Abstract
We describe an HIV-infected patient with pre-treatment resistance to raltegravir, nucleoside reverse transcriptase inhibitors, non-nucleoside reverse transcriptase inhibitors, and protease inhibitors, and the ultimate ability to achieve viral suppression. Pre-treatment integrase resistance testing is not routinely performed because transmitted integrase mutations conferring resistance to raltegravir are currently thought to be negligible. We suggest obtaining a pre-treatment integrase genotype in patients with transmitted multiclass drug resistance in order to create an optimal first regimen and increase the chance for virologic suppression.
Introduction
Transmitted drug resistance occurs at a reported rate of 8–15% of HIV-infected patients in the United States and commonly involves mutations affecting nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), and to a lesser extent protease inhibitors (PIs) [1–3]. As a result, treatment guidelines recommend obtaining a standard genotype prior to starting antiretroviral therapy (ART) [4]. Raltegravir, the first FDA-approved HIV integrase strand transfer inhibitor (INSTI), commonly used in salvage regimens, is now an option for use as part of first-line ART in treatment-naïve patients. Given the lack of evidence of transmitted raltegravir resistance to date, treatment guidelines currently do not recommend obtaining an integrase genotype prior to starting a raltegravir-based regimen [4]. Here we report a case of transmitted multi-class drug resistance, including raltegravir, PIs, NNRTIs, and NRTIs, and the difficulty in achieving viral suppression.
Case
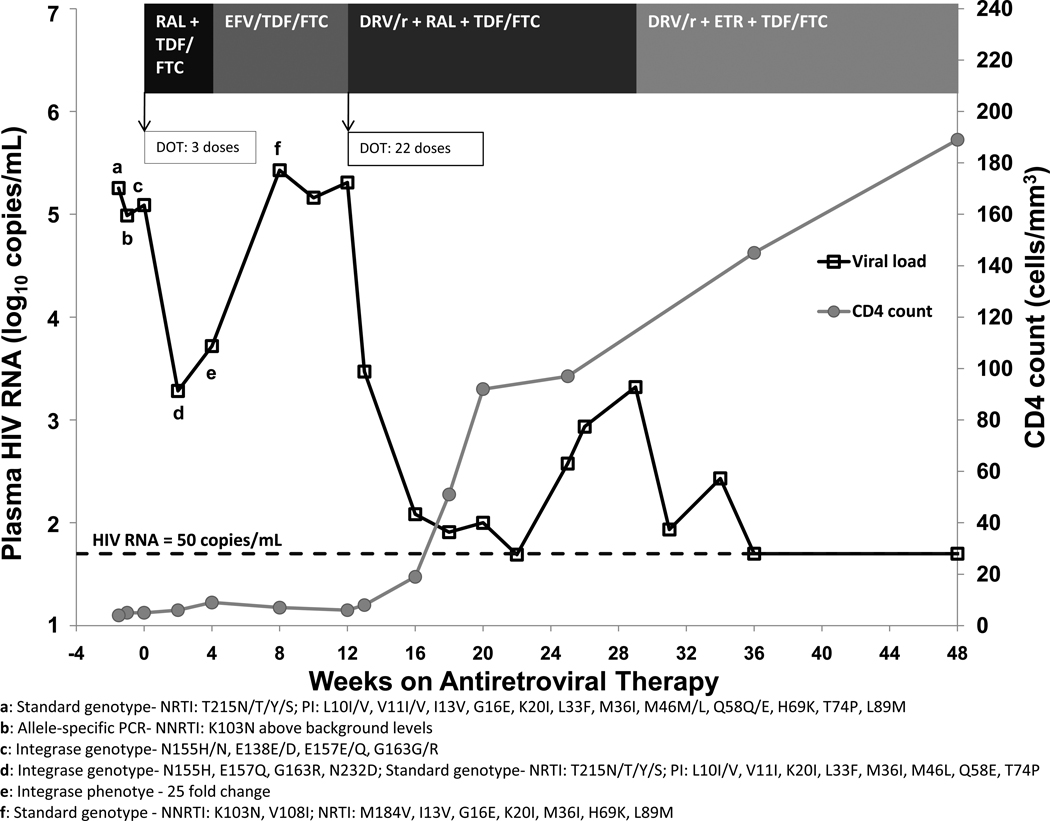
The patient is a 47 year-old female who immigrated to the U.S. from Cameroon in 2001 and was well until September 2009 when she was hospitalized with cerebral toxoplasmosis and found to be infected with HIV-1 CRF_AG with an HIV RNA of 179,826 copies/mL and a CD4 count of 4 cells/mm3. She was treated with sulfadiazine, pyrimethamine, and leucovorin for 3 weeks during hospitalization and 4 weeks after discharge, with radiographic improvement of the toxoplasmosis lesions. Her course was complicated by a new onset seizure, which was treated with phenytoin and subsequently changed to levetiracetam five days prior to ART initiation to avoid potential cytochrome P450 (CYP)-mediated drug interactions. Because of the desire to avoid efavirenz and PIs until washout of CYP induction effects of phenytoin and based on the pre-treatment genotype test (TRUGENE™ HIV-1, Siemens Healthcare Diagnostics, Deerfield, IL), which noted the presence of major and minor protease resistance mutations and one thymidine analogue mutation, the patient initiated raltegravir and tenofovir/emtricitabine. Figure 1 depicts the patient’s antiretroviral regimens, HIV RNA, CD4 count, and resistance testing results up to week 48 of therapy.
Figure 1. Antiretroviral regimens, HIV RNA, CD4 count, and resistance testing results through week 48 of treatment.
DOT = directly observed therapy, DRV/r = darunavir plus low-dose ritonavir, EFV = efavirenz, ETR = etravirine, FTC = emtricitabine, RAL = raltegravir, TDF = tenofovir
Two weeks after initiating ART, the patient’s HIV RNA declined 100-fold from baseline to 1,915 copies/mL. At week 4, however, her HIV RNA remained relatively unchanged at 5,228 copies/mL. In order to better assess adherence, a stored sample from week 2 was sent to determine a raltegravir concentration (HPLC assay, University of Florida – College of Pharmacy, Infectious Disease Pharmacokinetics Laboratory, Gainesville, FL), which was 300 ng/mL. The therapeutic trough of raltegravir, although not definitively established, has widely varied, ranging <20 – 2,470 ng/mL with a median of 90 ng/mL [5]. Based on a therapeutic raltegravir concentration, pill count, refill history, and self-report, the patient appeared to be adherent to her medications. At week 4, raltegravir was changed to efavirenz for patient convenience. By week 8, however, the patient’s HIV RNA returned to pre-treatment level (268,036 copies/mL), and a standard genotype revealed K103N and M184V mutations. Although resistance to NNRTIs was not seen on the pre-treatment standard genotype, an allele specific PCR (NCI Drug Resistance Program Core Laboratory, Frederick, MD) performed on a pre-treatment stored sample suggested low-level K103N was present above background levels. Pre-treatment NNRTI resistance mutations Y181C and G190A and the NRTI mutation M184V were not detected by allele specific PCR.
At week 12, the patient’s regimen was switched to darunavir/ritonavir (600mg/100mg twice daily) with re-initiation of raltegravir and continuation of tenofovir/emtricitabine. Her HIV RNA declined but failed to remain <50 copies/mL. Adherence was again confirmed by pill count, refill history, pillbox check, self-report, and family report. Darunavir trough levels (1,320 ng/mL and 1,740 ng/mL) (HPLC assay, University of Florida – College of Pharmacy, Infectious Disease Pharmacokinetics Laboratory, Gainesville, FL) measured 10 weeks apart were at the lower end of the therapeutic trough range seen in treatment-experienced clinical trials (1,255 – 7,368 ng/mL) [6], prompting an increase in darunavir dose to 800mg twice daily. At the same time, raltegravir was switched to etravirine, and her HIV RNA consequently declined to <50 copies/mL.
An integrase genotype (HIV GenoSURE, LabCorp, Research Triangle Park, NC) performed on a stored sample from week 2 revealed a major raltegravir mutation, N155H, and minor mutations, E157Q and G163R. A raltegravir phenotype (PhenoSense Integrase, Monogram Biosciences, Inc., San Francisco, CA) from a week 4 stored sample showed a 25-fold increase in resistance, indicating substantial reduction in susceptibility to raltegravir, and 36% replication capacity, suggesting a virus with reduced replication compared with pNL43 laboratory strain. These results prompted us to obtain an integrase genotype (HIV-1 Integrase Genotype, Quest Diagnostics, Nichols Institute, San Juan Capistrano, CA) on a pre-ART stored sample, which revealed that the raltegravir resistance mutations were present before raltegravir exposure. The patient repeatedly denied receiving any antiretroviral medications prior to entering into care at our clinic and admitted to potential HIV exposure in the United States as late as December 2008 with a male partner from Cameroon. Her history and pre-treatment genotypes, including a number of mutations in the surveillance drug resistance mutation list [7] (Table 1), strongly suggest she acquired resistance to four antiretroviral drug classes, including INSTI, via transmission.
Table 1.
Designation of the patient’s pre-treatment genotypic mutations as drug resistance mutations by three sources, and prevalence of each mutation in treatment-naïve CRF_AG HIV-infected patients.
| Patient’s Pre-treatment Mutations |
IAS-USA Drug Resistance Mutation [11] |
Stanford University HIVdb Mutation [12] |
Surveillance Drug Resistance Mutation [7] |
Prevalence in Treatment- Naïve CRF_AG Patients [7] |
|---|---|---|---|---|
| NRTI | ||||
| T215N | 0% | |||
| T215T | 0% | |||
| T215Y | X | X | X | 0% |
| T215S | X | X | 0.1% | |
| NNRTI | ||||
| K103N | X | X | X | 0.2% |
| PI | ||||
| L10I | X | X | 4.9% | |
| L10V | X | X | 10% | |
| V11I | X | X | 2% | |
| I13V | X | 92% | ||
| G16E | X | X | 21% | |
| K20I | X | X | 93% | |
| L33F | X | X major | 0.4% | |
| M36I | X | X | 97% | |
| M46L | X major | X major | X | 0.3% |
| Q58E | X major | X | 0% | |
| H69K | X | 97% | ||
| T74P | X major | X | 0.1% | |
| L89M | X | 96% | ||
Abbreviations: IAS-USA: International AIDS Society-USA, HIVdb: HIV Drug Resistance Database, NRTI: nucleoside reverse transcriptase inhibitor, NNRTI: non-nucleoside reverse transcriptase inhibitor, PI: protease inhibitor
Discussion
To our knowledge, this case is the first report of transmitted raltegravir resistance mutations in a treatment-naïve patient with HIV-1 CRF_AG. Raltegravir transmitted resistance has been reported in the U.S. [8]; as raltegravir use and subsequent failure increases, the number of patients with integrase mutations is likely to increase. The frequency of raltegravir resistance and risk of transmitting raltegravir-resistant HIV is unclear, and whether the prevalence of transmitted integrase resistance will be similar to NRTIs and NNRTIs or relatively low as with PIs remains to be seen. Our case adds to the evidence that raltegravir resistance can be acquired during primary infection, and furthermore, transmission of four-class drug resistance is possible.
Transmitted resistance mutations affecting NRTIs, NNRTIs, and to a lesser extent PIs have been reported in patients with CRF_AG [9–10], similar to patients with other HIV subtypes and recombinant forms. Although many of our patient’s pre-treatment protease mutations are highly prevalent in treatment-naïve CRF_AG patients, 2–3 are major protease mutations [11–12] prevalent in less than 0.5% of treatment-naïve CRF_AG patients [7] (Table 1). Many of her pre-treatment mutations are considered drug resistance mutations by the International AIDS Society-USA and the Stanford University database [11–12], but the observation that she harbored surveillance drug resistance mutations T215Y/S, K103N, and M46L (Table 1) supports the notion that she had transmitted drug resistance [7].
Transmitted integrase resistance regardless of subtype or recombinant form has not yet been widely reported given that raltegravir is a relatively new drug with a novel mechanism of action. Integrase genotypic and phenotypic studies in INSTI-naïve patients have shown that E157Q and G163R can occur naturally, particularly in CRF_AG viruses, but cause little to no raltegravir resistance [13–15]. In contrast, the major mutation N155H has not been routinely detected in INSTI-naïve patients, even by allele-specific PCR [16], and primarily occurs as a result of raltegravir treatment failure [17]. Therefore, it is highly likely that our patient acquired N155H through infection but possible that the minor mutations occurred naturally.
Current standard commercial genotyping/phenotyping assays do not detect resistance mutations below a threshold of 10–20% and do not routinely include analysis of integrase resistance. Based on the pre-treatment standard genotype test result, our patient’s first regimen of raltegravir plus tenofovir/emtricitabine presumably contained three fully active drugs and was a currently preferred regimen for treatment-naïve patients based on the DHHS guidelines [4]. Pre-existing raltegravir resistance was not found until a stored sample was sent for integrase genotype testing. Furthermore, the baseline standard genotype did not detect the NNRTI resistance mutation K103N, which was later found at very low levels. A lack of these test results prior to treatment initiation, neither of which are current standard of care, contributed to failure of the patient’s first two regimens.
Our determination of her lack of prior therapy is based on her denial of having received any medical care from 2001 until late 2008. She has been a resident of the U.S. for many years, and she reports sexual contact with a man from Cameroon since arriving in the U.S. from whom she may have been infected. Specifically, she resides in Maryland, where the rate of non-subtype B infection has been reported around 13% of which CRF_AG is most common [18]. The peripheral CD4 count measured in this patient was profoundly suppressed, suggesting that she was infected for a prolonged period or had a rapid loss of CD4 cells as a consequence of infection. The patient’s HLA type was B*07, which has not been associated with rapid progression to AIDS [19]. If she was initially infected prior to raltegravir availability, she may have been re-infected with a resistant virus more recently. Despite lack of information of HIV transmission in this patient, these data highlight the potential for multidrug resistance even in non-B subtypes circulating in the U.S.
HIV RNA decline at weeks 2 and 4 in the presence of an adequate raltegravir concentration and raltegravir mutations may represent residual activity of raltegravir despite the major mutation N155H. Upon restarting raltegravir at week 12 in a salvage-like regimen, HIV RNA reached <50 copies/mL, which was likely because of the high potency of darunavir/ritonavir and activity of tenofovir rather than residual activity of raltegravir. Subsequent increase of HIV RNA by more than 10-fold by week 26 confirmed insufficient raltegravir activity in the presence of N155H and increased risk of virologic failure. Raltegravir was discontinued after virologic rebound since continuation of a raltegravir-based regimen in the face of raltegravir resistance with ongoing viral replication may select for additional integrase mutations [20], which may or may not impact the use of future, second generation integrase inhibitors [21–22]. First generation integrase inhibitors, raltegravir and elvitegravir, appear to share major resistance pathways, but second generation integrase inhibitors may have activity in the presence of some integrase mutations [21–22]. Despite multiclass resistance, it was still possible to construct a suppressive regimen with PI, NNRTI, and NRTI components for this patient.
Integrase inhibitors represent a new class of antiretroviral drugs with increasing usage in both treatment-naïve and treatment-experienced patients. This case provides evidence that raltegravir resistance mutations can be acquired at the time of HIV transmission. Although pre-treatment integrase genotypes are not current standard of care, practitioners may consider obtaining one before starting a raltegravir-based regimen in a treatment-naïve patient if significant resistance mutations are present on the standard genotype.
Acknowledgements
This work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. It is also funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
None of the authors have any conflicts of interest.
Contributor Information
Sarita D. Boyd, SAIC-Frederick, Inc., National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Bethesda, Maryland.
Frank Maldarelli, HIV Drug Resistance Program, National Cancer Institute (NCI), NIH, Bethesda, Maryland
Irini Sereti, NIAID, NIH, Bethesda, MD
G. Laissa Ouedraogo, Formerly with SAIC-Frederick, Inc., NIAID, NIH, Bethesda, Maryland.
Catherine A. Rehm, NIAID, NIH, Bethesda, Maryland
Valerie Boltz, HIV Drug Resistance Program, NCI, NIH, Frederick, Maryland.
Diana Shoemaker, NIAID, NIH, Bethesda, Maryland
Alice K. Pau, NIAID, NIH, Bethesda, Maryland
References
- 1.Ross L, Lim ML, Liao Q, et al. Prevalence of antiretroviral drug resistance and resistance-associated mutations in antiretroviral therapy-naïve HIV-infected individuals from 40 United States cities. HIV Clin Trials. 2007;8:1–8. doi: 10.1310/hct0801-1. [DOI] [PubMed] [Google Scholar]
- 2.Wheeler WH, Ziebell RA, Zabina H, et al. Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006. AIDS. 2010;24:1203–1212. doi: 10.1097/QAD.0b013e3283388742. [DOI] [PubMed] [Google Scholar]
- 3.Weinstock HS, Zaidi I, Heneine W, et al. The epidemiology of antiretroviral drug resistance among drug-naive HIV-1-infected persons in 10 US cities. J Infect Dis. 2004;189:2174–2180. doi: 10.1086/420789. [DOI] [PubMed] [Google Scholar]
- 4.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services; [Accessed 18 October 2010]. (Updated 1 December 2009). [Google Scholar]
- 5.Baroncelli S, Villani P, Weimer LE, et al. Raltegravir plasma concentrations in treatment-experienced patients receiving salvage regimens based on raltegravir with and without maraviroc coadministration. Ann Pharmacother. 2010;44:838–843. doi: 10.1345/aph.1M688. [DOI] [PubMed] [Google Scholar]
- 6.Package insert. Yardley, PA, USA: Tibotec, Inc.; 2010. Prezista (darunavir) [Google Scholar]
- 7.Bennett DE, Camacho RJ, Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug resistance: 2009 update. PLoS One. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fransen S, Young B, Frantzell A, et al. Control of viral replication following transmission of HIV-1 exhibiting resistance to reverse transcriptase, protease and integrase inhibitors [abstract A56] Antivir Ther. 2010;15:A67. [Google Scholar]
- 9.Adje-Toure C, Bile CE, Borget MY, et al. Polymorphism in protease and reverse transcriptase and phenotypic drug resistance of HIV-1 recombinant CRF02_AG isolates from patients with no prior use of antiretroviral drugs in Abidjan, Côte d'Ivoire. J Acquir Immune Defic Syndr. 2003;34:111–113. doi: 10.1097/00126334-200309010-00016. [DOI] [PubMed] [Google Scholar]
- 10.Burda ST, Viswanath R, Zhao J, et al. HIV-1 reverse transcriptase drug-resistance mutations in chronically infected individuals receiving or naïve to HAART in Cameroon. J Med Virol. 2010;82:187–196. doi: 10.1002/jmv.21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009;17:138–145. [PubMed] [Google Scholar]
- 12.Liu TF, Shafer RW. Web Resources for HIV type 1 Genotypic-Resistance Test Interpretation. Clin Infect Dis. 2006;42:1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceccherini-Silberstein F, Baelen K, Armenia D, et al. Secondary integrase resistance mutations found in HIV-1 minority quasispecies in integrase therapy-naïve patients have little or no effect on susceptibility to integrase inhibitors. Antimicrob Agents Chemother. 2010;54:3938–3948. doi: 10.1128/AAC.01720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low A, Prada N, Topper M, et al. Natural polymorphisms of human immunodeficiency virus type 1 integrase and inherent susceptibilities to a panel of integrase inhibitors. Antimicrob Agents Chemother. 2009;53:4275–4282. doi: 10.1128/AAC.00397-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee SY, Liu TF, Kiuchi M, et al. Natural variation of HIV-1 group M integrase: implications for a new class of antiretroviral inhibitors. Retrovirology. 2008;5:74. doi: 10.1186/1742-4690-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charpentier C, Laureillard D, Piketty C, et al. High frequency of integrase Q148R minority variants in HIV-infected patients naive of integrase inhibitors. AIDS. 2010;24:867–873. doi: 10.1097/QAD.0b013e3283367796. [DOI] [PubMed] [Google Scholar]
- 17.Cooper DA, Steigbigel RT, Gatell JM, et al. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Eng J Med. 2008;359:355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- 18.Carr JK, Osinusi A, Flynn CP, Gilliam BL, Maheshwari V, Zhao RY. Two independent epidemics of HIV in Maryland. J Acquir Immune Defic Syndr. 2010;54:297–303. doi: 10.1097/QAI.0b013e3181e0c3b3. [DOI] [PubMed] [Google Scholar]
- 19.Gao X, O'Brien TR, Welzel TM, et al. HLA-B alleles associate consistently with HIV heterosexual transmission, viral load, and progression to AIDS, but not susceptibility to infection. AIDS. 2010;24:1835–1840. doi: 10.1097/QAD.0b013e32833c3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatano H, Lampiris H, Fransen S, et al. Evolution of integrase resistance during failure of integrase inhibitor-based antiretroviral therapy. J Acquir Immune Defic Syndr. 2010;54:389–393. doi: 10.1097/QAI.0b013e3181c42ea4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goethals O, Vos A, Van Ginderen M, et al. Primary mutations selected in vitro with raltegravir confer large fold changes in susceptibility to first-generation integrase inhibitors, but minor fold changes to inhibitors with second-generation resistance profiles. Virology. 2010;402:338–346. doi: 10.1016/j.virol.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi M, Nakahara K, Seki T, et al. Selection of diverse and clinically relevant integrase inhibitor-resistant human immunodeficiency virus type 1 mutants. Antiviral Res. 2008;80:213–222. doi: 10.1016/j.antiviral.2008.06.012. [DOI] [PubMed] [Google Scholar]