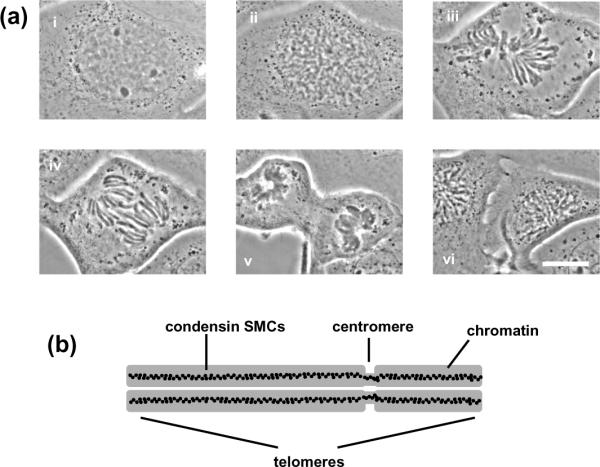
Mitotic chromosome organization.
(a) Cell cycle (newt Notophthalmus viridescens lung epithelial cell) showing interphase (i); prophase (ii); metaphase (iii); anaphase (iv); telophase (v); and interphase following division of daughter cells (vi) (pictures courtesy M. Poirier). Our chromosome-stretching experiments are carried out at “prometaphase”, just before capture of the mitotic chromosomes by the mitotic “spindle” apparatus (iii). Bar is 20 μm.
(b) Sketch of metaphase chromosome showing side-by-side chromatids and centromere; chromatid-axial condensin distibution inferred from experiments [4, 7] are indicated by black dots.
Present understanding of mitotic chromosome structure is based on the idea that chromatin fiber, itself a flexible polymer, must be folded, or structured, into the spatially ordered mitotic form, where two essentially segregated duplicate chromatids are attached side-by-side to form a “double-noodle” structure (figure 1(b)). At metaphase, the point of cell division when the chromosomes are fully condensed, it is well established that genes are in approximately linear order along mitotic chromosomes. However, whether this is achieved through formation of chromatin loops linked back to an organizing complex along the axis of each chromatid [4] or by a hierarchical folding scheme [5] is controversial.
Whatever the folding scheme, chromatin must somehow be secured into the mitotic form. This is widely thought to be due to the action of “condensin” SMC (Structural Maintenance of Chromosomes) complexes, large (≈50 nm long) ATP-utilizing protein complexes known to be important for formation of stable mitotic chromosomes in vitro [6] and in vivo [7, 8, 9]. Although condensins are known to condense naked DNA in vitro [10], exactly how they act and are coordinated on large scales during chromosome condensation in vivo is unknown. What role condensins play between cell divisions is unclear; other SMC complexes such as cohesin SMCs, which play a sister-chromatid-tethering role during mitosis [11], are increasingly appreciated for their chromosome-organizing and gene-regulation roles during interphase [12]. It is also thought in some quarters that other as yet undiscovered proteins may regulate large-scale mitotic chromosome structure [13, 14]. To add further uncertainty to the picture, recent cryo-electron-microscopy studies (cryo is regarded by many as the EM prep least disturbing for ultrastructural studies of biomolecules) suggest that chromatin itself is unfolded into a “melt of nucleosomes” inside mitotic chromosomes [15].
Measurement of chromosome mechanics provides a powerful tool for analysis of chromosome organization. Methods have been developed for single chromosome micromanipulation with force measurement using glass microneedles [16, 17] and glass micropipettes [18, 19]. Nicklas made the first measurement of the mechanical properties of meiotic metaphase chromosomes, using microneedles to manipulate single chromosomes inside grasshopper cells, and found that approximately nanonewton forces were needed to stretch chromosomes to double their native length [16, 17]. Micropipette-based manipulation showed that single, isolated mitotic chromosomes from newt and Xenopus cells were also doubled in length by roughly nanonewton forces [18, 19, 20]. Similar stretching elasticity results were obtained in experiments on chromatids (unreplicated Xenopus chromosomes) assembled in vitro using Xenopus egg extracts [23, 21]. Notably, the unreplicated in vitro reconstitued chromatids have far weaker bending rigidity than somatic Xenopus chromosomes [22], likely reflecting appreciable differences in internal organization.
Micromechanics experiments can be used to study how modification or removal of specific molecules impacts chromosome mechanics. Previously, our group has used this approach to probe the internal structure of mitotic chromosomes [20, 24]. Effects of structural changes made using enzymes are read out using force-extension measurements on whole mitotic chromosomes. An example important to this paper are experiments using blunt-cutting restriction enzymes with 4-base specificity, which led to complete dissolution of newt mitotic chromosomes [25]. This result shows that non-histone proteins such as condensin SMC complexes, and topoisomerase II, cannot be connected together to form a “chromosome scaffold” to which chromatin is attached. Instead, nonhistone proteins must act as isolated “crosslinkers” between chromatin segments. This experiment illustrates how our biochemical-biophysical approach can probe molecular connectivity of chromatin in chromosomes.
The close relationship between chromosome condensation and cell division suggests that mitotic chromosome organization may be modified in rapidly growing tumorigenic cells. Indeed, evidence exists suggesting that mitotic chromosome structure is distinct in non-tumorigenic and tumorigenic cells [26]. Our group's role in our Physical Science and Oncology Center (PS-OC) is to study differences in chromosome organization that arise in cancer cells. One of the techniques we will use to study changes in chromosome organization is study of mechanical properties of individual mitotic chromosomes. Previously our experiments have been focused on studies of relatively large chromosomes from amphibian species (the newt Notophthalmus viridescens, and the toad Xenopus laevis). However, many genetic tools and techniques that exist for human cells are unavailable for the amphibian cells we have been studying. Furthermore, to address questions concerning chromosome reorganization associated with human diseases, experiments on human cells are indicated.
These factors have driven us to learn how to carry out experiments on human chromosomes, which are appreciably smaller than amphibian chromosomes: the largest human chromosome contains about 250 Mbp, while the average Notophthalmus viridescens (newt) chromosome contains about 2 Gbp. Put another way, a typical newt chromosome is roughly 2 μm thick and 20 μm long; the largest human chromosome is about 10 μm long and only a little more than 1 μm thick. Despite being much smaller, it is possible to do micromanipulation of individual human chromosomes using micron-size glass pipettes. In this paper we report results of our initial studies, aimed mainly at determining whether the basic elasticity and response to nuclease and protease of mammalian chromosomes is similar to that of amphibian chromosomes. The Conclusion briefly discusses prospects for further experiments using genetic tools such as small-RNA gene interference (siRNA) in order to study roles played by specific proteins in folding mitotic chromosomes, as well as prospects for studying cell lines with mutations of chromosome-associated proteins associated with specific genetic diseases.
2. Methods
2.1. Cell culture
Sample dishes were prepared using #1 microscope glass onto which 25 mm-diameter rubber O-rings were affixed using wax. Human HeLa cells were grown in open dishes in a 37°C 5% CO2 atmosphere incubator, in DMEM (Cellgro) with 10% added FBS and 1% penn-strep antibiotic (Invitrogen). Medium was replaced every three days. Experiments were performed on 70% confluent samples.
2.2. Microscopy, micromanipulation, and force measurement
Micromanipulation experiments were performed on the stage of an inverted microscope (IX-70; Olympus) using a 60× 1.4 NA oil immersion objective as previously described [19, 20, 27, 28] (figure 2). We maintained the temperature of the slide containing the living cells at 30°C using an objective heater (20–20 Technologies). We found that using 30°C instead of the 37°C at which the cells were grown improved human cell viability in room air, allowing cultures to be used in room air for several hours and eliminating the need for CO2 atmosphere incubation on the microscope. All imaging was performed in the cell culture medium using a CCD camera (WV-BP310; Panasonic) with images acquired by a frame grabber (IMAQ PCI-1408; National Instruments) to a PC. Micromanipulation and reagent microspraying was done using glass micropipettes mounted on micromanipulators.
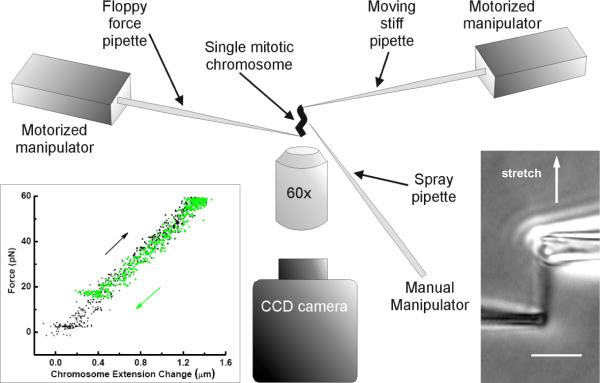
Figure 2.
Chromosome stretching experiment.
A single mitotic chromosome following its removal from a dividing cell is suspended between two pipettes on motorized manipulators in the aqueous tissue culture medium in which the cells are grown. Inset at lower right shows a phase-contrast micrograph of chromosome and pipettes. Bar is 5 μm. Inset at lower left shows force-extension data for a human chromosome (black: extension, gray: retraction) showing its linear, reversible elasticity.
In brief, samples were placed on the microscope stage, prometaphase cells were identified, and a spray pipette filled with 0.05% Triton X-100 in PBS (Thermo Fisher Scientific) positioned by a motorized micromanipulator (MP-285; Sutter Instrument Co.) was used to destabilize the cell membrane via microspraying. After the chromosomes flowed out of the cell, one pipette was used to grab one end of one chromosome using aspiration; then, a second pipette was attached to the other chromosome end (figure 2, lower right).
One of the two pipettes was pulled with a rather short taper so that it was very stiff, while the other was pulled with a long taper so as to have a soft end deflection spring constant of ≈50 pN/μm. During stretching experiments, the stiffer pipette was moved at a rate of 5 μm/min, slowly enough to avoid viscoelastic effects [19] and bending of the softer pipette was measured and recorded. Each extension-relaxation measurement was repeated two or three times to ensure reproducibility and to verify that changes in chromosome sti ness caused by previous enzyme exposures had ceased. For study of human chromosomes, the major difference in methods from those used in our previous experiments with newt chromosomes is that the pipettes must be fabricated to have ≈ 1 μm diameter end openings, about half the opening size used for newt experiments, and the soft pipette has a lower spring constant.
During stretching experiments, micromechanical data were collected automatically using online image analysis software written in Labview (National Instruments). Pipette position data for both the stiff (moved) and floppy (deflected) pipettes were recorded. The spring constant of the floppy, force-measuring pipette was calibrated following the experiment following methods of Ref. [19]. The chromosome extension and force were then computed from relative pipette positions after the experiment using separate Labview programs. We estimate experimental errors in individual spring constant measurements to be < 10%, on the basis of observed systematic errors in spring constant calibration [19]. The images of each chromosome were analyzed using ImageJ (National Institutes of Health, http://rsbweb.nih.gov/ij). The cross-sectional diameter of each chromosome was determined by averaging several different diameters measured along its length; an error estimate was obtained from the variance of those measurements.
2.3. Enzyme microspray experiments
Solutions to be sprayed onto chromosomes were loaded into glass pipettes which were pulled and cut so as to have a ≈5 μm-diameter aperture. The pipettes were mounted on a three-axis manual manipulator (Taurus, World Precision) used to position them in the sample dish. Pressure between 10 and 100 Pa was used to drive spraying of the enzyme mix onto chromosomes. Microspray reactions were performed for 20 min onto unstretched chromosomes. The flow of enzyme was stopped for several minutes before stretching to allow the small amounts of reagents introduced into the ≈1.5 ml sample cell to diffuse away from the chromosome. Previous experiments indicated that enzyme reactions do not continue after spraying is finished and that successive stretch-release cycles after enzyme exposures are all the same [25, 27, 28].
Proteinase K (Fermentas) was diluted to 300 nM in PK buffer (100 mM potassium acetate, 20 mM Hepes, pH 7.9) for spray experiments. Trypsin (Sigma) was diluted to 100 nM in PBS for microspray experiments. Restriction enzymes (AluI, RsaI, HaeIII, SnaBI, PvuII, BmgBI, New England Biolabs) were diluted to 0.5 units/μl for microspray experiments, in the buffers recommended by New England Biolabs (NEB): PvuII uses NEB Buffer 2 (50 mM NaCl, 10 mM Tris-HCl, 10mM MgCl2, 1 mM DTT, pH 7.9); BmgBI uses NEB Buffer 3 (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.9); AluI, RsaI, HaeIII, SnaBI use NEB Buffer 4 (20 mM Tris-acetate, 50 mM potassium acetate, 10 mM Mg acetate, 1 mM DTT, pH 7.9). All enzyme spray experiments were carried out for 20 min, except for cases where chromosomes were completely dissolved in less than 20 min.
3. Results
3.1. Chromosome elasticity
A series of chromosome-stretching experiments were carried out to determine the elastic properties of human chromosomes. Figure 3(a) shows images collected during a stretching experiment. As the stiff (right) pipette is moved progressively to the right, tension is applied to the attached chromosome, causing its extension and bending of the floppy (left) pipette. The force as a function of extension and relaxation for this series of pictures is shown in the lower left of figure 2. As is the case for amphibian chromosomes, we have found human chromosomes to show reversible elasticity, with force depending nearly linearly on extension for extensions smaller than 100%.
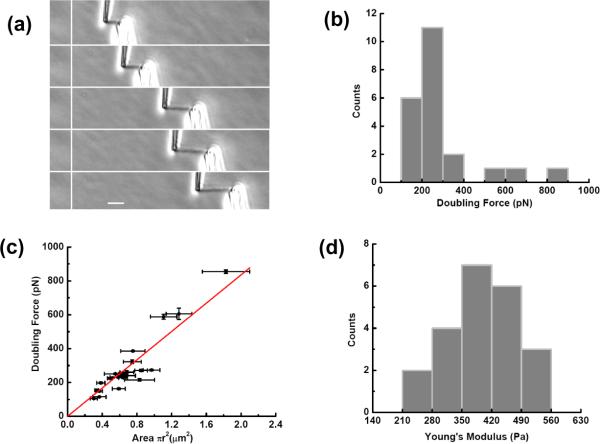
Figure 3.
Human chromosomes display linear elasticity over a wide range that can be described by a Young's modulus of ≈ 420 Pa.
(a) Images of a human chromosome during a stretching experiment. Bar is 5 μm.
(b) Distribution of force constants from a series of 22 human chromosome-stretching experiments.
(c) Plot of force constant vs cross-sectional area for the measurements of (b); the data fit well to a straight line with slope 420 ± 20 Pa.
(d) Distribution of stretching (Young) modulus measurements from the measurements of (b); note the tighter distribution compared to the force constant distribution of (b). Mean Young modulus is 400 ± 20 Pa.
From a series of 22 stretching experiments we obtained force-extension data. The initial slope of those force-extension curves, multiplied by the length of the initially relaxed chromosome segment being stretched (i.e., the unstretched length of the chromosome between the two pipettes) gives the “force constant” or “doubling force” for a chromosome. The force constant is the force that, if the initial linear force-extension behavior was extrapolated, would correspond to doubling of the length of a chromosome [19]. Figure 3(b) shows the distribution of doubling forces, which is broad, but well-peaked at near 250 pN; the average doubling force is 290±40 pN. The characteristic force required to double the length of a human mitotic chromosome is therefore approximately 300 pN, significantly less than the ≈ 1 nN forces needed to double the length of the thicker newt chromosomes [20].
The force constant for a chromosome can be expected to be larger for a thicker chromosome, for which more chromatin fibers need to be stretched. This argument suggests that the doubling force should scale roughly linearly with chromosome cross-sectional area. Figure 3(c) shows that indeed the measured doubling forces rise approximately linearly with the square of measured cross-sectional radius of each chromosome. The slope of this fit line gives an estimate of the Young modulus of 420 ± 20 Pa.
Alternately we can compute Young moduli by dividing the force constant by the cross sectional area for each chromosome. This is histogrammed in figure 3(d): the distribution is narrower than that of the force constant. The mean Young modulus of a human mitotic chromosome obtained from this distribution 400 ± 20 Pa, consistent with the Young modulus obtained from the slope in figure 3(c).
3.2. Restriction nuclease digestion
We carried out a series of experiments on human mitotic chromosomes where, following their extraction and initial force-extension measurement, they were exposed to a spray of “blunt-cutting” restriction enzymes, enzymes which cut at specific DNA sequences, with both cuts at the same base position. Blunt cuts (unlike the overhanging cuts made by most restriction enzymes) cannot rehybridize, so once a cut is made, the DNA is permanently disconnected at that point.
Figure 4(a) shows a series of pictures during digestion by the enzyme AluI, which cuts in the middle of the 4-base sequence AG|CT (the | indicates the cut site). The result of these experiments was first that chromosome elasticity was lost, and then the chromosome was entirely dissolved between the two pipettes. The same result was obtained for the 4-base blunt-cutters RsaI (GT|AC) and HaeIII (GG|CC) (data not shown). For human chromosomes, all 4-base cutters used led to complete dissolution of the chromosome segment held between the pipettes within a few minutes of nuclease exposure, in accord with experiments carried out for newt chromosomes [25].
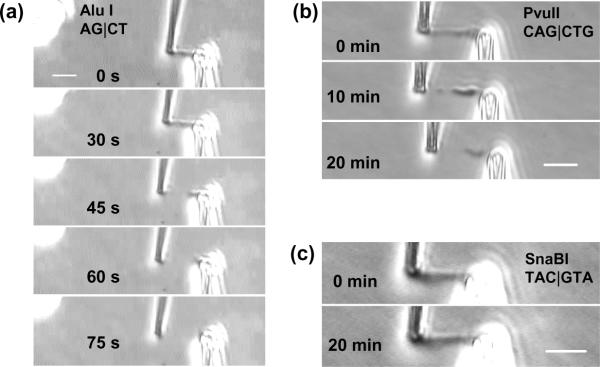
Figure 4.
Restriction enzyme digestion of a human mitotic chromosome. Bars are all 5 μm
(a) Photos top to bottom show cleavage and dissolution of a chromosome between the pipettes over the first 75 sec of spraying a chromosome with the blunt-cutter AluI (AG|CT). Cumulative enzyme spray time is indicated in each image.
(b) Photos top to bottom show partial digestion of a chromosome by the 6-base cutter PvuII (CAG|CTG) after 20 min spraying.
(c) Photos show no visible effect of the 6-base cutter SnaBI (TAC|GTA) on a human chromosome after 20 min spraying.
We also carried out experiments with 6-base blunt-cutting restriction enzymes, which have a higher sequence specificity than the 4-base cutters. Interestingly, and unlike the case for 4-base cutters, we observed strong variation in the results with cut sequence. Experiments with the enzyme PvuII (CAG|CTG) led to partial, and in some cases, inhomogeneous digestion of human chromosomes (figure 4(b)) over a 20 min time course. In this case, the force constant was strongly reduced by digestion (data not shown), but the chromosome was not completely disconnected. Therefore the PvuII cut sites are just frequent enough in some regions of the chromosome to permit appreciable unfolding.
However, experiments with other 6-base blunt-cutters SnaBI (TAC|GTA) and and BmgBI (CAC|GTC) were also carried out, with the result that there was no effect on chromosome morphology or force constant. Figure 4(c) shows the result of a digestion experiment using SnaBI; the force constant was unchanged by the nuclease digestion in this experiment (data not shown).
3.3. Protein digestion
Experiments were carried out with proteases to study how cleavage of proteins would impact chromosome structure, following the approach we used previously for newt mitotic chromosomes [27]. Figure 5(a)–(b) shows results for digestion of a native human chromosome by trypsin. Trypsin is a serine protease which cleaves peptide bonds on the carboxyl-terminal side of lysine and arginine amino acids, except when proline is on the carboxyl side of the cleavage site. Trypsin is therefore a protease with some degree of specificity in where it cuts a given protein.
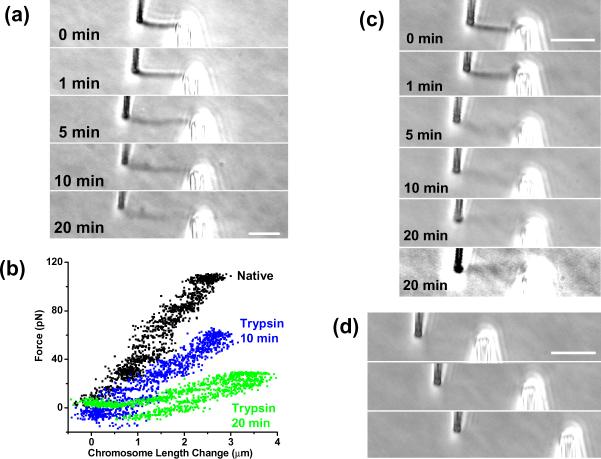
Figure 5.
Effect of protein digestion on a human mitotic chromosome. Bars are all 5 μm.
(a) Series of photos during digestion by trypsin showing gradual loss of phase contrast, lengthening, and thickening; cumulative enzyme spray time is indicated in each image.
(b) Force-extension curves for the experiment of (a) showing that the chromosome remains elastic during the digestion.
(c) Series of photos of mitotic chromosome during a digestion by proteinase K. The last two images show the same digestion time point (20 min); final image has higher contrast. Bar is 5 μm.
(d) Series of photos showing the digested chromosome of (c) subjected to a stretching experiment indicating that it still supports stress.
Figure 5(a) shows how progressive digestion by 100 nM trypsin in PBS gradually makes the chromosome longer and wider, with a gradual loss of phase contrast as protein is digested. However, at all times during this experiment, the chromosome remained connected to both pipettes, and was able to be reversibly stretched. Figure 5(b) shows force curves measured for the untreated chromosome, and after 10 min and 20 min of trypsin digestion. The chromosome force constant is gradually reduced, but the chromosome continues to behave like an elastic solid, unlike the case for 4-base blunt-cutting restriction enzyme treatment.
Figure 5(c)–(d) shows results from a digestion experiment using 300 nM proteinase K; this serine protease cleaves peptide bonds on the carboxyl-terminal side of aliphatic and aromatic amino acids, and thus digests proteins with less site-specificity than trypsin. Figure 5(c) shows that that proteinase K in PBS causes the mitotic chromosome to be progressively lengthened and thickened, and that it gradually loses its phase contrast as digestion proceeds. As is the case for trypsin, the nearly invisible chromosome still connects the pipette and can support stress, as evidenced by the deflection of the right “floppy” pipette in figure 5(d). Overall, the results for trypsin and proteinase K digestions are rather similar for human chromosomes and newt chromosomes [27].
4. Conclusions
We have reported glass micropipette micromanipulation experiments on human mitotic chromosomes. Previously, we had only carried out micromanipulation of relatively large amphibian chromosomes, but despite their smaller size, similar experiments on human chromosomes are possible. Our initial experiments have established that human chromosomes display a wide range of nearly linear elastic response similar to what we have observed for newt and Xenopus chromosomes. The force required to double the length of a human chromosome is broadly distributed, with an average of 300 pN, about a third of that observed for newt chromosomes. However, after taking into account variation in the thickness of the chromosomes, we find the Young modulus to be more narrowly distributed around an average of 400 Pa, a value close to that obtained for the much thicker newt chromosomes [20].
In a series of enzyme-spray experiments we found the same basic connectivity properties for human mitotic chromosomes observed in previous studies of amphibian chromosomes. Digestion by 4-base-specificity blunt-cutting restriction enzymes led to complete disconnection and dissolution of chromosome segments. In contrast to the situation for 4-base-cutter nucleases, digestion by trypsin and protease did not completely cut through human mitotic chromosomes, but instead led to gradual thickening and lengthening of the chromosome and a reduction of its force constant.
These experiments indicate that chromatin itself holds human mitotic chromosomes together, and rule out the possibility that human mitotic chromosome structure is defined by a protein “core” to which chromatin is merely attached. Instead, we conclude that human chromosomes have a “chromatin network” organization (figure 6), where the chromatin fiber inside each chromatid is attached to itself by chromatin-chromatin “crosslinks” or “interconnects”, most plausibly involving non-covalent protein-protein interactions.
Figure 6.
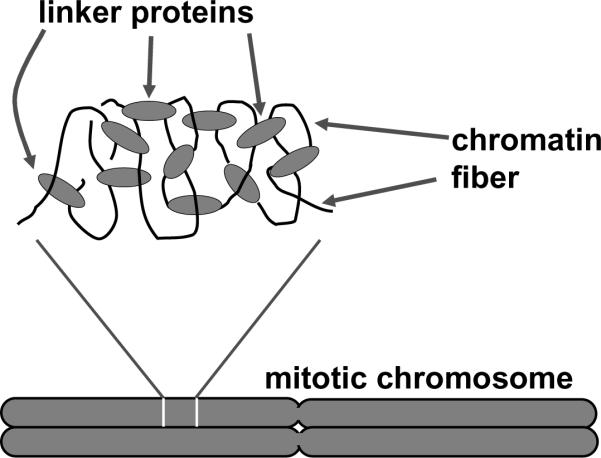
“Chromatin network” model of mitotic chromosome folding.
Chromatin segments (black curves) are connected together by protein-chromatin linkers (gray ovals). Cutting DNA (chromatin) disconnects the network, while cutting protein only lengthens and softens the network.
A network, or gel-like structure will be completely destroyed by sufficiently frequent cuts, (e.g., by 4-base specificity blunt-cutting restriction enzymes), but will not be as severely damaged by less frequent cuts. Indeed, some 6-base specificity blunt-cutting enzymes led to partial reduction of the force constant of chromosomes during a 20-minute digestion. The simplest interpretation of this result is that most 6-base cuts are too rare to completely disconnect a mitotic chromosome in a 20-min reaction, i.e., the distance between 6-base cutter cleavages is comparable to the distance between chromatin “crosslinks” or “interconnects”. The conclusion that chromatin itself is the main load-bearing element inside a human mitotic chromosome provides a straightforward explanation for the high degree of extensibility of mitotic chromosomes, since chromatin itself shows a high degree of extensibility [29].
Overall, our experiments thus far indicate that human mitotic chromosomes behave substantially like newt mitotic chromosomes as regards as their overall elasticity and response to nucleic acid and protein digestion. However, we have observed some minor differences. First, it appears that human chromosomes are more susceptible to being disconnected by 6-base-specificity restriction enzymes than were newt chromosomes. This may reflect the fact that chromatin in newt mitotic chromosomes is slightly more tightly packaged (more frequently crosslinked by chromatin-binding proteins) than in human chromosomes, or this may reflect differences in sequence distribution in newt and human chromosomes. Newt chromosomes are huge (roughly ten times longer than human chromosomes) and contain much more repetitive (non-gene) DNA.
For 6-base-specificity blunt-cutting restriction enzyme, we observed strong variation in the degree of chromosome digestion: PvuII resulted in partial digestion; while SnaBI and BmgBI did not change chromosome elasticity. One possible explanation is that the frequencies of cutting sites of those enzymes are different. Given that the human genome sequence is known, it is possible to determine all possible cut sites for any restriction enzyme. However, not all the sites are accessible due to their being blocked by nucleosomes and other proteins bound to the DNA: it has been shown that nucleosomes protect roughly 90% of the DNA they are bound to from restriction enzyme digestion [30, 31]. Nevertheless, the statistical frequency of targets in human chromosomal DNA is likely to indicate the frequency of cuts in this experiment. A preliminary survey of sequences for human chromosome 1 indicates that indeed PvuII sites are several-fold more numerous than SnaBI sites. A more systematic study of effects of restriction enzymes on human chromosomes would be desirable, but this will require identification of the chromosome being studied in a given experiment. It is possible to engineer fluorescent tags onto specific chromosomes in mammalian cell lines[32, 33], so such a study is within the realm of possibility.
An alternative explanation to the variation of different 6-base cutters is that SnaBI and BmgBI activities are both blocked by CpG methylation, while PvuII is insensitive to methylation. Approximately 70% of all CpGs are methylated in HeLa cells [34], which could be the reason why we observed little or no change on human chromosomes by SnaBI or BmgBI.
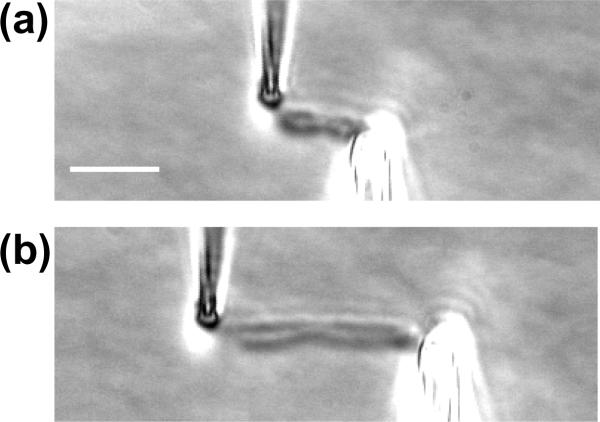
A second difference between human and newt chromosomes that we have noted is illustrated in figure 7. In most of our experiments on human mitotic chromosomes, we observe the chromosome to have a cylindrical shape, without clear distinction of the two side-by-side chromatids. However, in a small subset of our experiments, we have observed mitotic chromosomes where the two side-by-side chromatids are clearly discernible, with an apparent gap between them. Separation of sister chromatids by micromanipulation has been done by Paliulis et al [35] in grasshopper metaphase cells, where microneedles were used to pull centromeric regions of sister chromatids apart from one another, to study cohesion release during the late stages of the cell cycle. For isolated human chromosomes it appears that we should be able to at least in some cases attach a pipette to the ends of individual chromatids of a metaphase chromosome, and pull on the ends of sister chromatids with controlled force (see figure 7) to carry out similar experiments, but probing sister chromatid cohesion starting from the telomeres of chromosomes. This type of experiment may be useful for studying the strength of inter-chromatid cohesion which is thought to be due to a combination of cohesin SMCs [11] and inter-chromatid entanglements generated by DNA replication.
Figure 7.
“X-shaped” human chromosome attached to pipettes.
(a) Relaxed chromosome; the two-chromatid structure of the chromosome is evident in the image. Bar is 5 μm
(b) Stretched chromosome; the left pipette is attached to the upper chromatid and is extending it more than the lower chromatid.
We are still left with the rather basic question of the nature of the chromatinchromatin linkers (figure 6). It appears likely that at least some of the linkers are condensin SMC complexes, which are known to be necessary for establishment and maintenance of condensed mitotic chromosomes [6]. Chromosome manipulation experiments of Almagro et al [21] who used antibody-coated pipettes to generate attachments between pipettes and condensins in unreplicated chromatids, also suggest a key structural role for condensin SMCs in mitotic chromosomes. However, some researchers have suggested that there may be other as yet undiscovered “regulators of chromosome structure” [13, 14]. Even less well understood is how chromatin-linking proteins generate the distinctive large-scale folded cylindrical structure of mitotic chromosomes, e.g., via DNA-sequence-dependent protein-DNA interactions. Our recent experiments with topoisomerases suggest that DNA entanglement topology may play a role in mitotic chromosome folding [28].
The experiments of this paper establish micromechanical experimental methods for human chromosomes, and open the door to new experiments aimed at studying roles of specific proteins in mitotic chromosome structure. Human cells can be genetically controlled by a variety of means, and can have specific genes modified or deleted, allowing studies of e ects of removal of specific proteins. A particularly powerful approach which is even simpler than development of mutant cells is the use of small interfering RNA (siRNA) molecules to temporarily halt synthesis of specific proteins. Previous experiments have shown that mitotic chromosome structure can be disrupted by use of siRNA against condensin subunits [7, 9]. We are at present working on siRNA experiments targeted at condensin and cohesin SMC subunits, in order to diagnose the roles these molecules play in controlling mitotic chromosome mechanical properties. Also, we plan to use chemically inducible SMC subunit knockouts [8, 36] to do similar SMC-knockdown experiments without the need for transfection of RNAs.
Using human cells also will allow us to study cell lines with mutations corresponding to genetic disease states. One of our main objectives is studies of mutations associated with cancers, especially those which lead to changes in proteins which play a key role in chromosome structure. Given that cancer cells rapidly proliferate in response to genetic damage, it is possible that tumorigenic cells may have peculiar large-scale mitotic chromosome structure and mechanics. One of the labs in our PS-OC has recently discovered that mutations leading to overexpression of the MMSET histone methylase are associated with certain kinds of blood cancers. Those mutations are likely to lead to changes in large-scale chromatin structure, so cell lines carrying MMSET mutations are of interest to us [37].
In addition, it is increasingly being appreciated that mutations in chromosome-associated proteins are connected with genetic diseases. We are particularly interested in SMC mutations [12, 38] likely to cause disruption of large-scale chromosome structure, as well as nuclear lamin mutations associated with large-scale nucleus reorganization [39, 40, 41, 42] It is possible that modification of lamin gene expression may allow us to “soften” the normally very stiff nuclear envelope [39], allowing micromanipulation study of interphase and prophase chromosomes. Access to chromosomes inside the nuclear envelope may allow studies of the coupling between chromatin condensation and topological resolution of chromosomes from one another [2, 24].
Acknowledgements
This work was supported by NSF Grants DMR-0715099 and MCB-1022117, by NIH Grant 1U54CA143869-01 (NU-PS-OC), and by a predoctoral fellowship from the American Heart Association (M.S.).
References
- [1].Hirano T. Biochemical and genetic dissection of mitotic chromosome condensation. Trends Biol. Sci. 1995;20:357–61. doi: 10.1016/s0968-0004(00)89076-3. [DOI] [PubMed] [Google Scholar]
- [2].Marko JF, Siggia ED. Polymer models of meiotic and mitotic chromosomes. Mol. Biol. Cell. 1997;8:2217–31. doi: 10.1091/mbc.8.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shintomi K, Hirano T. Sister chromatid resolution: a cohesin releasing network and beyond. Chromosoma. 2010;119:459–67. doi: 10.1007/s00412-010-0271-z. [DOI] [PubMed] [Google Scholar]
- [4].Maeshima K, Laemmli UK. A two-step scaffolding model for mitotic chromosome assembly. Dev. Cell. 2003;4:467–80. doi: 10.1016/s1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- [5].Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS. Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J. Cell Biol. 2004;166:775–85. doi: 10.1083/jcb.200406049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–58. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- [7].Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–21. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- [8].Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell. 2003;5:323–36. doi: 10.1016/s1534-5807(03)00199-0. [DOI] [PubMed] [Google Scholar]
- [9].Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci. 2004;117:6435–45. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- [10].Strick TR, Kawaguchi T, Hirano T. Real-time detection of single-molecule DNA compaction by condensin I. Curr. Biol. 2004;14:874–80. doi: 10.1016/j.cub.2004.04.038. [DOI] [PubMed] [Google Scholar]
- [11].Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 2009;43:525–58. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- [12].Wendt KS, Peters JM. How cohesin and CTCF cooperate in regulating gene expression. Chrom. Res. 2009;17:201–14. doi: 10.1007/s10577-008-9017-7. [DOI] [PubMed] [Google Scholar]
- [13].Vagnarelli P, Hudson DF, Ribeiro SA, Trinkle-Mulcahy L, Spence JM, Lai F, Farr CJ, Lamond AI, Earnshaw WC. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat. Cell Biol. 2006;8:1133–42. doi: 10.1038/ncb1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hudson DF, Marshall KM, Earnshaw WC. Condensin: Architect of mitotic chromosomes. Chrom. Res. 2009;17:131–44. doi: 10.1007/s10577-008-9009-7. [DOI] [PubMed] [Google Scholar]
- [15].Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc. Natl. Acad. Sci. USA. 2008;105:19732–7. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nicklas RB. A quantitative study of chromosomal elasticity and its influence on chromosome movement. Chromosoma. 1963;14:276–95. doi: 10.1007/BF00326816. [DOI] [PubMed] [Google Scholar]
- [17].Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–8. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Houchmandzadeh B, Marko JF, Chatenay D, Libchaber A. Elasticity and structure of eukaryote chromosomes studied by micromanipulation and micropipette aspiration. J. Cell Biol. 1997;139:1–12. doi: 10.1083/jcb.139.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Poirier M, Eroglu S, Chatenay D, Marko JF. Reversible and irreversible unfolding of mitotic newt chromosomes by applied force. Mol. Biol. Cell. 2000;11:269–76. doi: 10.1091/mbc.11.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Poirier MG, Marko JF. Micromechanical studies of mitotic chromosomes. Curr. Top. Dev. Biol. 2003;55:75–141. doi: 10.1016/s0070-2153(03)01002-0. [DOI] [PubMed] [Google Scholar]
- [21].Almagro S, Riveline D, Hirano T, Houchmandzadeh B, Dimitrov S. The mitotic chromosome is an assembly of rigid elastic axes organized by structural maintenance of chromosomes (SMC) proteins and surrounded by a soft chromatin envelope. J Biol. Chem. 2004;279:5118–26. doi: 10.1074/jbc.M307221200. [DOI] [PubMed] [Google Scholar]
- [22].Poirier MG, Eroglu S, Marko JF. The bending rigidity of mitotic chromosomes. Mol. Biol. Cell. 2002;13:2170–9. doi: 10.1091/mbc.01-08-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Houchmandzadeh B, Dimitrov S. Elasticity measurements show the existence of thin rigid cores inside mitotic chromosomes. J Cell Biol. 1999;145:215–23. doi: 10.1083/jcb.145.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Marko JF. Micromechanical studies of mitotic chromosomes. Chrom. Res. 2008;16:469–97. doi: 10.1007/s10577-008-1233-7. [DOI] [PubMed] [Google Scholar]
- [25].Poirier MG, Marko JF. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc. Natl. Acad. Sci. USA. 2002;99:15393–97. doi: 10.1073/pnas.232442599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Maniotis AJ, Valyi-Nagy K, Karavitis J, Moses J, Boddipali V, Wang Y, Nunez R, Setty S, Arbieva Z, Bissell MJ, Folberg R. Chromatin organization measured by AluI restriction enzyme changes with malignancy and is regulated by the extracellular matrix and the cytoskeleton. Am. J. Pathol. 2005;166:1187–203. doi: 10.1016/S0002-9440(10)62338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pope LH, Xiong C, Marko JF. Proteolysis of mitotic chromosomes induces gradual and anisotropic decondensation correlated with a reduction of elastic modulus and structural sensitivity to rarely cutting restriction enzymes. Mol Biol Cell. 2006;17:104–13. doi: 10.1091/mbc.E05-04-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kawamura R, Pope LH, Christensen MO, Sun M, Terekhova K, Boege F, Mielke C, Andersen AH, Marko JF. Mitotic chromosomes are constrained by topoisomerase II-sensitive DNA entanglements. J. Cell. Biol. 2010;188:653–63. doi: 10.1083/jcb.200910085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kruithof M, Chien FT, Routh A, Logie C, Rhodes D, van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 2009;16:534–40. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- [30].Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995;254:130–49. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- [31].Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000;296:979–87. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- [32].Belmont AS. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 2001;11:250–7. doi: 10.1016/s0962-8924(01)02000-1. [DOI] [PubMed] [Google Scholar]
- [33].Strukov YG, Belmont AS. Mitotic chromosome structure: reproducibility of folding and symmetry between sister chromatids. Biophys. J. 2009;96:1617–28. doi: 10.1016/j.bpj.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ehrlich M, Gama Sosa MA, Huang L-H, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res. 1982;10:2709–21. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Paliulis LV, Nicklas RB. Kinetochore rearrangement in meiosis II requires attachment to the spindle. Curr Biol. 2004;14:2124–9. doi: 10.1007/s00412-005-0330-z. [DOI] [PubMed] [Google Scholar]
- [36].Ribeiro SA, Gatlin JC, Dong Y, Joglekar A, Cameron L, Hudson DF, Farr CJ, McEwen BF, Salmon ED, Earnshaw WC, Vagnarelli P. Condensin regulates the stiffness of vertebrate centromeres. Mol. Biol. Cell. 2009;20:2371–80. doi: 10.1091/mbc.E08-11-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Marango J, Shimoyama M, Meyer JA, Min DJ, Sirulnik A, Martinez-Martinez Y, Chesi M, Bergsagel PL, Zhou MM, Waxman S, Leibovitch BA, Walsh MJ, Licht JD. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111:3145–54. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mannini L, Menga S, Musio A. The expanding universe of cohesin functions: a new genome stability caretaker involved in human disease and cancer. Hum. Mutat. 2010;31:623–30. doi: 10.1002/humu.21252. [DOI] [PubMed] [Google Scholar]
- [39].Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006;281:25768–80. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- [40].Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2004;101:8963–8. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–21. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Taimen P, Pfleghaar K, Shimi T, Moller D, Ben-Harush K, Erdos MR, Adam SA, Herrmann H, Medalia O, Collins FS, Goldman AE, Goldman RD. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc. Natl. Acad. Sci. USA. 2009;106:20788–93. doi: 10.1073/pnas.0911895106. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]