Abstract
Objective
HIV infection of CD4 T cells can lead to HIV protease-mediated cleavage of procaspase 8 generating a novel, HIV-specific peptide called Casp8p41. Casp8p41 has at least two biologic functions: induction of cell death via mitochondrial depolarization and release of cytochrome C, as well as activation of nuclear factor kappa B (NFκB). We have previously shown that Casp8p41-induced NFκB activation enhances HIV LTR transcription and consequently increases HIV replication. Herein, we questioned whether Casp8p41-induced NFκB activation impacts the cytokine profile of cells expressing Casp8p41.
Design
Analysis of cells expressing Casp8p41 and HIV-infected T cells.
Methods
We assessed whether host genes are transcriptionally activated following Casp8p41 production, using microarray analysis, cytokine quantification, followed by western blot and flow cytometry.
Results
Microarray analysis identified 259 genes significantly upregulated following expression of Casp8p41. Furthermore, Casp8p41 expression in primary CD4 T cells results in increased production of interleukin (IL)-2, IL-15 and tumor necrosis factor (TNF), as well as IL-1RA; whereas levels of granulocyte macrophage colony-stimulating factor and interferon (IFN)-γ were reduced in the Casp8p41 expressing cells. Intra-cellular flow cytometry confirmed the co-association of Casp8p41 with elevated TNF in HIV-infected cells.
Conclusion
These data indicate that the expression of Casp8p41 in HIV-infected CD4 T cells in addition to promoting apoptosis and enhancing HIV replication also promotes a proinflammatory cytokine milieu, which is characteristic of untreated HIV infection.
Keywords: apoptosis, Casp8p41, HIV, inflammation, protease, tumor necrosis factor
Introduction
Untreated HIV infection results in a dysregulated cytokine milieu in infected individuals as well as continuous depletion of CD4 T cells such that coordinated immune responses become impossible. Indeed, the earliest reports of this disease included descriptions of anergy, lymphopenia, reduced CD4 T cell numbers [1] and elevated serum tumor necrosis factor (TNF) levels [2]. The ensuing decades of research have shed considerable insight into the underlying mechanisms which promote such immune dysregulation. Foremost among our understanding of HIV immunopathogenesis are the central concepts of an accelerated death of both HIV-infected and HIV-uninfected cells that occurs principally within lymphoid tissues at all stages of HIV disease, a shift in the cytokine and chemokine production of immune cells in HIV-infected patients and inappropriate and excessive polyclonal immune activation which exacerbates the already abnormal rates of T-cell death, proinflammatory cytokine production, HIV replication and T-cell and B-cell unresponsiveness.
We have recently described a novel pathway by which HIV infection can result in the death of an infected cell whereby HIV protease, which is active within the cytosolic fraction, can proteolytically cleave procaspase 8 producing a novel protein called Casp8p41 [3,4]. Casp8p41 production only occurs in HIV-infected but not in uninfected bystander cells [3]. Casp8p41 is biologically active and rapidly induces cell death via translocating to mitochondria and inducing depolarization, cytochrome C release and caspase 9 activity [5]. A separate unrelated activity of Casp8p41 is the ability to drive HIV replication via nuclear factor kappa B (NFκB)-mediated transactivation of the HIV LTR in a manner that does not depend on caspase activation, as it is not inhibited by Z-VAD (data not shown) [6]. As NFκB is pleiotropic and impacts both HIV replication as well as inflammation, we questioned whether NFκB activation by Casp8p41 would impact the transcriptome and cytokine production of cells in which it is present, resulting in a proinflammatory state.
Methods
Microarray analysis
Microarray analysis was conducted according to the manufacturer’s instructions for the Affymetrix One Cycle Target Labeling and Control Reagents kit (Santa Clara, California, USA). Briefly, cDNA was generated from 2 μg total RNA using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, California, USA) and T7 Oligo (dT) primer. Subsequently, the products were column-purified (Affymetrix) and then in-vitro transcribed to generate biotin-labeled cRNA. The in-vitro transcription and translation products were then column-purified, fragmented and hybridized onto Affymetrix U133 plus 2.0 GeneChip arrays at 45°C for 16 h. Subsequent to hybridization, the arrays were washed and stained with streptavidin-phycoerythrin, then scanned in an Affymetrix GeneChip Scanner 3000. All control parameters were confirmed to be within normal ranges before normalization and data reduction was initiated.
Flow cytometric determination of Casp8p41 and tumor necrosis factor content
For dual staining of intracellular Casp8p41 and TNF, primary human CD4+ T cells were phytohemoglutinin (PHA)/interleukin (IL)-2 stimulated and infected with HIV IIIB virus. On day 3 of infection, cells were harvested, fixed with 2% paraformaldehyde overnight, permeabilized with 0.1% NP40 for 2 min, then blocked at 4°C for 1 h with 2% fetal bovine serum (FBS) and 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS). Cells were stained with 0.25 μg/ml of fluorescein isothiocyanate (FITC) conjugated anti-Casp8p41 antibody [3] and 1 μg/ml of rabbit anti-TNF antibody (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) or matched isotype controls followed by goat antirabbit PE conjugated secondary antibody (Santa Cruz Biotechnology Inc.) for 1 h at 4°C. Cells were washed twice and analyzed by flow cytometry.
Multiplex cytokine assay
Cytokines were measured using a multiplexed microsphere cytokine assay (Human 27-Plex) as per the manufacturer’s direction (BioRad, San Diego, California, USA). Briefly, 100 μl of Bio-Plex assay buffer were added to each well of a MultiScreen MABVN 1.2 μm microfiltration plate followed by the addition of 50 μl of the multiplex bead preparation. Following washing of the beads with 100 μl of wash buffer, 50 μl of the samples (i.e. cell culture supernatants) or the standards were added to each well and incubated with shaking for 30 min at room temperature. Standard curves were generated with a mixture of cytokine standards and eight serial dilutions ranging from 0 to 32 000 pg/ml. The plate was washed three times followed by incubation of each well with 25 μl of premixed detection antibodies for 30 min with shaking. The plate was washed and 50 μl of streptavidin solution was added to each well and incubated for 10 min at room temperature with shaking. The beads were given a final washing and resuspended in 125 μl of Bio-Plex assay buffer. Cytokine levels in the supernatants were quantitated by analyzing 100 μl of each well on a Bio-Plex using Bio-Plex Manager software (version 4).
Cell culture
Using RossetteSep Human CD4 T-Cell Enrichment Cocktail (StemCell Technologies, Vancouver, British Columbia, Canada), CD4 T cells were isolated from the blood of healthy donors following informed consent. This protocol was reviewed and approved by the Mayo Institutional Review Board. Cells were either used directly or seeded at 2 × 106 cells/ml in Roswell Park Memorial Institute (RPMI) 1640 with 10% FBS (Atlanta Biologicals, Norcross, Georgia, USA), 2 mmol/l L-glutamine and 1 μg/ml PHA. Cells were used 48 h later.
Plasmids and transfections
Plasmids for Casp8p41 and mutations there of have been previously described [6]. CD4 T cells were transfected using the Human T-cell Nucleofector Kit as described in the manufacturer’s protocol (Amaxa, Walkersville, Mary-land, USA). Briefly, 107 cells were mixed with 5 μg highly purified plasmid DNA and 100 μl Human T-cell Nucleofector solution, transfected using program U-014 for fresh cells or T-023 for stimulated cells, transferred into prewarmed RPMI 10% FBS and incubated at 37°C. When measuring intracellular TNF levels, 10 μg/ml Brefeldin A (Sigma, St Louis, Missouri, USA) was added 1 h after transfection. At the specified times, cells were counted and centrifuged at 12 000 g for 10 min at 4°C. The resulting extracellular supernatant was used in the Human 27-Plex Cytokine Assay Panel (BioRad, Hercules, California, USA) as described below. The pelleted cells were checked for green fluorescent protein (GFP) expression or intracellular TNF expression by flow cytometry, or intracellular TNF expression by cellular lysis and western blot analysis.
Caspase 8-deficient cells
The caspase 8-deficient cell line was stably transfected with either wild-type caspase 8, or caspase 8 with FF:RN substitution at amino acids 355 and 356, as we have previously described [3,7]. HIV protease was cloned into the tetracycline responsive vector pTRE2hyg2-HA and the D25G mutant created by site directed mutagenesis, and sequences confirmed by sequencing.
Transfection of the I9.2 cells containing wild-type caspase 8 or caspase 8 FF:RN is accomplished using a square wave electrogenerator and the tetracycline responsive vector containing protease or protease D25G is co-transfected with the tetracycline responsive element (BD Biosciences, Franklin Lakes, New Jersey, USA) to induce expression. Twenty-four hours following transfection, cells are permealized with 0.1% NP40 and intracellularly stained for Casp8p41 and TNF-α, as described above, except using an Alexa-647 conjugated anti-Casp8p41 monoclonal antibody.
Cellular lysis and western blot analysis
To obtain total cellular proteins, cells were resuspended in 20 mmol/l Tris–HCl (pH 7.5) 150 mmol/l NaCl, 0.1% Triton X-100, 0.2% saponin, 2 μg/ml aprotinin, 10 μg/ml leupeptin, 2 μg/ml pepstatin and 1 mmol/l phenylmethanesulfonylfluoride, incubated on ice for 10 min and centrifuged at 12 000 g for 10 min at 4°C. The amount of cellular protein in the resultant supernatant was measured using the Bio-Rad protein assay.
For western blotting, equal amounts of protein were loaded and separated by 10% SDS-PAGE and transferred to Immobilon-P membranes (Millipore, Billerica, Massachusetts, USA). Immunoblotting was performed with anti-TNF-α (Santa Cruz Biotechnology, Santa Cruz, California, USA), anti-HA Peroxidase–High Affinity (eF10) (Roche, Indianapolis, Indiana, USA) and antiα-tubulin (Invitrogen, Eugene, Oregon, USA) and visualized using the HyGlo detection reagent (Denville Scientific Inc., Metuchen, New Jersey, USA).
Statistics
The transcriptome profiles of CD4 T cells transfected by Casp8p41, or procaspase 8, or empty transfection vectors were measured using Affymetrix HG-U133 plus 2.0 GeneChip arrays. The raw intensity files from microarray experiment were preprocessed using GCRMA’s background subtraction [8] and less normalization [9]. The expression values of each probe set were summarized from multiple probe pairs using median polish method. The absent, marginal and present calls for each probe set were calculated by dChip (http://biosun1.harvard.edu/complab/dchip/) using MAS 5.0 algorithm (Statistical Algorithms Description Document. 2002; Affymetrix).
The transcripts with ‘absent’ calls in all compared samples were excluded from further analysis. In addition, we consider transcripts whose expressions in all four samples below 50 percentile of all expression values were not expressed. Two samples transfected with Casp8p41 were compared to two control samples (empty vector and full-length procaspase 8) using t-test. The 13 666 transcripts that passed the two filtering steps described above were used in hierarchical clustering of the four samples (control, procaspase 8, 8p41 transfected for 2 and 4 h). The two samples with 8p41 transfection clustered together and away from control and procaspase 8 samples.
For cytokine analyses, normalization was performed using total cell counts. For the missing data beyond detection range, the 97.5 percentile of all observed values was used. For the missing data below detection sensitivity, the 2.5 percentile of all observed values was used. For each cytokine assay, the comparisons between GFP or HA tagged full-length procaspase 8 and GFP/HA, GFP or HA tagged 8p41 and GFP/HA, GFP or HA tagged 8p41 mutant and GFP/HA were performed using paired t-test, with correction for multiple comparisons.
Results
Casp8p41 expression alters the transcriptional profile of T cells
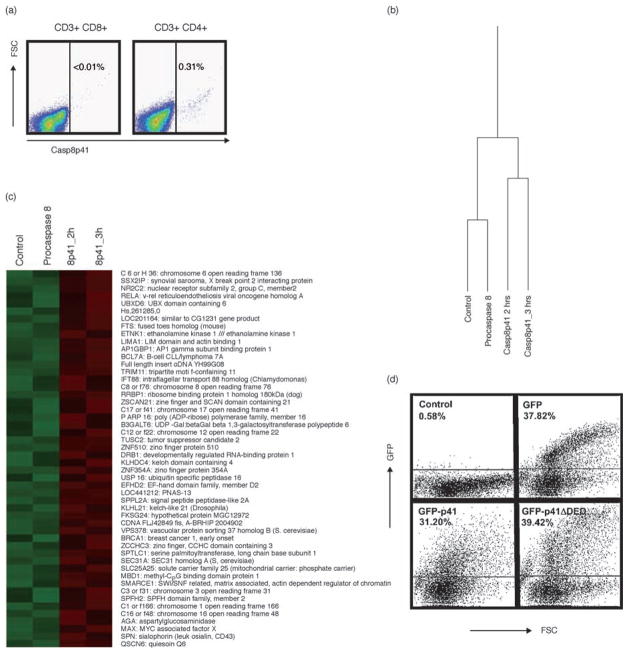
We have previously demonstrated that HIV protease can cleave procaspase 8 to produce Casp8p41 and that HIV-infected cells contain Casp8p41 [3,4]. We have observed in more than 50 patients infected with HIV that expression of Casp8p41 is restricted to the CD4 T-cell subset [3] as shown in this representative patient with suppressed viral replication (Fig. 1a). We next tested whether Casp8p41 expression alters the transcriptional profile of T cells. As Casp8p41 expression rapidly induces apoptosis and the molecular events of apoptosis include activation of nucleases, viable RNA was not available in cells in which apoptosis had begun. Therefore, instead, we chose a time point for microarray analysis which was before apoptosis was initiated, and therefore before executionary nucleases were activated and might limit our ability to detect newly produced transcripts. Jurkat T cells expressing Casp8p41 for 2 or 3 h were compared to cells expressing GFP empty vector and cells expressing GFP (full length) procaspase 8. The experiment was repeated three times. Of the more than 54 613 transcripts analyzed, 13 666 passed the noise filtering and were considered ‘expressed’. The transcript profile of these 13 666 transcripts for the cells expressing Casp8p41 were different from that for the cells expressing either full-length procaspase 8 or GFP empty vector control (Fig. 1b). The changes in transcription profile reached significance (P <0.05) in the case of 259 genes (Fig. 1c). The gene symbols and gene titles of the top 50 upregulated genes are displayed to the right of the figure. These genes clustered into the following families:
Fig. 1. Casp8p41 expression in T cells alters transcription.
(a) Peripheral blood mononuclear cell (PBMC) from an HIV-infected patient were analyzed by flow cytometry for CD3, CD4, CD8, CD27, CD45 and Casp8p41 (representative of >50 patients). (b) Affymetrix HG-U133 plus 2.0 GeneChip analysis of Jurkat T cells transfected with control vector, FL caspase 8, procaspase 8 or Casp8p41 for 2 or 3 h with 54 613 transcripts analyzed. After filtering, 13 666 transcripts were differentially regulated. Samples Casp8p41 (2 h) and Casp8p41 (3 h) were similar. Two hundred and fifty-nine transcripts were upregulated (P <0.05) in the Casp8p41 samples compared to control or procaspase 8, with the top 50 upregulated genes shown. Experiments were performed in triplicate (c). Primary CD4 T cells were transfected with green fluorescent protein (GFP) empty vector, GFP Casp8p41 or GFP Casp8p41ΔDED and analyzed for GFP content by flow cytometry to confirm fusion protein expression (d).
Transcription.
Spliceosome/RNA binding/RNA processing.
Endoplasmic reticulum/protein transport.
Regulation of apoptosis.
Protein modification.
Mitochondria.
Casp8p41 expression alters cytokine production of primary CD4 T cells
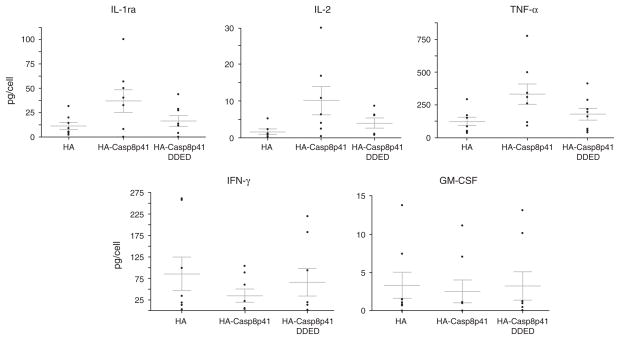
The transcriptional profiling of cells expressing Casp8p41 revealed significant increases the level of transcripts for genes affecting transcription. We, therefore, opted to assess whether cytokine production was different in cells expressing Casp8p41 compared with control cells. Analysis of cells 2 h after transfection did not cause any change in cytokine production. Therefore, unlike the microarray experiments in which we sampled cells before apoptosis began, in these experiments, we analyzed cytokine production 12 h after transfection. For this, we used primary CD4 T cells transfected with Casp8p41 and analyzed the production of the following cytokines: IL-1b, IL-1RA, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-15, IL-17, Eotaxin, FGF-basic, granulocyte colony-stimulating factor (G-CSF), granulocyte macrophage CSF (GM-CSF), interferon (IFN)-γ, IP-10, MCP-1, MIP-1a, MIP-1b, platelet-derived growth factor (PDGF)-BB, Rantes, TNF-α and VEGF using a multiplexed micro-sphere cytokine assay. We performed these experiments eight separate times with eight different healthy blood donors to ensure validity and consistency of results. As controls, we used empty vector, as well as Casp8p41 missing the amino terminal tandem death effector domain (Casp8p41ΔDED), which we have previously shown to be necessary for NFκB activation by Casp8p41 [6] and other groups have shown to be necessary for NFκB activation by c-FLIP [10]. First, we confirmed our ability to express GFP-Casp8p41 in primary CD4 T cells by flow cytometry (Fig. 1d). Most cytokines were unaffected by the expression of Casp8p41 (Table 1); however, production of IL-1RA, IL-2, IL-15 and TNF were significantly (P <0.05) increased in the cells expressing Casp8p41, whereas production of GM-CSF and IFN-γ were decreased in cells expressing Casp8p41 (Fig. 2).
Table 1.
Effect of Casp8p41 expression on selected cytokine production by primary CD4 T cells.
| Cytokine | HA | HACasp8p41 | HACasp8p41ΔDED | P |
|---|---|---|---|---|
| IL-1b | 0.110191 | 0.145075 | 0.13917596 | NS |
| IL-1RA | 11.30055 | 36.976 | 16.58867749 | 0.01 |
| IL-2 | 1.23846 | 8.822 | 2.997907704 | 0.05 |
| IL-4 | 0.096106 | 0.1244 | 0.09045644 | NS |
| IL-5 | 0.378586 | 0.149585 | 0.177616927 | NS |
| IL-6 | 1.08943 | 0.344145 | 0.637830346 | NS |
| IL-7 | 0.0250765 | 0.07095 | 0.03242824 | 0.08 |
| IL-8 | 47.34095 | 35.7715 | 61.4376208 | NS |
| IL-9 | 2.33828 | 2.49515 | 2.166054085 | NS |
| IL-10 | 18.5535 | 21.516 | 16.83512521 | NS |
| IL-12p70 | 0.59533 | 0.30721 | 0.394373528 | NS |
| IL-13 | 5.38405 | 1.9784 | 3.006429702 | NS |
| IL-15 | 0.03536 | 0.08541 | 0.051772573 | 0.03 |
| IL-17 | 5.98745 | 3.62965 | 4.95092198 | NS |
| Eotaxin | 0.2799795 | 0.47365 | 0.348581184 | NS |
| FGF-basic | 0.3101295 | 0.7034 | 0.306851611 | NS |
| GCFS | 0.2037355 | 0.42731 | 0.215285777 | NS |
| GM-CSF | 3.35514 | 2.5599 | 3.268170728 | 0.05 |
| IFN-γ | 85.9545 | 35.554 | 66.5147637 | 0.08 |
| IP-10 | 818.12 | 571.95 | 551.2636505 | NS |
| MCP-1 | 0.07225 | 0.117345 | 0.08004584 | NS |
| MIP-1a | 64.2785 | 119.045 | 70.45258705 | NS |
| MIP-1b | 160.4445 | 147.87 | 113.2748765 | NS |
| PDGF-BB | 1.2001 | 2.789 | 1.351000266 | NS |
| Rantes | 358.2155 | 328.055 | 249.9843041 | NS |
| TNF-α | 123.9105 | 331.24 | 178.5089302 | 0.005 |
| VEGF | 15.0265 | 7.2425 | 10.50087123 | NS |
Mean cytokine amounts (pg/cell) produced by cells expressing empty vector (HA), HACasp8p41 or Casp8p41 missing the tandem death effector domains (HACaspp8p41ΔDED) are shown.
Fig. 2. Casp8p41 expression in primary CD4 T cells alters cytokine production.
Primary CD4 T cells were transfected with either empty vector (HA), Casp8p41 (HACasp8p41) or with Casp8p41 missing the amino terminal tandem death effector domain (HACasp8p41ΔDED) and analyzed for cytokine production as indicated. Cytokines significantly upregulated or downregulated by Casp8p41 expression are shown. Each point is a separate blood donor.
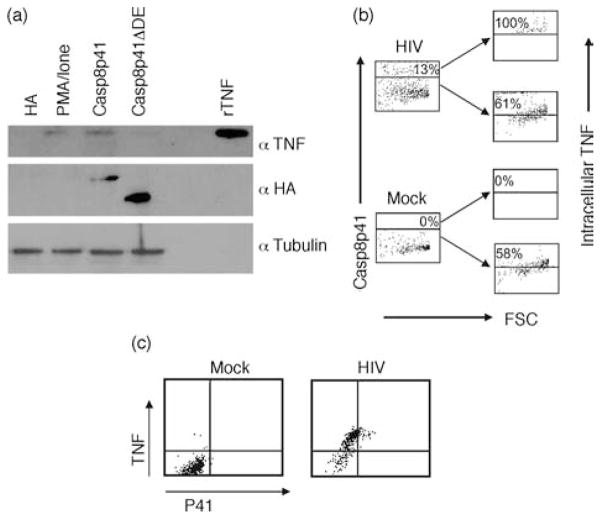
To confirm the cytokine production data, we next analyzed Casp8p41 expressing cells to confirm that these cells responsible for upregulated cytokine production. We chose TNF as a representative cytokine. The multiplex cytokine results showing increased levels of TNF were confirmed independently using western blot (Fig. 3a). These results were confirmed using intracellular flow cytometry for TNF, with primary CD4 T cells expressing HACasp8p41 containing significantly more TNF than primary CD4 T cells expressing either empty vector or HACasp8p41ΔDED.
Fig. 3. Casp8p41 expression increases tumor necrosis factor.
(a) Primary CD4 T cells were transfected with HA, HACasp8p41 or HACasp8p41ΔDED and analyzed by western blot for tumor necrosis factor (TNF) [recombinant TNF (rTNF)]. Control blotting with anti-HA and antitubulin was performed. (b) Primary human CD4 T cells were infected with HIVIIIb and 3 days after infection were analyzed for Casp8p41 and TNF by flow cytometry. Isotype control antibodies were used to determine TNF positivity. (c) The specificity of TNF production in Casp8p41-positive cells was assessed by simultaneous analysis of TNF and Casp8p41. Results are representative of four separate experiments.
Casp8p41 and tumor necrosis factor colocalize in HIV-infected cells
As Casp8p41 is present in a minority of HIV-infected cells, it represents an ideal situation to discriminate the impact of Casp8p41 expression on cytokine production in cells from the same donor. We, therefore, compared TNF content in cells that contained Casp8p41 and TNF content in cells negative for Casp8p41. For these experiments, purified primary CD4 T cells were infected with HIVIIIb and analyzed by intracellular flow cytometry for Casp8p41 and TNF. As expected, mock infected cells contained no Casp8p41. In the HIV-infected cells, Casp8p41-positive cells were compared to Casp8p41-negative cells, and consistently in four independent experiments, intracellular TNF content was higher in the former (Fig. 3b). Analyzing Casp8p41 and TNF simultaneously demonstrates no Casp8p41 and decreased TNF in mock-infected cells. In HIV-infected cells, all of the Casp8p41-positive cells express TNF, and interestingly a proportion of the Casp8p41-negative cells do as well (Fig. 3c). Whether the expression of TNF in these Casp8p41-negative cells is due to paracrine effects of TNF produced by the Casp8p41 co-expressing cells is worthy of further examination.
Enhanced tumor necrosis factor expression is specific to cells in which HIV protease has produced Casp8p41
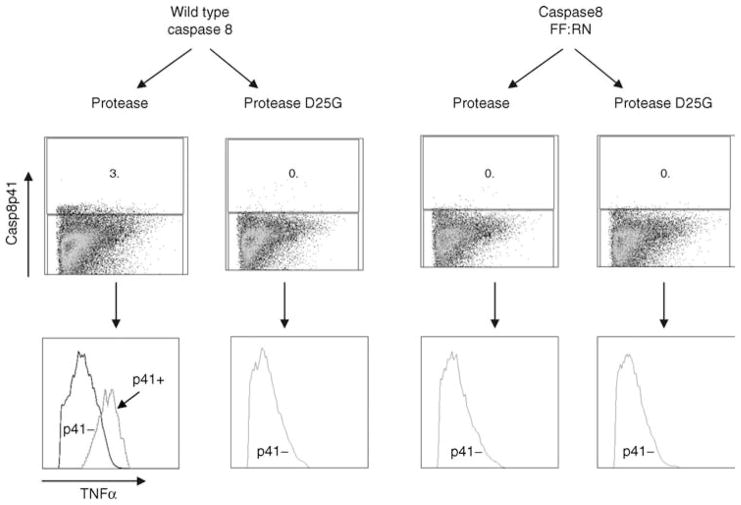
The above observation suggests that TNF expression is causatively related to production of Casp8p41. We next opted to specifically assess that association by using the caspase 8-deficient cell line I9.2 where we have stably reconstituted either wild-type, caspase 8, or caspase 8 with mutations at positions 355:356 which render procaspase 8 noncleavable by HIV protease [3,7]. Both of these transfected cell lines signal normally through caspase 8 [7]. We then transfected wild-type caspase 8-reconstituted I9.2 cells or I9.2 cells reconstituted with the noncleavable procaspase 8 (caspase 8 FF:RN) with either wild-type HIV protease or HIV protease with a D-to-G mutation at residue 25 rendering it catalytically inactive. As depicted in Fig. 4, when either I9.2 cell line was transfected with active site dead protease, minimal TNF production occurred. Similarly, I9.2 cells reconstituted with caspase 8 FF:RN and transfected with HIV protease did not result in Casp8p41 production and these cells produced very low levels of TNF. By contrast, I9.2 cells reconstituted with wild-type caspase 8 responded to HIV protease expression by producing Casp8p41. Analysis of TNF expression in these Casp8p41-containing cells compared with the Casp8p41-negative cells in the same transfection demonstrated a significantly increased production of TNF by the Casp8p41-positive cells. Altogether, therefore, these experiments demonstrate the specificity of TNF production in cells where HIV protease has resulted in Casp8p41 production (Fig. 4).
Fig. 4. Tumor necrosis factor production is specifically increased in cells where HIV protease has created Casp8p41.
The caspase 8-deficient cell line I9.2 was stably transfected with either wild-type caspase 8 or caspase 8 containing an FF:RN mutation at positions 355:356, which render it noncleavable by HIV protease. These cells were either transfected with HIV protease or with protease containing the active site dead mutation, D25G. Following transfection, the cells were analyzed for intracellular Casp8p41 content and for TNF by intracellular flow cytometry.
Discussion
Since apoptosis activates endogenous nucleases, we were unable to assess microarray changes after Casp8p41-induced apoptosis had begun. However, at an early time point of 2–3 h after transfection, significant changes in transcriptional profiles were present, notably in genes affecting transcription. At such an early time point, no changes in cytokine profile were observed; however, 12 h after transfection, significant proinflammatory changes were apparent in the Casp8p41-treated samples.
The particular cytokines which are altered following Casp8p41 expression are worth examining. TNF-α was the most consistently upregulated cytokine tested. Its ability to recruit monocytes, efficient replicators and reservoirs of the HIV virus to the site of infection may be advantageous for viral promotion. In addition, TNF is a powerful stimulator for NFκB activation and consequent HIV replication. The production of IL-2 and IL-15, whose principal roles are to promote proliferation and expansion of T cells, while promoting antiapoptotic regulatory molecules, may represent an attempt of a dying cell to maintain itself in the face of a potent apoptosis inducer such as Casp8p41. Such a teleologic argument might also apply to the downregulation of IFN-γ As IFN-γ is a potent antiviral defense, it would stand to reason that it would be in the interest of a virally infected cell to downregulate this cytokine in order to favor its own survival. Casp8p41-containing cells are, by definition, virally infected because Casp8p41 is produced only by HIV protease which is present only in HIV-infected cells [3,4]. Likewise, it would not favor an infected T-cell to produce factors which promote the expansion of granulocytes or macrophages; consequently, GM-CSF production is reduced. It is also worth noting that IL-2, IL-15 and TNF all promote HIV replication.
Much has been learned about the cytokine dysregulation that occurs during the progression of HIV disease, and it has been the subject of many excellent reviews [11]. It is notable that many cytokine alterations that occur in HIV-infected patients are mimicked by the cytokine alterations produced by Casp8p41-expressing cells. Indeed, elevated levels of TNF-α, IL-15 and IL-1RA are seen in both HIV-infected patients [2,12–14] and Casp8p41-expressing primary CD4 T cells. In addition, IFN-α is generally decreased in both infected patients [15] and Casp8p41-containing cells. Although IL-2 levels are generally decreased in HIV-infected patients, this does not occur until T-cell numbers (the only physiologic source of IL-2) are significantly low and the amount of IL-2 is tightly correlated with CD4 T-cell number [13,14]. On a per cell basis, T cells from HIV-infected patients produce normal or even elevated levels of IL-2 compared with controls [16]. Such findings are consistent with our observations that the rare Casp8p41-containing cells produce elevated levels of IL-2 compared with Casp8p41-negative uninfected cells.
The mechanisms of action attributed to Casp8p41 increase our understanding of apoptosis of HIV-infected CD4 T cells and the role of immune dysregulation on generation of anergy and death of uninfected CD4 T cells. The production of Casp8p41 is restricted to the HIV-infected T-cell subset and promotes mitochondrial-dependent death. In addition, it causes enhanced HIV replication and proinflammatory cytokine production which favor new rounds of viral infection, viral replication and immune activation.
Acknowledgments
A.D.B. is supported by the following grants: NIH R01AI62261. He has a patent filed on Casp8p41.
We would like to thank the Mayo Clinic Advanced Genomic Technology Center.
Footnotes
Author contributions: J.A.T., G.D.B., J.C.K., Y.W.A. and A.D.B. designed the research; J.A.T., G.D.B., N.W.C., S.A.R., C.P.K., M.D.B. and K.L.K. performed the research; J.C.K., Y.W.A. and A.D.B. analyzed the data; and A.D.B. wrote the manuscript.
There are no other conflicting or financial interests by the authors.
References
- 1.Gottlieb MS, Schroff R, Schanker HM, Weisman JD, Fan PT, Wolf RA, et al. Pneumocystis carinii pneumonia and mucosal candidiasis in previously healthy homosexual men: evidence of a new acquired cellular immunodeficiency. N Engl J Med. 1981;305:1425–1431. doi: 10.1056/NEJM198112103052401. [DOI] [PubMed] [Google Scholar]
- 2.Reddy MM, Sorrell SJ, Lange M, Grieco MH. Tumor necrosis factor and HIV P24 antigen levels in serum of HIV-infected populations. J Acquir Immune Defic Syndr. 1988;1:436–440. [PubMed] [Google Scholar]
- 3.Nie Z, Bren GD, Vlahakis SR, Schimnich AA, Brenchley JM, Trushin SA, et al. Human immunodeficiency virus type 1 protease cleaves procaspase 8 in vivo. J Virol. 2007;81:6947–6956. doi: 10.1128/JVI.02798-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nie Z, Phenix BN, Lum JJ, Alam A, Lynch DH, Beckett B, et al. HIV-1 protease processes procaspase 8 to cause mitochondrial release of cytochrome c, caspase cleavage and nuclear fragmentation. Cell Death Differ. 2002;9:1172–1184. doi: 10.1038/sj.cdd.4401094. [DOI] [PubMed] [Google Scholar]
- 5.Algeciras-Schimnich A, Belzacq-Casagrande AS, Bren GD, Nie Z, Taylor JA, Rizza SA, et al. Analysis of HIV protease killing through caspase 8 reveals a novel interaction between caspase 8 and mitochondria. Open Virol J. 2007;1:39–46. doi: 10.2174/1874357900701010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bren GD, Whitman J, Cummins N, Shepard B, Rizza SA, Trushin SA, et al. Infected cell killing by HIV-1 protease promotes NF-kappaB dependent HIV-1 replication. PLoS One. 2008;3:e2112. doi: 10.1371/journal.pone.0002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nie Z, Bren GD, Rizza SA, Badley AD. HIV protease cleavage of procaspase 8 is necessary for death of HIV-infected cells. Open Virol J. 2008;2:1–7. doi: 10.2174/1874357900802010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Z, Irizarry R, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 9.Ballman KV, Grill DE, Oberg AL, Therneau TM. Faster cyclic loess: normalizing RNA arrays via linear models. Bioinformatics. 2004;20:2778–2786. doi: 10.1093/bioinformatics/bth327. [DOI] [PubMed] [Google Scholar]
- 10.Golks A, Brenner D, Krammer PH, Lavrik IN. The c-FLIP-NH2 terminus (p22-FLIP) induces NF-kappaB activation. J Exp Med. 2006;203:1295–1305. doi: 10.1084/jem.20051556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kedzierska K, Crowe SM. Cytokines and HIV-1: interactions and clinical implications. Antivir Chem Chemother. 2001;12:133–150. doi: 10.1177/095632020101200301. [DOI] [PubMed] [Google Scholar]
- 12.Aukrust P, Muller F, Lien E, Nordoy I, Liabakk NB, Kvale D, et al. Tumor necrosis factor (TNF) system levels in human immuno-deficiency virus-infected patients during highly active antiretroviral therapy: persistent TNF activation is associated with virologic and immunologic treatment failure. J Infect Dis. 1999;179:74–82. doi: 10.1086/314572. [DOI] [PubMed] [Google Scholar]
- 13.Haas DW, Geraghty DE, Andersen J, Mar J, Motsinger AA, D’Aquila RT, et al. Immunogenetics of CD4 lymphocyte count recovery during antiretroviral therapy: an AIDS Clinical Trials Group study. J Infect Dis. 2006;194:1098–1107. doi: 10.1086/507313. [DOI] [PubMed] [Google Scholar]
- 14.Kirkpatrick CH, Davis KC, Horsburgh CR, Jr, Cohn DL, Penley K, Judson FN. Interleukin-2 production by persons with the generalized lymphadenopathy syndrome or the acquired immune deficiency syndrome. J Clin Immunol. 1985;5:31–37. doi: 10.1007/BF00915166. [DOI] [PubMed] [Google Scholar]
- 15.Meyaard L, Hovenkamp E, Keet IP, Hooibrink B, de Jong IH, Otto SA, et al. Single cell analysis of IL-4 and IFN-gamma production by T cells from HIV-infected individuals: decreased IFN-gamma in the presence of preserved IL-4 production. J Immunol. 1996;157:2712–2718. [PubMed] [Google Scholar]
- 16.Burkes RL, Abo W, Levine AM, Linker-Israeli M, Parker JW, Gill PS, et al. Characterization of immunologic function in homosexual men with persistent, generalized lymphadenopathy and acquired immune deficiency syndrome. Cancer. 1987;59:731–738. doi: 10.1002/1097-0142(19870215)59:4<731::aid-cncr2820590412>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]