Abstract
Excessive vascularization is a hallmark of many diseases including cancer, rheumatoid arthritis, diabetic nephropathy, pathologic obesity, age-related macular degeneration, and asthma. Compounds that inhibit angiogenesis represent potential therapeutics for many diseases. Karagiannis and Popel (PNAS, 2008) used a bioinformatics approach to idenify more than 100 peptides with sequence homology to known angiogenesis inhibitors. The peptides could be grouped into families by the conserved domain of the proteins they were derived from. The families included type IV collagen fibrils, CXC chemokine ligands, and type I thrombospondin domain-containing proteins. The relationships between these families have received relatively little attention. To investigate these relationships, we approached the problem by placing the families of proteins in the context of the human interactome including >120,000 physical interactions among proteins, genes, and transcripts. We built on a graph theoretic approach to identify proteins that may represent conduits of crosstalk between protein families. We validated these findings by statistical analysis and analysis of a time series gene expression dataset taken during angiogenesis. We identified six proteins at the center of the angiogenesis-associated network including three syndecans, MMP9, CD44 and versican. These findings shed light on the complex signaling networks that govern angiogenesis phenomena.
Introduction
Excessive vascularization is a hallmark of many diseases including cancer, rheumatoid arthritis, diabetic nephropathy, pathologic obesity, age-related macular degeneration, and asthma. Compounds that inhibit angiogenesis represent potential therapeutics for many diseases. Judah Folkman performed pioneering research in the field of angiogenesis;1 his work led to the identification of a number of proteins and polypeptides with anti-angiogenic activity.2
Karagiannis and Popel3 used a bioinformatics approach to group the peptides with anti-angiogenic activity into families by the conserved domain of the proteins they are derived from. The families included type IV collagens, CXC chemokines, and type I thrombospondin domain TSP1-containing proteins. Karagiannis and Popel identified conserved domains within each family by performing a multiple sequence alignment. They ran BLAST for each conserved domain against the proteome to identify other peptides with sequence homology. Their work revealed more than 100 peptides derived from over 80 proteins with sequence homology to known angiogenesis inhibitors. We will refer to this set of proteins throughout the rest of the article as angiogenesis-associated proteins. We extended the series of work from Karagiannis and Popel3 to investigate the collection of interactions surrounding the angiogenesis-associated proteins. In this study, we selected three families: type IV collagen, CXC chemokines and TSP1-containing proteins, for which we identified interactions with other proteins, thus building a protein-protein interaction (PPI) network. Note that the grouping of these angiogenesis-associated proteins into families only indicates that they share one or more conserved domains.
Karagiannis and Popel experimentally validated in vitro inibition of endothelial cell (EC) proliferation and migration by peptides derived from type IV collagens,4 thrombospondin domain-containing proteins,5,6 and CXC chemokines.7 These studies showed that a large fraction of the peptides had anti-angiogenic potential. Using EC proliferation assays, they also revealed synergy between the peptides derived from the CXC chemokines and TSP1-containing protein families,3 thus suggesting a possible crosstalk between the signaling networks. A greater understanding of the signaling pathways associated with the peptides is an important step in understanding their mechanisms of action. In vivo experiments with selected peptides demonstrated anti-angiogenic activity in tumor xenografts8 and ocular models.9
While the functional relationships between these protein families and angiogenesis have been catalogued by the gene ontology,10 the relationships between pairs of protein families are not well characterized. To better understand the relationships within and between type IV collagens, CXC chemokines, and TSP1-containing proteins, we placed each family of proteins in the context of the human interactome including 126,763 physical protein-protein, protein-DNA, or protein-RNA interactions accumulated in the Michigan Molecular Interactions database (MiMI).11 We used graph diffusion (see Methods) to identify those proteins that are in close topological proximity with multiple angiogenesis-associated protein families. The proteins that are well connected to multiple protein families represent potential mediators of crosstalk. We verified their statistical significance by repeatedly rewiring the human protein-protein interaction network. We found that many of these proteins had perturbed gene expression during time course measurements of VEGF-stimulated angiogenesis in endothelial cells.
Materials and Methods
Data sets
The interaction dataset was taken from the Michigan Molecular Interaction database (MiMI)11 (Feb 2009 version). The dataset is composed of 13,491 genes, proteins, and RNA connected by 126,763 physical interactions. The interaction types include protein-protein, protein-DNA, protein-RNA, and RNA-RNA. As a result, the dataset captures diverse aspects of biomolecular interactions including protein complexation, transcriptional regulation, and RNA interference. The dataset consists of interactions curated from reputable online databases such as Reactome,12 BIND, BioGrid,13 HPRD.14 This network of physical interactions forms the basis for crosstalk discovery. Gene Ontology (GO)10 annotations were used for verification (6/2010 version). For additional verification, we used a time series gene expression dataset of VEGF-induced capillary endothelial tube formation in a 3D collagen matrix in vitro.15 The dataset included 8 time points: 15 min, and 1, 3, 6, 9, 12, 18, and 24 h of VEGF stimulation.
Graph diffusion
By treating a biomolecular interaction network as a graph where nodes correspond to biomolecules and edges represent physical interactions between those biomolecules, we can efficiently find topological associations between protein families. Diffusion kernel algorithms have proven to be powerful tools for identifying topological associations between a node and a seed set of nodes. The method can be thought of in terms of repeated random walks originating at the seed nodes. A parameter γ controls the length of the random walks. A lower value for γ results in longer random walks. Nodes are then assigned a diffusion kernel score (DKS) based on the fraction of random walks that pass through the node. While many values of γ will suffice, we selected γ such that all nodes has some non-zero DKS.
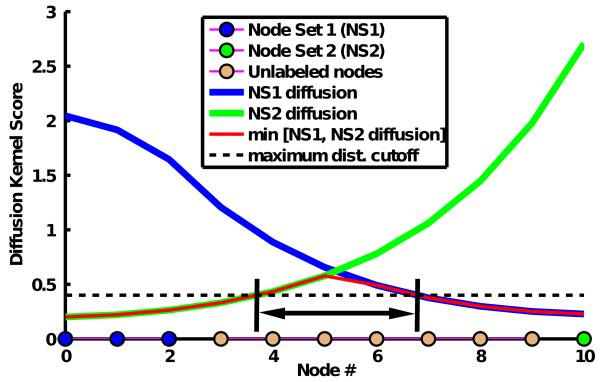
Figure 1 illustrates the principle of graph diffusion on a simple network consisting of a chain of 11 nodes connected by 10 edges. The nodes lie along the x-axis. Node set 1 (NS1) consists of nodes 1 through 3. The DKS of all the nodes with respect to NS1 is given by the blue line. Node set 2 (NS2) consists of only node 10 in green. The DKS with respect to NS2 is given by the green line. To identify the crosstalk proteins with respect to NS1 and NS2, we identify the intersection of the minimum of NS1 and NS2 diffusion (shown in red) and a minimum DKS threshold. The curves intersect before node 4 and after node 6. As a result, nodes 4, 5, and 6 would be labelled crosstalk nodes with respect to NS1 and NS2.
Figure 1. Graph diffusion on a simple linear network.
Graph diffusion on a network consisting of a chain of 11 nodes connected by 10 edges. The nodes lie along the x-axis. Node set 1 (NS1) consists of nodes 1 through 3. The DKS of all the nodes with respect to NS1 is given by the blue line. Node set 2 (NS2) consists of only node 10 in green. The DKS with respect to NS2 is given by the green line. To identify the crosstalk proteins with respect to NS1 and NS2, we identify the intersection of the minimum of NS1 and NS2 diffusion (shown in red) and a minimum DKS threshold (black line). The curves intersect before node 4 and after node 6. As a result, nodes 4, 5, and 6 would be labelled crosstalk nodes with respect to NS1 and NS2.
High confidence associations are established through multiple short length paths. Even if a single path is found to be incorrect, alternate paths through the network will still support the associations. This aggregation of evidence from multiple paths leads to a more stable result from potentially unreliable data. As the DKS is additive, we normalize the DKS by the number of query nodes. The software for performing this operation is provided through our website (sysbio.bme.jhu.edu).
For a weighted undirected graph G(V,E) with vertex set V and edge set E, let A be the symmetric adjacency matrix representing G. Let qi be 1 if node i is in the query set or zero otherwise. We express the time derivative of the diffusion kernel score si(q) for node i∈V as
| (1) |
Let D be the degree weighted diagonal matrix of A. In matrix notation, we have
| (2) |
Our goal is to identify the values of s at steady-state. We set and solve for s.
| (3) |
Crosstalk proteins
We define crosstalk proteins as topologically close to multiple node sets. A protein p is a crosstalk protein if the normalized DKS is greater than a threshold for multiple node sets. A maximum distance (minimum DKS) threshold φ marks the annotation boundary for a node set. The parameter φ is constant across all node sets. In this study, the parameter φ is set at 0.018. We use statistics to verify the significance of the results found using our fixed values of φ. We define a crosstalk protein p relative to node sets q1, q2, … , qn, if
| (4) |
We computed the normalized DKS for each protein to each node sets. We normalized by the number of proteins in the node set. In this way, DKS are comparable for different node set sizes. DKS results are given in Table 2. Then for a given protein, we could identify all potential crosstalk with other node sets.
Table 2. Normalized diffusion kernel scores (DKS) and statistical significance for proteins in Figure 3.
The proteins presented in this study are grouped by category and sorted by statistical significance. The normalized DKS for each protein is also given. The table shows the 126 proteins discussed in this study. The original 31 angiogenesis-associated proteins are indicated as seed proteins (p-value column). The statistical significance of the seed proteins is not evaluated. The additional 95 proteins are all statistically significant (p < 0.05) with respect to their category (e.g. CXC associated proteins, COL4 associated proteins, and CXC, COL4 crosstalk proteins). A p-value of 0.004 may indicate that a pseudocount was added to avoid a fitted probability of zero. The fourth column gives the trajectory of gene expression change during a time series of VEGF-induced angiogenesis. We measure the trajectory of gene expression change by the covariance between the gene expression and the time series. Missing or unreliable measurements were marked not available (na).
| Symbol | DKS | p-value | cov | Symbol | DKS | p-value | cov | Symbol | DKS | p-value | cov |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CXC, COL4, TSP1 crosstalk | RNF10 | 0.039 | 0.004 | 0.00 | LRP2BP | 0.047 | 0.004 | na | |||
| MMP9 | 0.134 | 0.004 | na | SAA1 | 0.09 | 0.004 | 0.14 | MAGI3 | 0.027 | 0.004 | −0.21 |
| SDC1 | 0.04 | 0.004 | −1.11 | SAA2 | 0.03 | 0.004 | na | MGEA5 | 0.041 | 0.004 | −1.21 |
| SDC2 | 0.048 | 0.004 | −1.80 | SERPINE2 | 0.167 | 0.004 | 0.00 | MMP13 | 0.097 | 0.004 | −0.44 |
| SDC4 | 0.051 | 0.004 | −6.19 | TGFBI | 0.096 | 0.004 | −0.25 | PACSIN3 | 0.037 | 0.004 | na |
| VCAN | 0.037 | 0.009 | na | USH2A | 0.064 | 0.004 | na | PTPN3 | 0.026 | 0.004 | −0.13 |
| CD44 | 0.022 | 0.013 | −0.74 | SAA4 | 0.038 | 0.009 | na | SCARB2 | 0.024 | 0.004 | 1.50 |
| COL4, TSP1 crosstalk proteins | COL4A1 | 0.486 | seed | 8.63 | SH3D19 | 0.06 | 0.004 | −0.08 | |||
| ACAN | 0.109 | 0.004 | na | COL4A2 | 0.528 | seed | 3.33 | SHANK2 | 0.033 | 0.004 | 0.00 |
| APP | 0.05 | 0.004 | 1.55 | COL4A3 | 0.496 | seed | na | SNX9 | 0.049 | 0.004 | 4.73 |
| BCAN | 0.045 | 0.004 | 0.00 | COL4A4 | 0.553 | seed | 0.00 | TLN1 | 0.034 | 0.004 | −1.22 |
| BGN | 0.053 | 0.004 | −0.18 | COL4A5 | 0.626 | seed | −2.81 | VEGFA | 0.042 | 0.004 | na |
| CD36 | 0.051 | 0.004 | 0.00 | COL4A6 | 0.601 | seed | na | VWF | 0.099 | 0.004 | 0.41 |
| DCN | 0.099 | 0.004 | na | CXC associated proteins | BAIAP2 | 0.026 | 0.009 | 0.70 | |||
| FBLN2 | 0.085 | 0.004 | 1.15 | IL8RA | 0.217 | 0.004 | na | CD47 | 0.026 | 0.009 | −0.66 |
| FN1 | 0.049 | 0.004 | −2.58 | IL8RB | 0.161 | 0.004 | na | FBLN1 | 0.027 | 0.009 | 0.00 |
| GP6 | 0.044 | 0.004 | na | CXCL1 | 0.716 | seed | −7.72 | ITGA9 | 0.048 | 0.009 | na |
| HSPG2 | 0.022 | 0.004 | 8.67 | CXCL3 | 0.844 | seed | 0.00 | LTBP1 | 0.035 | 0.009 | −0.33 |
| SDC3 | 0.05 | 0.004 | 1.52 | CXCL5 | 0.719 | seed | na | TNFAIP6 | 0.047 | 0.009 | na |
| THBS3 | 0.043 | 0.004 | 3.76 | CXCL6 | 0.78 | seed | na | UGCGL1 | 0.062 | 0.009 | −0.36 |
| MFAP2 | 0.038 | 0.009 | −0.03 | TSP1 associated proteins | ITGA3 | 0.022 | 0.013 | −3.94 | |||
| COL5A1 | 0.018 | 0.013 | −4.77 | AP3D1 | 0.025 | 0.004 | −0.79 | ITGA4 | 0.022 | 0.013 | −0.05 |
| COL7A1 | 0.021 | 0.017 | 0.15 | BAIAP3 | 0.159 | 0.004 | na | MAD2L2 | 0.026 | 0.013 | na |
| CXC, COL4 crosstalk proteins | C3 | 0.033 | 0.004 | na | MT2A | 0.03 | 0.013 | na | |||
| AGPAT5 | 0.041 | 0.004 | −0.29 | C5 | 0.039 | 0.004 | 0.51 | SHANK1 | 0.024 | 0.013 | na |
| ENO1 | 0.038 | 0.004 | −6.51 | C7 | 0.042 | 0.004 | 0.00 | ADAM12 | 0.575 | seed | na |
| IL8 | 0.03 | 0.004 | 0.00 | C8A | 0.044 | 0.004 | 1.98 | ADAM9 | 0.596 | seed | na |
| LDLR | 0.028 | 0.004 | −2.01 | C8B | 0.035 | 0.004 | 0.48 | ADAMTS1 | 0.738 | seed | −1.38 |
| SRGN | 0.036 | 0.004 | na | C8G | 0.035 | 0.004 | na | ADAMTS13 | 0.835 | seed | na |
| CXCR3 | 0.031 | 0.009 | na | C9 | 0.039 | 0.004 | na | ADAMTS4 | 0.652 | seed | na |
| CXCL2 | 0.783 | seed | −1.07 | CFB | 0.097 | 0.004 | na | ADAMTS5 | 0.757 | seed | na |
| PF4 | 0.608 | seed | na | SH3GL2 | 0.02 | 0.004 | −3.75 | BAI1 | 0.638 | seed | na |
| CXC, TSP1 crosstalk proteins | CORO1A | 0.019 | 0.004 | 0.06 | BAI2 | 0.632 | seed | na | |||
| CCL5 | 0.024 | 0.004 | 0.00 | EPB41L1 | 0.029 | 0.004 | −0.67 | C6 | 0.589 | seed | na |
| PROC | 0.074 | 0.004 | 0.00 | FSTL3 | 0.038 | 0.004 | −14.64 | CFP | 0.745 | seed | na |
| THBD | 0.032 | 0.004 | −3.74 | FURIN | 0.035 | 0.004 | na | CTGF | 0.678 | seed | −7.63 |
| COL4 associated proteins | GABPB2 | 0.026 | 0.004 | 2.77 | CYR61 | 0.861 | seed | −3.92 | |||
| ANTXR2 | 0.108 | 0.004 | 4.54 | HDHD2 | 0.029 | 0.004 | 0.95 | NOV | 0.678 | seed | na |
| BMP3 | 0.082 | 0.004 | na | HP | 0.044 | 0.004 | na | SPON1 | 0.907 | seed | −0.91 |
| CD93 | 0.086 | 0.004 | 5.19 | IGF2 | 0.025 | 0.004 | 2.73 | THBS1 | 0.414 | seed | −5.73 |
| COL4A3BP | 0.027 | 0.004 | −0.59 | IGF2R | 0.063 | 0.004 | 1.68 | THBS2 | 0.647 | seed | −0.44 |
| HABP2 | 0.151 | 0.004 | na | IGFBP5 | 0.028 | 0.004 | −3.30 | THSD1 | 0.909 | seed | na |
| MATN2 | 0.106 | 0.004 | na | ITGAV | 0.055 | 0.004 | 4.57 | WISP1 | 0.734 | seed | −0.39 |
| OSM | 0.137 | 0.004 | na | ITGB5 | 0.045 | 0.004 | −0.73 | WISP2 | 0.83 | seed | −2.46 |
Statistical significance
We computed the statistical significance of crosstalk proteins by permutation testing. We tested the null hypothesis that the DKS of a protein is equal to the DKS of a protein in rewired networks. The alternative hypothesis is that the DKS of a protein lower in rewired networks. To test these hypotheses, we generated 300 randomly edge swapped networks. The probability of the null hypothesis is given by the fraction of rewired networks where the DKS of the protein exceeds the DKS of the protein in the real network to a fixed set of seed proteins.
The statistical significance calculation controls for node set size and node degree. Crosstalk proteins can be compared and ranked based on their statistical significance. By computing statistical significance we eliminate a bias towards hub proteins in the network. The computation of statistical significance gives a global measure of the important of the associations that we identify through crosstalk proteins. We do not evaluate the statistical significance of the seed nodes (i.e. the angiogenesis-annotated proteins). Seed nodes were selected for the study and as such they are inherently biased.
Functional enrichment
We identified enriched functions for sets of crosstalk proteins using Ontologizer 2.0.16 Results are shown using default settings with Parent-Child-Union association and the Benjamini-Hochberg method of multiple hypothesis correction. The background set consisted of all human proteins in the interactome according to MiMI.17 All network images were produced using the Cytoscape18 network visualization software.
Results
We aimed to (i) identify proteins that may be mediators of crosstalk between angiogenesis-associated protein families and (ii) characterize their association with angiogenesis. We accomplished the first aim by application of graph diffusion on the human molecular interaction network followed by verification of statistical significance. We accomplished the second aim using a previously reported time series gene expression experimental dataset taken during angiogenesis.15
The search for angiogenesis-associated proteins and crosstalk
We used the human physical interactome as a basis for the analysis. We used a graph theoretic technique called graph diffusion to quantify the distance between proteins in the interactome19 (see Methods). The graph diffusion method also known as the diffusion kernel allowed us to quantify the distance between a single protein and a protein family. We referred to the distance between a protein and a protein family as the diffusion kernel score (DKS). A protein with a high DKS interacts closely with the protein family. For example, consider the family of type IV collagen fibrils, a protein that physically interacts with all type IV collagens would receive a high DKS, while a protein that only indirectly interacts with type IV collagens would receive a relatively lower DKS. We use the DKS to estimate the association between a single protein and a family of proteins.
To locate those proteins that potentially mediate crosstalk between families, we define crosstalk proteins that are highly associated with multiple protein families (i.e. the proteins have a DKS which is greater than a threshold for multiple families). For example, a crosstalk protein for type IV collagens and CXC chemokines would have many direct and indirect interactions with both protein families. We evaluate the statistical significance of a crosstalk protein by considering hundreds of rewired networks. We create each rewired network by repeatedly swapping interactions. The statistical test that we use for crosstalk proteins controls for the size of the protein families and the degree of protein interaction.
Using this approach, we found 126 proteins that were topologically close to the angiogenesis-associated protein families. We evaluated the quality of the protein annotations by their statistical significance and functional enrichment in angiogenesis. To put this network in context with the rest of the known human interactome, these are less than 1% of proteins (i.e. 0.93%) and interactions (0.25%). The analysis pointed to many proteins whose role in angiogenesis is well known, which serves as a validation of the approach. There are 194 human proteins that have angiogenesis as part of a GO annotation (as of 6/2010). The likelihood that a protein is annotated with angiogenesis by chance is 0.014. Excluding 31 seed proteins, our analysis of the human protein-protein interaction network identifies 4 proteins that have angiogenesis as part of their GO annotation. The probability that the 95 (i.e. 126 associated − 31 seeds) proteins contained 4 angiogenesis annotated proteins by chance is 0.045. We calculated the p-value using Fisher’s exact test. Our analysis suggests new or understudied modulators of angiogenesis. These centrally located proteins may be attractive targets due to their potential to minipulate multiple protein families.
In Figure 2, we show a Venn diagram to illustrate the associations of the 126 proteins. These proteins are topologically close to the type IV collagens, CXC chemokines or TSP1-containing proteins or some combination of families, as indicated by the figure. The figure gives the putative crosstalk between three angiogenesis-associated protein families: type IV collagens (blue), CXC chemokines (red), and TSP1-containing proteins (green). The crosstalk proteins are shown for CXC chemokines and type IV collagens (purple), CXC chemokines and TSP1-containing proteins (tan), type IV collagens and TSP1-containing proteins (yellow), and between all three (orange). The number of angiogenesis-associated proteins is shown in parentheses. In the results, we focus on the proteins associated with multiple families. First, we discuss crosstalk proteins between type IV collagen and TSP1-containing proteins. Then, we highlight six proteins identified as crosstalk proteins between all three families. These six proteins: three syndecans, MMP9, CD44 and versican may be important mediators of crosstalk for these angiogenesis-associated protein families.
Figure 2. Venn diagram of the putative crosstalk.
We show the associations of 126 proteins that are topologically close to type IV collagens, CXC chemokines, TSP1-containing proteins, or some combination, as indicated by the Venn diagram. A Venn diagram between three angiogenesis-associated protein families: type IV collagens (blue), CXC chemokines (red), and TSP1-containing proteins (green). The crosstalk proteins are shown for CXC chemokines and type IV collagens (purple), CXC chemokines and TSP1-containing proteins (tan), type IV collagens and TSP1-containing proteins (yellow), and between all three (orange). While crosstalk proteins may not be directly part of multiple protein families, they are in close topological proximity to multiple protein families. The number of angiogenesis-annotated proteins (i.e. seed proteins) is shown in parentheses.
Method for comparison of topology-based annotation
Crosstalk between pathways is an important concept in biology. There have been both computational20 and experimental21 efforts to identify crosstalk between pathways. Some of these approaches are not suitable in this context because they rely on overlapping pathways to identify crosstalk. Alternate approaches might consider “first neighbors” or “second neighbors” to identify association between pathways or modules. These rigid approaches have the inherent disadvantage of being unable to identify crosstalk between modules of distance 2 for “first neighbors” or distance 3 for “second neighbors. Other studies used shortest paths to help define crosstalk proteins.20,22 These methods borrow from concepts such as betweenness centrality. Because graph diffusion considers all paths, our method has inherent advantages over those that only consider shortest paths between proteins.
To motivate the use of the graph diffusion method, we performed a systematic comparison of three alternative methods in a head-to-head comparison with graph diffusion. The we compared graph diffusion with methods based on first neighbors, second neighbors, and betweenness centrality. In Table 1, we show the results of this comparison. We found that graph diffusion identified more statistically significant proteins at both the 0.01 and 0.05 levels. The graph diffusion method identified a more functionally cohesive set of proteins as demonstrated by the number of GO term enrichments at the 0.001 and 0.0001 levels.
Table 1. Head-to-head comparison of topological annotation methods.
We compared four methods including graph diffusion, betweenness centrality, first neighbors, and second neighbors. The table shows the number of statistically significant crosstalk proteins at the 0.05 and 0.01 levels. We show the number of significant proteins excluding seed proteins. Seed proteins are excluded because they were selected for this study. We show the significance of the enrichment in angiogenesis terms for the crosstalk proteins found using each method. The table also shows the number of GO term enrichments at the 0.01, 0.001, and 0.0001 levels.
| Comparison Type | Graph Diffusion |
Betweenness Centrality |
First Neighbors |
Second Neighbors |
|---|---|---|---|---|
| Proteins (p < 0.05) out of 13491 | 877 | 145 | 143 | 66 |
| Proteins (p < 0.01) | 398 | 87 | 69 | 44 |
| Proteins (p < 0.01) & seed (out of 31) | 10 | 5 | 7 | 10 |
| GO enrichments (p < 0.01 ) | 117 | 90 | 57 | 40 |
| GO enrichments (p < 0.001) | 85 | 49 | 34 | 34 |
| GO enrichments (p < 0.0001) | 65 | 36 | 22 | 28 |
Gene expression validates crosstalk proteins as angiogenesis-associated
To further validate the role of the crosstalk proteins in angiogenesis we reanalysed a time series gene expression dataset taken during VEGF-induced angiogenesis. We expected that many crosstalk proteins would have perturbed gene expression during angiogenesis. If this proved to be the case, the microarray dataset would provide additional evidence of the role of crosstalk proteins in angiogenesis.
A research team led by Claesson-Welsh took measurements from a gene expression time series of VEGF-induced capillary endothelial tube formation in a 3D collagen matrix in vitro.15 The dataset included 8 time points: 15 min, and 1, 3, 6, 9, 12, 18, and 24 h of VEGF stimulation. We reanalysed these data to identify the transcription profiles that are significantly increasing or decreasing during tube formation (that we refer to as angiogenesis). To accomplish this, we ranked transcripts by the absolute value of the covariance between the transcript measurements and the time points. We tested the null hypothesis that the crosstalk proteins are uniformly distributed among the ranked list of genes. We computed the family-wise error rate (FWER) p-value using gene set enrichment analysis23 which is based the Kolmogorov-Smirnov test followed by permutation testing. We found the crosstalk proteins significantly enriched at the head of the ranked list of perturbed genes (p=3·10−4). In Table 2, we give the trajectory of gene expression during VEGF-induced angiogenesis. We measure the trajectory of gene expression change by the covariance between the gene expression and the time points. The statistical test indicates that many of the crosstalk proteins have either increasing or decreasing gene expression during angiogenesis. This analysis helped confirm the importance of these crosstalk proteins in VEGF-induced angiogenesis and serves as a validation of our bioinformatics analysis.
Putative crosstalk between type IV collagens and TSP1-containing proteins
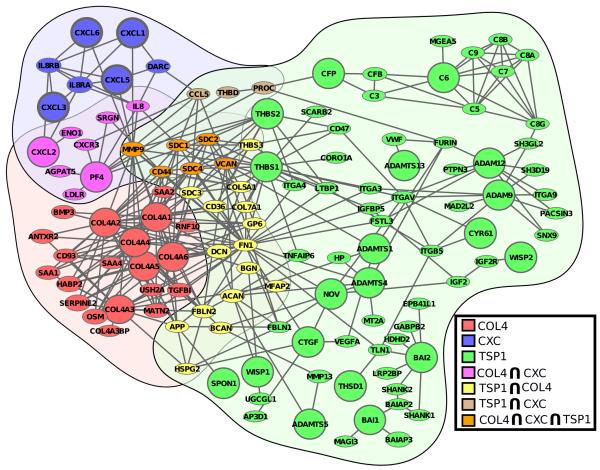
We studied the association between type IV collagens and TSP1-containing proteins to reveal the mediators of crosstalk between these two families. In Figure 3, the crosstalk proteins between type IV collagens and TSP1-containing proteins are highlighted in yellow. A significant number of these proteins bind collagen and associate with the vesicle lumen (Table 3). CD36 is also known to interact with type IV collagens.24 The identification of CD36 as a crosstalk protein for type IV collagens and TSP1-containing proteins helps confirm our approach. Decorin (DCN) is another proteoglycan that we identify as a crosstalk protein. Decorin interacts with collagens and extracellular matrix (ECM) and promotes angiogenesis.25 Fibronectin 1 (FN1) is an important connective molecule in the extracellular space. FN1 has domains for collagens, fibulin 1, heparin, and syndecan binding.26 We identify FN1 as a crosstalk protein between type IV collagens and TSP1-containing proteins. FN1 connects extracellular collagens with membrane-bound integrins (Figure 3). As such, FN1 has a central role in endothelial cell adhesion to the ECM. Another important conduit of information between TSP1-containing proteins and type IV collagens is through aggrecan (ACAN) and brevican (BCAN) through fibulin 2 (FBLN2).27,28 The crosstalk between type IV collagens and TSP1-containing proteins through ACAN, BCAN, and FBLN2 has not been reported in the context of angiogenesis, although it is known that FBLN2 inhibits tumor angiogenesis.28 The crosstalk between type IV collagens and TSP1-containing proteins may be significantly influenced by fibronectin 1, aggrecan, brevican, and fibulin 2. The amyloid beta (A4) precursor protein (APP) is also annotated as a crosstalk protein between type IV collagen and TSP1-containing proteins. Figure 3 shows the direct interaction between APP and COL4A1, COL4A2, COL4A5, COL4A6 and TSP1-containing spondin 1 (SPON1). APP is known to be associated with Alzheimer’s disease.29 It is also known that Alzheimer’s disease is related to angiogenesis.30 This study suggests angiogenesis might influence Alzheimer’s disease through the association between APP and type IV collagens and TSP1-containing proteins.
Figure 3. Network of association between type IV collagens, CXC chemokines and TSP1-containing proteins.
The network of association between three angiogenesis-associated protein families: type IV collagens (blue), CXC chemokines (red), and TSP1-containing proteins (green). The crosstalk proteins are shown for CXC chemokines and type IV collagens (purple), CXC chemokines and TSP1-containing proteins (tan), type IV collagens and TSP1-containing proteins (yellow), and between all three (orange).
Table 3. Crosstalk protein functional enrichment.
Interesting and statistically significant functional enrichments from the sets of crosstalk proteins. (A) Functions of crosstalk proteins from TSP1-containing proteins, CXC chemokines, and type IV collagens. (B) Functions of crosstalk proteins from TSP1-containing proteins and type IV collagens. (C) Functions of crosstalk proteins from CXC chemokines and type IV collagens.
| Annotation | Adj p-value | Type | Genes |
|---|---|---|---|
| (A) TSP1, CXC, and COL4 Crosstalk Protein Enrichment | |||
| cell surface | 0.0103 | component | CD44, SDC1, SDC4 |
| collagen binding | 0.0273 | function | CD44, MMP9 |
| (B) COL4 and TSP1 Crosstalk Protein Enrichment | |||
| vesicle lumen | 0.0046 | component | APP, FN1, THBS3 |
| peptide cross-linking | 0.00109 | process | BGN, FN1, DCN |
| collagen binding | 0.00072 | function | FN1, GP6, DCN |
| thrombospondin receptor activity | 0.0952 | function | CD36 |
| (C) COL4 and CXC Crosstalk Protein Enrichment | |||
| locomotion | 0.0274 | process | CXCL2, CXCR3, IL8, PF4 |
| cytoplasmic vesicle | 0.0288 | component | LDLR, PF4, SRGN |
| G-protein receptor binding | 0.0967 | function | CXCL2, IL8, PF4 |
Putative crosstalk between type IV collagens, CXC chemokines and TSP1-containing proteins
We were also interested in identifying the potential avenues of crosstalk between type IV collagens, CXC chemokines, and TSP1-containing proteins. We identified six proteins that are well connected to all three families of angiogenesis-associated proteins. In Figure 3, we show the crosstalk proteins between all three families in orange. A significant number of these proteins bind collagen and are localized on the cell surface (Table 3). MMP9 was identified as a crosstalk protein between the three families of angiogenesis-associated proteins. MMP9 is known to degrade type IV collagens31 and CXC chemokines like PF4.32 Thrombospondins are known to regulate the amount of MMP9.33 These functions outline the pivotal role of MMP9 in association with angiogenesis. Although MMP9 degrades many proteins, the interaction between MMP9 and the angiogenesis-associated protein families is highly significant (Table 2, p=0.004).
Our work highlights syndecan 1 (SDC1), syndecan 2 (SDC2), syndecan 4 (SDC4) at the centre of crosstalk between type IV collagens, CXC chemokines, and TSP1-containing proteins. Syndecans have been previously implicated in angiogenesis.34 Endothelial CD44 plays an important role in tube formation during angiogenesis.35 Our study suggests that CD44 may operate as a mediator of crosstalk between type IV collagens, CXC chemokines, and TSP1-containing proteins. Note that WISP-1, a TSP1-containing protein, is connected to the type IV collagen family through Bone Morphogenetic Protein 3 (BMP-3). An anti-angiogenic peptide derived from WISP-1 with relatively low anti-proliferative and anti-migratory in vitro activity identified in,5 showed a significant in vivo activity in corneal and laser-induced choroidal neovascularization mouse models.9
Versican (VCAN) is the last protein in the set of centrally located proteins. VCAN is involved in the attachment of endothelial cells to the extracellular matrix. The importance of VCAN in angiogenesis could easily be missed by other methods that only consider the direct interactions. VCAN has only a few physical protein-protein interactions, and it has only one direct interaction with the angiogenesis-associated proteins (i.e. ADAMTS1). Still, our analysis highlights VCAN as a potential component of crosstalk between type IV collagens, CXC chemokines, and TSP1-containing proteins. Using the quantitative comparison shown in Table 1, we confirmed that local approaches like first neighbors (p=0.046) and second neighbors (p=0.11) would have missed VCAN, while non-local approaches like graph diffusion (p=0.008) and betweenness centrality (p=0.006) would have identified the significance of VCAN at the 0.01 level. We identify six proteins at the center of the type IV collagen, CXC chemokine, and TSP1-containing protein network. These proteins, SDC1, SDC2, SDC4, MMP9, CD44, and VCAN, appear to be important components of angiogenesis, based on their position within the angiogenesis-associated network.
Discussion
Figure 3 reflects the three families of angiogenesis-associated proteins and the putative crosstalk identified between each family. We identified proteins that either directly or indirectly interact with many of proteins from individual families. The association of proteins to angiogenesis-associated families was computed using graph diffusion. By identifying proteins that are well connected to multiple protein families, we identified proteins that are likely to represent conduits of crosstalk between these important angiogenesis-associated families. Statistical analysis and the incorporation of a time series gene expression dataset helped confirm the role of these proteins in angiogenesis.
In our study of type IV collagens, CXC chemokines, and TSP1-containing proteins, we identified many classes of proteins that are known to be associated with angiogenesis such as vascular endothelial growth factor A (VEGFA) as well as other families that receive less attention such as proteoglycans decorin (DCN), aggrecan (ACAN), brevican (BCAN), and versican (VCAN). We also identified six proteins that appear to be at the center of the network between type IV collagens, CXC chemokines, and TSP1-containing proteins. Those proteins are syndecan 1 (SDC1), syndecan 2 (SDC2), syndecan 4 (SDC4), versican (VCAN), CD44, and matrix metalloproteinase 9 (MMP9). These proteins may facilitate crosstalk between type IV collagens, CXC chemokines, and TSP1-containing proteins.
We examined protein-protein interactions (PPI) that are related to three angiogenesis-associated protein families: type IV collagen fibrils, CXC chemokine ligands and TSP-1 domain-containing proteins. To our knowledge, this work represents the first integrated network analysis of these angiogenesis-associated protein families. We identified several proteins that appear to be important mediators of crosstalk, and yet they have received relatively little attention such as the proteoglycans decorin (DCN), aggrecan (ACAN), brevican (BCAN), and versican (VCAN). We identified syndecans at the centre of the network associating type IV collagens, CXC chemokines, and TSP1-containing proteins.
Acknowledgements
The work was supported by NIH grants R01 HL101200 and R01 CA138264. The authors would like to thank Emmanouil Karagiannis for helpful discussions at the initial stage of the project. We would also like to thank Sofie Mellberg and Lena Claesson-Welsh for use of their time series gene expression dataset.
Footnotes
Competing interests The authors declare no competing interests.
Authors’ contributions CGR implemented the method, performed the analysis, generated the images and wrote the paper. ASP and JSB designed the study and edited the paper.
References
- 1.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285(21):1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. Angiogenesis: an organizing principle for drug discovery? Nat Rev Drug Discov. 2007;6(4):273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 3.Karagiannis ED, Popel AS. A systematic methodology for proteome-wide identification of peptides inhibiting the proliferation and migration of endothelial cells. Proc Natl Acad Sci U S A. 2008;105(37):13775–13780. doi: 10.1073/pnas.0803241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karagiannis ED, Popel AS. A theoretical model of type I collagen proteolysis by matrix metalloproteinase (MMP) 2 and membrane type 1 MMP in the presence of tissue inhibitor of metalloproteinase 2. J Biol Chem. 2004;279(37):39105–39114. doi: 10.1074/jbc.M403627200. [DOI] [PubMed] [Google Scholar]
- 5.Karagiannis ED, Popel AS. Anti-angiogenic peptides identified in thrombospondin type I domains. Biochem Biophys Res Commun. 2007;359(1):63–69. doi: 10.1016/j.bbrc.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karagiannis ED, Popel AS. Peptides derived from type I thrombospondin repeat-containing proteins of the CCN family inhibit proliferation and migration of endothelial cells. Int J Biochem Cell Biol. 2007;39(12):2314–2323. doi: 10.1016/j.biocel.2007.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karagiannis ED, Popel AS. Novel anti-angiogenic peptides derived from ELR-containing CXC chemokines. J Cell Biochem. 2008;104(4):1356–1363. doi: 10.1002/jcb.21712. [DOI] [PubMed] [Google Scholar]
- 8.Koskimaki JE, Karagiannis ED, Rosca EV, et al. Peptides derived from type IV collagen, CXC chemokines, and thrombospondin-1 domain-containing proteins inhibit neovascularization and suppress tumor growth in MDA-MB-231 breast cancer xenografts. Neoplasia. 2009;11(12):1285–1291. doi: 10.1593/neo.09620. [DOI] [PMC free article] [PubMed] [Google Scholar]; Koskimaki JE, Karagiannis ED, Tang BC, et al. Pentastatin-1, a collagen IV derived 20-mer peptide, suppresses tumor growth in a small cell lung cancer xenograft model. BMC Cancer. 10:29. doi: 10.1186/1471-2407-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cano Mdel V, Karagiannis ED, Soliman M, et al. A peptide derived from type 1 thrombospondin repeat-containing protein WISP-1 inhibits corneal and choroidal neovascularization. Invest Ophthalmol Vis Sci. 2009;50(8):3840–3845. doi: 10.1167/iovs.08-2607. [DOI] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayapandian M, Chapman A, Tarcea VG, et al. Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic Acids Res. 2007;35(Database issue):D566–571. doi: 10.1093/nar/gkl859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vastrik I, D’Eustachio P, Schmidt E, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8(3):R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breitkreutz BJ, Stark C, Reguly T, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36(Database issue):D637–640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasad T. S. Keshava, Goel R, Kandasamy K, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mellberg S, Dimberg A, Bahram F, et al. Transcriptional profiling reveals a critical role for tyrosine phosphatase VE-PTP in regulation of VEGFR2 activity and endothelial cell morphogenesis. FASEB J. 2009;23(5):1490–1502. doi: 10.1096/fj.08-123810. [DOI] [PubMed] [Google Scholar]
- 16.Bauer S, Grossmann S, Vingron M, et al. Ontologizer 2.0--a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics. 2008;24(14):1650–1651. doi: 10.1093/bioinformatics/btn250. [DOI] [PubMed] [Google Scholar]
- 17.Tarcea VG, Weymouth T, Ade A, et al. Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic Acids Res. 2009;37(Database issue):D642–646. doi: 10.1093/nar/gkn722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Killcoyne S, Carter GW, Smith J, et al. Cytoscape: a community-based framework for network modeling. Methods Mol Biol. 2009;563:219–239. doi: 10.1007/978-1-60761-175-2_12. [DOI] [PubMed] [Google Scholar]
- 19.Tsuda K, Noble WS. Learning kernels from biological networks by maximizing entropy. Bioinformatics. 2004;20(Suppl 1):i326–333. doi: 10.1093/bioinformatics/bth906. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Agarwal P, Rajagopalan D. A global pathway crosstalk network. Bioinformatics. 2008;24(12):1442–1447. doi: 10.1093/bioinformatics/btn200. [DOI] [PubMed] [Google Scholar]
- 21.Natarajan M, Lin KM, Hsueh RC, et al. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8(6):571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 22.Zielinski R, Przytycki PF, Zheng J, et al. The crosstalk between EGF, IGF, and Insulin cell signaling pathways--computational and experimental analysis. BMC Syst Biol. 2009;3:88. doi: 10.1186/1752-0509-3-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo HJ, Maslen CL, Keene DR, et al. Type VI collagen anchors endothelial basement membranes by interacting with type IV collagen. J Biol Chem. 1997;272(42):26522–26529. doi: 10.1074/jbc.272.42.26522. [DOI] [PubMed] [Google Scholar]
- 25.Schonherr E, Sunderkotter C, Schaefer L, et al. Decorin deficiency leads to impaired angiogenesis in injured mouse cornea. J Vasc Res. 2004;41(6):499–508. doi: 10.1159/000081806. [DOI] [PubMed] [Google Scholar]
- 26.Ruoslahti E. Fibronectin. J Oral Pathol. 1981;10(1):3–13. doi: 10.1111/j.1600-0714.1981.tb01242.x. [DOI] [PubMed] [Google Scholar]
- 27.Silva R, D’Amico G, Hodivala-Dilke KM, et al. Integrins: the keys to unlocking angiogenesis. Arterioscler Thromb Vasc Biol. 2008;28(10):1703–1713. doi: 10.1161/ATVBAHA.108.172015. [DOI] [PubMed] [Google Scholar]
- 28.Albig AR, Neil JR, Schiemann WP. Fibulins 3 and 5 antagonize tumor angiogenesis in vivo. Cancer Res. 2006;66(5):2621–2629. doi: 10.1158/0008-5472.CAN-04-4096. [DOI] [PubMed] [Google Scholar]
- 29.McCarty MF. Toward prevention of Alzheimers disease--potential nutraceutical strategies for suppressing the production of amyloid beta peptides. Med Hypotheses. 2006;67(4):682–697. doi: 10.1016/j.mehy.2006.04.067. [DOI] [PubMed] [Google Scholar]; Suo Z, Humphrey J, Kundtz A, et al. Soluble Alzheimers beta-amyloid constricts the cerebral vasculature in vivo. Neurosci Lett. 1998;257(2):77–80. doi: 10.1016/s0304-3940(98)00814-3. [DOI] [PubMed] [Google Scholar]
- 30.Tony JC. Alzheimer’s disease and angiogenesis. Lancet. 2003;361(9365):1300. doi: 10.1016/S0140-6736(03)13002-4. [DOI] [PubMed] [Google Scholar]
- 31.Gioia M, Monaco S, Van Den Steen PE, et al. The collagen binding domain of gelatinase A modulates degradation of collagen IV by gelatinase B. J Mol Biol. 2009;386(2):419–434. doi: 10.1016/j.jmb.2008.12.021. [DOI] [PubMed] [Google Scholar]; Mira E, Lacalle RA, Buesa JM, et al. Secreted MMP9 promotes angiogenesis more efficiently than constitutive active MMP9 bound to the tumor cell surface. J Cell Sci. 2004;117(Pt 9):1847–1857. doi: 10.1242/jcs.01035. [DOI] [PubMed] [Google Scholar]
- 32.Opdenakker G, Van den Steen PE, Dubois B, et al. Gelatinase B functions as regulator and effector in leukocyte biology. J Leukoc Biol. 2001;69(6):851–859. [PubMed] [Google Scholar]
- 33.Nagase H, Woessner JF., Jr. Matrix metalloproteinases. J Biol Chem. 1999;274(31):21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 34.Tkachenko E, Rhodes JM, Simons M. Syndecans: new kids on the signaling block. Circ Res. 2005;96(5):488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- 35.Cao G, Savani RC, Fehrenbach M, et al. Involvement of endothelial CD44 during in vivo angiogenesis. Am J Pathol. 2006;169(1):325–336. doi: 10.2353/ajpath.2006.060206. [DOI] [PMC free article] [PubMed] [Google Scholar]