SUMMARY
Dexras1 is a 30kDa G-protein in the Ras subfamily whose discovery was based on its pronounced inducibility by the glucocorticoid dexamethasone. It binds to neuronal nitric oxide synthase (nNOS) via the adaptor protein CAPON, eliciting S-nitrosylation and activation of Dexras1. We report that Dexras1 binds to the Peripheral Benzodiazepine Receptor Associated Protein (PAP7), a protein of unknown function which binds to cyclic AMP dependent protein kinase and the peripheral benzodiazepine receptor. PAP7 in turn binds to the Divalent Metal Transporter (DMT1), an iron import channel. We have identified a novel signaling cascade in neurons whereby stimulation of NMDA receptors activates nNOS, leading to S-nitrosylation and activation of Dexras1 which, via PAP7 and DMT1, physiologically induces iron uptake. As selective iron chelation prevents NMDA neurotoxicity in cortical cultures, the NMDA-NO-Dexras1-PAP7-DMT1-iron uptake signaling cascade also appears to mediate NMDA neurotoxicity.
INTRODUCTION
It is increasingly appreciated that nitric oxide (NO) may not simply diffuse freely to reach its physiological targets but may be conveyed to these sites by interactions of NO synthase (NOS) with other proteins. One of the best known examples involves neuronal NOS (nNOS) binding to the adapter protein PSD95/93 which in turn binds to NMDA receptors (Brenman et al., 1996; Brenman et al., 1996). This ternary complex enables NO to S-nitrosylate NMDA receptors and alters their signaling (Lipton et al., 1993). Earlier, we identified CAPON, a 55 kDa protein that contains a C-terminal domain that binds to the PDZ domain of nNOS as well as an N-terminal phosphotyrosine binding (PTB) domain (Jaffrey et al., 1998). CAPON interacts with Dexras1, a brain enriched member of the Ras family of small G proteins which is selectively induced by dexamethasone (Fang et al., 2000; Kemppainen and Behrend, 1998). Dexras1 shares about 35% homology with the Ras subfamily of proteins, and contains all of the conserved domains of typical GTPases such as the GTP binding domain, the Mg2+ binding domain, and a C-terminal prenylation site. Unlike conventional GTPases, Dexras1 contains an extended 7 kDa C-terminal tail. Dexras1 has also been designated Activator of G-protein Signaling 1 (AGS1) or RASD1 (Blumer et al., 2005). The term AGS1 refers to the ability of Dexras1 to selectively bind to Gαi2, increasing GTPγS binding to Gi and Go and activating extracellular signal-regulated kinases 1,2 (ERK1,2)(Cismowski et al., 2000; Cismowski et al., 2001; Graham et al., 2002).
NMDA receptor stimulation of nNOS activates Dexras1. Glutamate via NMDA receptors triggers cellular calcium entry with calcium-calmodulin activating nNOS (Bredt and Snyder, 1994), whose binding to CAPON provides a mechanism for NO delivery to Dexras1 leading to S-nitrosylation of Dexras1 on cysteine-11 (Fang et al,, 2000; Jaffrey et al,, 1998; Jaffrey et al., 2002). The selective lessening of Dexras1 activation in the brains of mice with targeted deletion of nNOS and the existence of a ternary complex of nNOS, CAPON and Dexras1, establish that neuronally derived NO physiologically serves as a guanine nucleotide exchange factor to activate Dexras1 by S-nitrosylation (Fang et al., 2000).
Thus far, the physiological function of S-nitrosylated Dexras1 has not been determined. In the present study, to characterize downstream targets of Dexras1, we have conducted yeast two-hybrid analysis and discovered interactions of Dexras1 with PAP7, a protein which was discovered on the basis of its binding to peripheral benzodiazepine receptors and cAMP dependent protein kinase (Li et al., 2001). We demonstrate that PAP7 binds to DMT1, the only known physiological import channel for iron. We show that activation of glutamate-NMDA receptors stimulates nNOS resulting in S-nitrosylation and activation of Dexras1 which physiologically induces iron uptake via interactions with PAP7 and DMT1. This signaling cascade participates in NMDA neurotoxicity which is prevented by selective iron chelation.
RESULTS
Dexras1, PAP7 and DMT1 physiologically interact
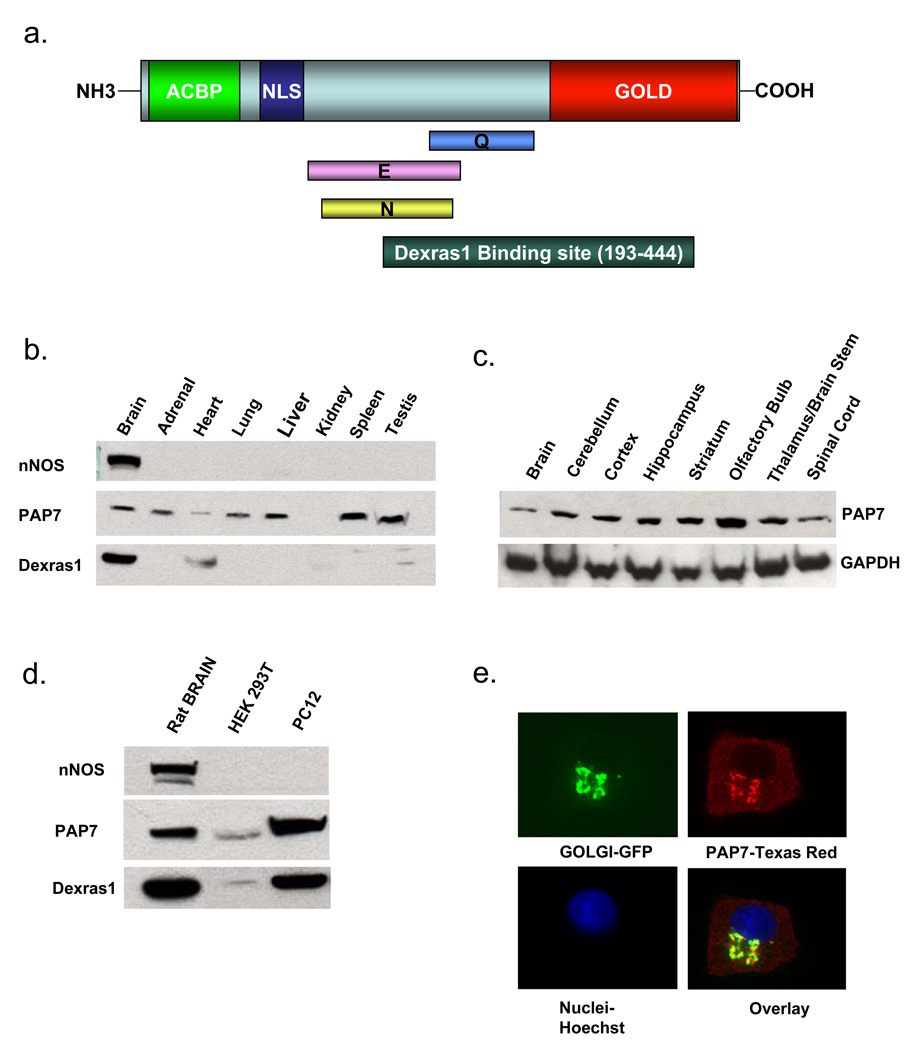
To elucidate physiologic roles for Dexras1, we conducted yeast two-hybrid analysis utilizing full length Dexras1 fused to the GAL4 DNA binding domain, and a rat whole brain cDNA library fused to the GAL4 DNA activation domain. A single colony identified PAP7 as a potential interactor (data not shown). PAP7 is a 62 kDa protein and includes several domains: an acylcoenzymeA-binding protein signature and a bipartite nuclear localization signal at the N-terminus, a Golgi dynamics (GOLD) domain at the C-terminus, and a glutamate(E)-, glutamine(Q)- and an asparagine(N)-rich region in the middle of the protein (Li et al., 2001) (Figure 1a). Western blot analysis reveals substantial levels of PAP7 in multiple mouse tissues including brain, adrenal, heart, lung, liver, spleen and testes but no detectable protein in the kidney, whereas Dexras1 is brain-selective with modest levels in heart and testes (Figure 1b). PAP7 is expressed at similar levels in several areas of the brain (Figure 1c). Both PAP7 and Dexras1 occur in high levels in undifferentiated PC12 cells with very low levels in HEK293T cells (Figure 1d).
Figure 1. Characterization of PAP7.
(a) Schematic diagram of PAP7. An Acyl-CoA Binding Protein Domain (ACBP) and bipartite nuclear localization signal (NLS) are located at the N-terminus, while a Golgi Dynamics (GOLD) Domain resides at the C-terminus. The middle of the protein contains asparagine (N), glutamate (E) and glutamine (Q)-rich domains. The Dexras1 binding domain is located between amino acids 193–444 as determined by yeast two-hybrid analysis. (b) PAP7 is occurs in all major tissues except kidney, while nNOS is brain-specific, and Dexras1 is predominantly in the brain, with moderate levels in the heart and testis (c) PAP7 is present at similar levels in different regions of the brain. (d) Dexras1 and PAP7 are present in high levels in PC12 cells, with small levels in HEK 293T cells. (e) Undifferentiated PC12 cells were transfected with a Golgi marker and stained for endogenous PAP7, revealing that PAP7 is localized to both the Golgi apparatus and the cytosol.
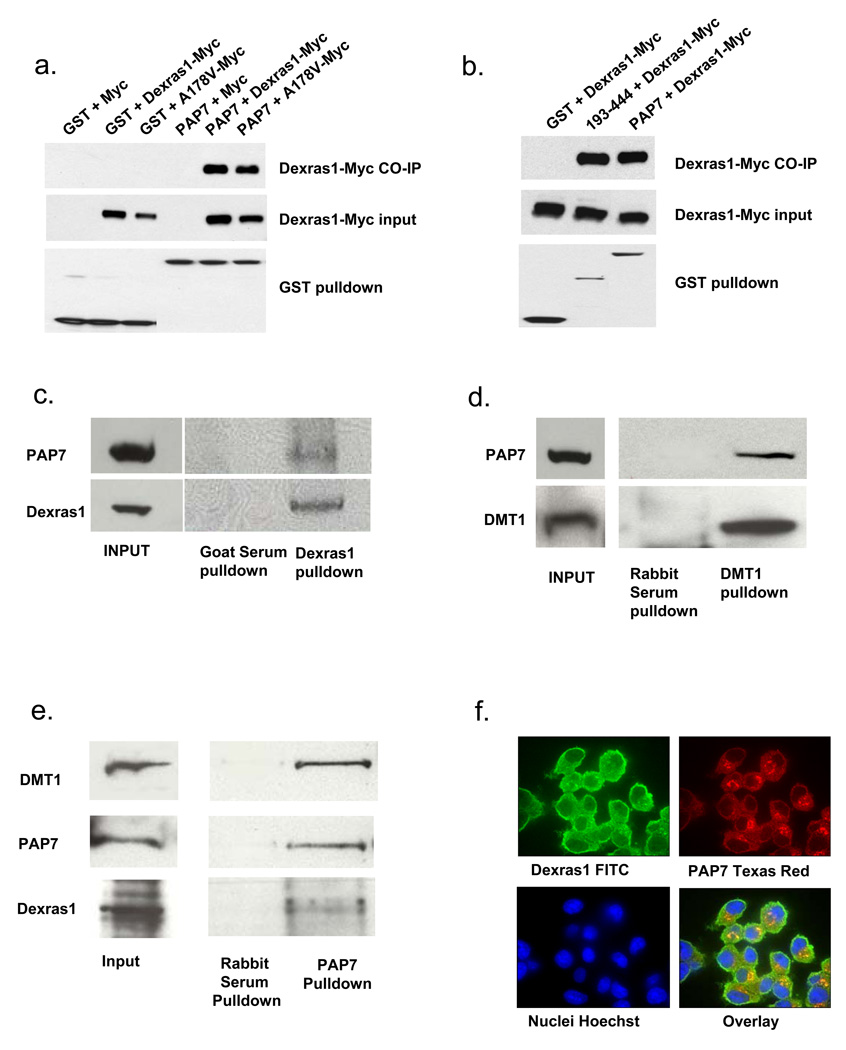
We have confirmed the Dexras1-PAP7 interaction by transient transfection of GST-PAP7 and Dexras1-Myc in HEK293T cells which reveals selective binding of Dexras1 and PAP7 (Figure 2a). PAP7 also binds to a constitutively active form of Dexras1 (A178V) (Graham et al., 2001) at about the same level as wild-type, indicating that the activity status of Dexras1 is not important for protein interaction. Yeast-two-hybrid analysis suggested that Dexras1 interacts with PAP7 between amino acids 193–444, encompassing parts of the E-, Q-, and N-rich domains, as well as the first part of the GOLD domain. To confirm the importance of PAP7-193–444 for Dexras1 binding, we conducted immunoprecipitation experiments in HEK293 cells containing over-expressed PAP7 and Dexras1 and observe co-precipitation of PAP7-193–444 with Dexras1 of comparable magnitude to the co-precipitation of wild-type PAP7 and Dexras1 (Figure 2b). To evaluate the interaction of endogenous Dexras1, PAP7 and DMT1, we first employed PC12 cells which contain high levels of both PAP7 and Dexras1. Utilizing an antibody to Dexras1, we observe co-immunoprecipitation with PAP7 (Figure 2c).
Figure 2. Dexras1, PAP7 and DMT1 physiologically interact.
(a) HEK 293T cells were transfected either with GST or GST-PAP7 and a Myc-tagged Dexras1 construct. A178V is a constitutively active form of Dexras1. Dexras1 and A178V specifically interact with PAP7. (b) HEK 293T cells were transfected either with GST or GST-PAP7 or GST-PAP7-193–444 and a Myc-tagged Dexras1 construct, showing that Dexras1 is able to bind to PAP7-193–444, as originally shown by yeast-two-hybrid analysis. (c) Immunoprecipitation experiments were performed using undifferentiated PC12 cells with a Dexras1 antibody, showing co-precipitation of PAP7. (d) Immunoprecipitation experiments were performed using undifferentiated PC12 cells with a DMT1 antibody, showing co-precipitation of PAP7. (e) Immunoprecipitation experiments were performed using mouse brain lysate with a PAP7 antibody, showing simultaneous immunoprecipitation of Dexras1 and DMT1. (f) Undifferentiated PC12 cells were fixed and stained for endogenous Dexras1 and PAP7. Dexras1 is shown to be localized to the cytosol and plasma membrane, while PAP7 is localized to the cytosol and Golgi apparatus.
An NCBI nucleotide sequence submission reported PAP7 as interacting with DMT1 (NCBI accession number NM_182843). DMT1, a twelve transmembrane channel localized to the plasma membrane, is the only known mammalian iron transporter associated with iron import into cells. It was previously identified as Natural Resistance Associated Macrophage Protein-2 (Nramp2) and Divalent Cation Transporter (DCT1) (Cellier et al., 1995; Fleming et al., 1997). Using an antibody to DMT1, we have confirmed the binding of PAP7 and DMT1 by co-immunoprecipitation in PC12 cells (Figure 2d). To examine a more physiologic preparation, we conducted immunoprecipitation experiments in whole mouse brain and examined for the presence of a ternary complex (Figure 2e). Utilizing an antibody to PAP7, we observe simultaneous co-precipitation with DMT1 and Dexras1. Thus, these three proteins appear to exist in a ternary complex in intact brain suggesting that they may interact physiologically. Immunohistochemical staining of undifferentiated PC12 cells reveals PAP7 is localized to the Golgi apparatus (Figure 1e) and to the cytosol, while Dexras1 is cytosolic and plasma membrane associated (Figure 2c). Localization of PAP7 in Golgi, where it interacts with gigantin and was designated GCP60, has been previously reported (Sohda et al., 2001).
Iron Uptake is Regulated by Dexras1-PAP7-DMT1 Interactions
Iron is transported into cells as either transferrin (Tf)-iron or Non-Transferrin Bound Iron (NTBI) (Hentze et al., 2004; Richardson and Ponka, 1997). Iron required for physiological processes, such as synthesis of heme, enters cells via the classical Tf-mediated iron uptake pathway wherein two molecules of diferric iron bind to Tf, which then binds to the transferrin receptor (TfR). Receptor-mediated endocytosis leads to internalization of TfR and DMT1 within an endocytic vesicle containing diferric-Tf. Acidification of the endosome leads to the liberation of iron from Tf, which then exits the endosome via DMT1 and enters the intracellular labile pool or mitochondria for heme biosynthesis (Picard et al., 2000). For NTBI uptake, DMT1 in the plasma membrane directly mediates iron import (Farcich and Morgan, 1992; Garrick et al., 1999; Richardson and Ponka, 1997). In the brain, the extracellular iron concentration exceeds that of serum transferrin, so that both Tf-mediated and NTBI uptake pathways import iron (Moos and Morgan, 1998; Rouault, 2001).
Since PAP7 binds to DMT1, we wondered whether Dexras1 and PAP7 influence iron homeostasis. In HEK293T cells, over-expression of Dexras1 very modestly augments both NTBI (Figure 3a) and Tf-associated (data not shown) iron uptake, which is substantially enhanced by co-transfection with PAP7. Thus PAP7 is required for a robust influence of Dexras1 on iron uptake. The action of Dexras1 on iron uptake derives from its GTPase actions, as constitutively active Dexras1, with a mutation of alanine-178 to valine (A178V), increases iron uptake more than native Dexras1 in the presence or absence of PAP7 (Figure 3a).
Figure 3. Dexras1 is S-nitrosylated and NTBI uptake is increased in PC12 cells after treatment with NO donors.
(a) HEK 293T cells were transfected with Dexras1 or Dexras1-A178V in the presence or absence of PAP7. NTBI uptake was measured as described in the Materials and Methods. (*, p<0.05, **, p<0.01) (b) Dexras1 in undifferentiated PC12 cells is S-nitrosylated by GSNO in a concentration-dependent manner (c) GSNO treatment for 3h leads to a concentration-dependent increase NTBI uptake in undifferentiated PC12 cells, as does (d) SNP for 1h and (e) DETA NONOate for 4h. (All NTBI uptake experiments were repeated three times, each sample in triplicate. Comparison with two-tailed student’s t-test; error bars represent SEM).
As NO S-nitrosylates and activates Dexras1 (Fang et al., 2000), we examined effects of NO donors on iron uptake. Treatment of undifferentiated PC12 cells with S-nitrosoglutathione (GSNO) results in S-nitrosylation of Dexras1 (Figure 3b) and enhances NTBI uptake (Figure 3c). Other NO donors, sodium nitroprusside (SNP) (Figure 3d) and DETANONOate (Figure 3e), elicit similar effects. As Dexras1 activation by S-nitrosylation involves cysteine-11 (Jaffrey et al., 2002), we examined effects of a Dexras1 cysteine mutant (C11S) on NO-mediated iron uptake. The C11S mutant still interacts with PAP7 (Figure 4a), however, in HEK 293T cells transfected with PAP7 and wild-type or C11S mutated Dexras1, the C11S mutation abolishes GSNO activation of NTBI uptake (Figure 4b).
Figure 4. NO-mediated NTBI uptake is mediated by S-nitrosylation of Dexras1.
(a) HEK 293T cells were transfected with GST or GST-PAP7 Dexras1-C11S-myc, a nitrosylation-dead mutant, which is able to bind specifically to PAP7. (b) HEK 293T cells were transfected with PAP7 and either wild-type Dexras1 or the C11S mutant. After transfection, the cells were treated with 100 µM GSNO for 3h and NTBI uptake was measured (*, p<0.01). GSNO treatment up-regulates NTBI uptake in cells containing wild-type Dexras1, but not the C11S mutant. (c) Undifferentiated PC12 cells were transfected with either control RNAi or Dexras1 RNAi (mock is untransfected). Dexras1 protein levels was fully depleted after transfection with Dexras1 RNAi, but not in control or mock-transfected cells (d) Undifferentiated PC12 cells were transfected with either control RNAi or Dexras1 RNAi, treated with 100 µM GSNO for 3h and NTBI uptake was measured (*, p<0.01). GSNO treatment up-regulates NTBI uptake in control cells, but not in cells depleted of Dexras1. (All NTBI uptake experiments were repeated three times, each sample in triplicate. Comparison with two-tailed student’s t-test; error bars represent SEM).
To determine whether Dexras1 is required for NO-mediated iron uptake, we sought to deplete Dexras1. Since it is difficult to transfect primary cortical neurons, we employed undifferentiated PC12 cells, which contain endogenous Dexras1, PAP7 and DMT1. We successfully depleted Dexras1 in PC12 cells using RNA interference (Figure 4c). While GSNO augments NTBI uptake in control PC12 cells, it fails to increase uptake in the Dexras1-depleted cells (Figure 4d).
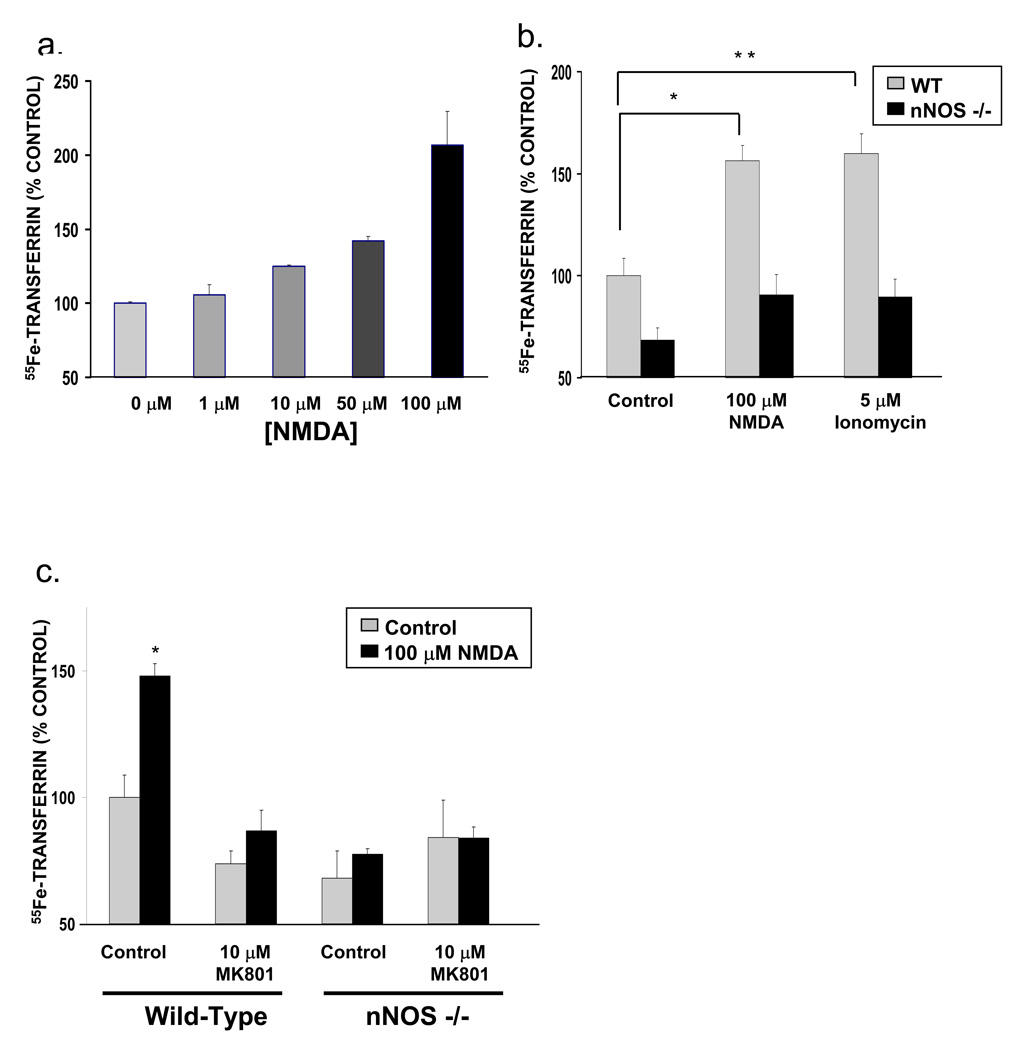
Since Dexras1 is activated by glutamate-NMDA neurotransmission acting via NO, we examined the influence of NMDA upon iron uptake in primary cortical neuronal cultures. NMDA treatment increases iron uptake in a concentration-dependent manner (Figure 5a) with the effects blocked by pretreatment with the NMDA antagonist MK801 (Figure 5c). NMDA fails to increase NTBI uptake in cultures from nNOS knockout mice (Figure 5b and 5c), showing that nNOS and NO are required for NMDA-mediated NTBI uptake. NMDA transmission augments calcium conductance which fits with the ability of the calcium ionophore, ionomycin, to stimulate NTBI uptake, an effect abolished in nNOS knockout cultures (Figure 5b).
Figure 5. Glutamate-NMDA neurotransmission increases NTBI uptake, which is abolished in nNOS knockout mice.
(a) NMDA stimulation increases NTBI uptake in primary cortical neurons in a concentration-dependent manner. (b) NTBI uptake in primary cortical neurons is increased by both NMDA stimulation and ionomycin treatment, a calcium ionophore. This up-regulation is not seen in primary cortical neurons from nNOS knockout mice (*, p<0.05, **, p<0.01) and (c) is abolished by pretreatment with MK801, an NMDA receptor antagonist (*, p<0.005). (All NTBI uptake experiments were repeated three times, each sample in triplicate. Comparison with two-tailed student’s t-test; error bars represent SEM).
Physiological and Pathophysiological Relevance of Dexras1 Regulation of Iron Uptake
Tf-mediated iron uptake is also augmented by treatment with NMDA (Figure 6a) or ionomycin (Figure 6b), effects abolished by pre-treatment with MK801(+) (Figure 6c) or in nNOS knockout cultures (Figure 6b and 6c). To assess the physiological relevance of NMDA-stimulated iron uptake, we monitored 55Fe incorporation into heme in cortical cultures treated with NMDA (Figure 7a). NMDA stimulation markedly increases the rate of heme biosynthesis.
Figure 6. Glutamate-NMDA neurotransmission increases Tf-iron uptake, which is abolished in nNOS knockout mice.
(a) Tf-mediated iron uptake is increased in a dose-dependent manner with NMDA treatment in primary cortical neurons. (b) Tf-mediated iron uptake is increased by both NMDA and ionomycin (*, p<0.01, **, p<0.01). This up-regulation is not seen in primary cortical neurons from nNOS knockout mice and (c) is abolished by pre-treatment with MK801 (*, p<0.005). No increase in uptake is seen in neurons from nNOS knockout mice. (All Tf uptake experiments were repeated three times, each sample in triplicate. Comparison with two-tailed student’s t-test; error bars represent SEM).
Figure 7. Physiological and Pathophysiological Relevance of Iron Uptake.
(a) Rate of heme biosynthesis was measured using incorporation of 55Fe into heme and was increased after NMDA treatment (*, p<0.01). (b) NTBI uptake is significantly increased after treatment with an excitotoxic concentration of NMDA. (c) An increase in HPF fluorescence was observed after treatment with NMDA, indicating an increase in ROS formation. This increase was abolished with pre-treatment of the neurons with SIH. (d) Treatment of primary cortical neurons with 300 µM NMDA elicits a high level of neurotoxicity, which is attenuated by the pre-treatment with either 10 µM MK801, an NMDA receptor antagonist, or SIH, an iron chelator.
We wondered whether the stimulation of iron uptake by NMDA neurotransmission might have pathophysiologic consequences, as overactivation of NMDA receptors leads to neurotoxicity which has been implicated in vascular stroke and neurodegenerative diseases (Choi, 1994). In cerebrocortical cultures a concentration of 300 µM NMDA is well established to elicit neurotoxicity (Dawson et al., 1991; Koh and Choi, 1988). We have verified that this concentration of NMDA elicits a major augmentation of iron uptake (Figure 7b). As neurotoxicity is associated with a pronounced increase in reactive oxygen species (ROS), we monitored ROS formation utilizing a dye that selectively detects hydroxyl free radicals. These predominantly arise from the Fenton reaction that typically reflects iron interacting with hydrogen peroxide to form hydroxyl free radicals. Treatment with 300 µM NMDA elicits a 3.5 fold augmentation in hydroxyl free radical formation (Figure 7c). To determine whether this increase is caused by the Fenton reaction elicited by iron, we employed SIH (salicylaldehyde isonicotinoyl hydrazone), a selective cell permeable iron chelator. SIH treatment abolishes the increase in hydroxyl free radicals caused by NMDA (Figure 7c), implying that the free radical formation arises from iron whose influx is stimulated by NMDA. To ascertain whether the prevention of free radical formation by SIH also prevents neurotoxocity, we measured neurotoxicity in the same cortical cultures utilizing propidium iodide staining to monitor dead cells (Figure 7D). In these preparations, 300 µM NMDA kills more than 90% of cells, while pretreatment with the NMDA antagonist MK801 (10µM) provides virtually complete protection. Pretreatment with 100 µM SIH also markedly protects the neurons from cell death. Thus, it appears that iron contributes both to the NMDA-elicited formation of free radicals and neuronal death.
DISCUSSION
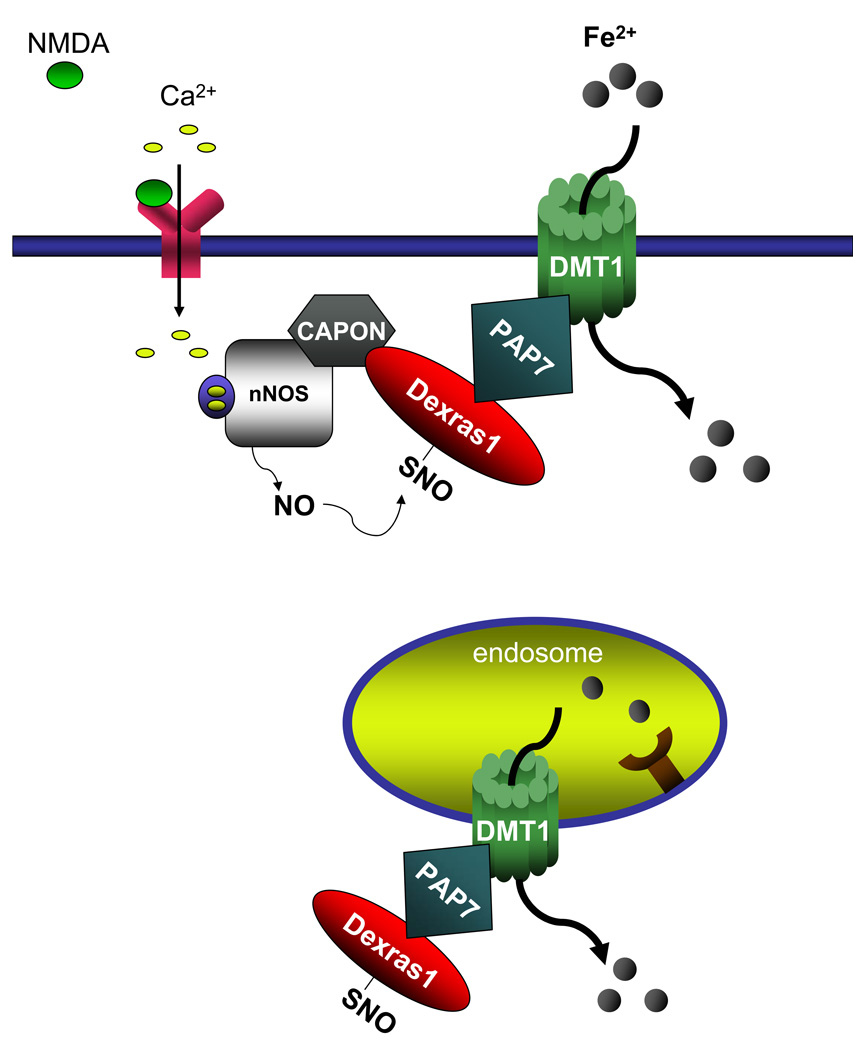
In the present study, we have identified a novel signaling cascade whereby neurotransmission regulates iron homeostasis (Figure 8). Glutamate, acting via NMDA receptors, activates nNOS to form NO (Bredt and Snyder, 1994), which leads to protein S-nitrosylation (Hess et al., 2005). This modification activates Dexras1 which, by its link to PAP7, augments both Tf-mediated and NTBI uptake. PAP7 does not appear to have a direct influence on iron homeostasis, as its over-expression does not affect iron uptake, though it does potentiate Dexras1-induced enhancement of iron uptake. Instead, it presumably serves as a scaffold bringing Dexras1 into proximity to DMT1. Stimulation of iron uptake by Dexras1 reflects its GTPase activity, as constitutively active Dexras1 manifests enhanced activity in stimulating iron uptake. The influence of Dexras1 upon iron uptake is evidently the first example of a G-protein regulating an iron transporter.
Figure 8. Dexras1, PAP7 and DMT1 mediate both NTBI and Tf-mediated iron uptake.
A model of a signaling cascade whereby glutamate-NMDA neurotransmission regulates cellular iron homeostasis. Glutamate-NMDA stimulation leads to the activation of nNOS and, via the scaffolding protein CAPON, to S-nitrosylation of Dexras1, which interacts with PAP7 and DMT1 both at the endosome and the plasma membrane, thus influencing cellular iron uptake in the cell.
Besides the role of the NMDA-NO-Dexras1-PAP7-DMT1 cascade in mediating physiologic influences of NMDA on iron uptake, our findings implicate this pathway in pathophysiologic actions of NMDA-glutamate neurotransmission. Thus, excessive NMDA receptor activation is thought to mediate the neurotoxicity that occurs in vascular stroke and other neurodegenerative conditions (Choi, 1994). While such neurotoxicity may involve multiple systems activated by the calcium entry associated with NMDA transmission, NO formation has been implicated. Inhibitors of NOS prevent NMDA neurotoxicity and stroke damage, while neurotoxicity and stroke damage are markedly reduced in brains of nNOS−/− mice (Huang et al., 1994). In our experiments, selective chelation of intracellular iron blocks formation of free radicals in brain cultures and also markedly attenuates NMDA neurotoxicity. This implies that iron uptake in response to NMDA-NO activation plays an important role in neurotoxicity.
The molecular mechanism whereby Dexras1 and its GTPase activity regulate signaling via PAP7 and DMT1 is unclear. We presume that Dexras1 influences DMT1 directly but cannot rule out some action upon PAP7. Conceivably, other reported activities of Dexras1 may participate. For instance, Dexras1 activates the ERK1, 2 pathway in a pertussis toxin sensitive fashion (Cismowski et al., 2000). It impairs activation of G-protein coupled receptors that act via Gi, inhibiting adenylyl cylase (Graham et al., 2004; Nguyen and Watts, 2005). Functional relevance of those effects is suggested by its involvement in regulating circadian rhythm. Dexras1 expression cycles in a circadian fashion in the suprachiasmatic nucleus (Takahashi et al., 2003). Its genetic deletion reduces photic entrainment of circadian responses to glutamate-NMDA transmission (Cheng et al., 2004).
Interestingly, two spontaneous mutations of DMT1 have been reported in the Belgrade (b/b) rat and microcytic anemia (mk) mice (Fleming et al., 1998; Fleming et al., 1997). Surprisingly, both of these spontaneous mutations involved the same amino acid, glycine 185 which is mutated to arginine. While both mutant rodents display systemic iron deficiency and anemia, they do survive. This implies that the G185R mutation confers some selective advantage which may reflect a new constitutive calcium permeability as reported by Xu et al (Xu et al., 2004).
A homologue of Dexras1 called Ras Homolog Enriched in Striatum (Rhes, Dexras2) is induced by thyroid hormone, an interesting contrast to the stimulation of Dexras1 by glucocorticoids (Falk et al., 1999; Vargiu et al., 2001). Rhes is selectively localized to the corpus striatum. It also binds to PAP7 (data not shown) and so might impact NMDA receptor influences on iron entry into the striatal neurons.
Iron is required for many physiological processes, such as heme synthesis, mitochondrial oxidation reactions and DNA synthesis, but is toxic in excess, so that its cellular disposition is tightly regulated (Hentze et al. 2004; Richardson and Ponka, 1997). Studies of iron in the brain have largely focused on its pathophysiological roles, with iron accumulation occurring in numerous neurodegenerative diseases (Ponka, 2004; Rouault, 2001; Shoham and Youdim, 2000; Zecca et al., 2004). Heretofore, no influence of neurotransmission upon iron uptake has been reported. Our findings establish a physiologic role for glutamate neurotransmission in regulating iron uptake in the brain in a novel signaling cascade. It is conceivable that misregulation of this physiological pathway participates in the iron accumulation in neurodegenerative disease.
METHODS AND MATERIALS
Cells and Reagents
HEK 293T cells were maintained in DMEM with 10% fetal bovine serum (FBS), 2 mM L-Glutamine and 100U/ml penicillin-streptomycin (PS) at 37°C with 5% CO2 atmosphere in a humidified incubator. PC12 cells were maintained in DMEM with 10% FBS, 5% horse serum, 2 mM L-glutamine and 100U/ml PS in the same environment. All chemicals were purchased from Sigma, unless otherwise indicated.
Generation of constructs
Rat PAP7 was cloned from EST #5621578 (Open Biosystems) into either pCMV-GST or pCMV-HA (Clontech). Rat Dexras1, wild-type and A178V or C11S mutants, were all cloned into pCMV-Myc (Clontech).
Characterization of PAP7
To examine the expression pattern of PAP7 throughout the body, a male C57/B6 mouse was dissected and each organ was homogenized in Buffer A (100 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 15% Glycerol, 1 mM PMSF, 25 µg/ml antipain, 50 µg/ml leupeptin, 50 µg/ml aprotinin, 25 µg/ml chymostatin and 25 µg/ml pepstatin). Total protein (100 µg) was loaded onto a Western blot and immunoblotted with a rabbit anti-nNOS antibody (previously generated in our laboratory), rabbit anti-PAP7 (V. Papadopolous) antibody and rabbit anti- Dexras1 antibody (Calbiochem).
To examine the expression pattern of PAP7 in the brain, a male Sprague-Dawley rat brain was dissected into several parts, lysed in Buffer A, and total protein (100 µg) loaded and a Western blot performed.
To examine the expression pattern of PAP7 and Dexras1 in different cell lines, 100 µg of total protein from HEK 293T and undifferentiated PC12 cells lysed in Buffer A were loaded onto a Western blot.
GST pull-down assay
GST or GST-tagged PAP7 constructs were co-transfected with Dexras1-Myc constructs into HEK 293T cells using PolyFect (Qiagen), with a transfection efficiency of greater than 90%. Cells were lysed 48h after transfection in Buffer A. Lysates were pre-cleared with pansorbin cells (Calbiochem), then 1 mg of total protein was incubated with glutathione-Sepharose beads overnight at 4°C. Beads were washed with Wash Buffer (50 mM Tris pH 7.4, 500 mM NaCl, 10 mM β-glycerophosphate) twice, then once with Buffer A. Beads were quenched in sample buffer (100 mM Tris, pH 6.8, 10% glycerol, 250 mM β-mercaptoethanol, 2% sodium dodecyl sulfate and bromophenol blue). Total protein (50 µg) was loaded as input. Dexras1-Myc binding was examined using an anti-myc antibody (Roche) followed by incubation with anti-mouse secondary conjugated to horseradish peroxidase(HRP) (Jackson Immunoresearch); blots were then stripped and probed with an anti-GST antibody conjugated to HRP to detect PAP7. Chemiluminescence (Pierce) was used to detect bands on the Western blot.
Co-immunoprecipitation
Undifferentiated PC12 cells were lysed in Buffer A and lysates were pre-cleared with protein A Sepharose. 1 mg of total protein was incubated with 2 µg of either goat anti-Dexras1 antibody (Abcam) or rabbit anti-NRAMP2 antibody (Alpha Diagnostics) overnight at 4°C, then protein A Sepharose beads were added for 1h. Beads were washed in Buffer A three times and quenched in sample buffer. Total PC12 lysate (100 µg) was loaded as input. PAP7 binding was detected using a rabbit anti-PAP7 antibody, followed by anti-rabbit Ig conjugated to HRP.
For the ternary complex interaction, a male C57/B6 mouse was sacrificed and the whole brain was dissected and homogenized in Buffer A. Lysates were pre-cleared with protein A Sepharose. 1 mg of total protein was incubated with 2 µg of rabbit anti-PAP7 antibody overnight at 4°C, then protein A Sepharose beads were added for 1h. Beads were washed in Buffer A three times and quenched in sample buffer. Total brain lysate (100 µg) was loaded as input. Dexras1 and DMT1 binding was detected using a rabbit anti-Dexras1 or anti-nRAMP2 antibody, followed by anti-rabbit Ig conjugated to HRP.
Immunofluorescence Staining
Undifferentiated PC12 cells were plated on poly-D-lysine coated glass coverslip dishes. For PAP7 localization to the Golgi, these cells were transfected with a plasmid containing a Golgi marker conjugated to YFP (pEYFP-Golgi, Clonetech) using Lipofectamine 2000 (Invitrogen). After 48 hours, cells were stained as below.
Cells were fixed in 4% paraformaldehyde in PBS, permeabilized in 1% Triton X-100 in PBS and blocked in PBS with 1% normal goat serum and 2% normal horse serum. Endogenous Dexras1 was detected using a goat anti-Dexras1 antibody; endogenous PAP7 was detected using a rabbit anti-PAP7 antibody. Anti-goat Ig conjugated to FITC and anti-rabbit Ig conjugated to rhodamine was obtained from Molecular Probes. Confocal microscropy images were obtained using a PerkinElmer UltraView LCI (Live Cell Imaging) System.
S-nitrosylation of Dexras1
PC12 cells were treated with various concentrations of GSNO (Alexis Biochemicals) for 3h. Cells were washed in PBS and harvested. S-nitrosylation of Dexras1 was monitored using the biotin-switch assay previously described (Jaffrey, and Snyder, 2001). Dexras1 was detected on the immunoblot using a rabbit anti-Dexras1 antibody (Calbiochem).
Iron Uptake Assays
In HEK 293T Cells
NTBI uptake assays were performed as previously described (Picard, and Govoni, et al, 2000). In brief, HEK 293T cells were transfected (greater than 90% efficiency) with PAP7-HA and Dexras1-Myc (or mutants) using Lipofectamine and PLUS Reagent (Invitrogen) for 3h in DMEM only, then supplemented with full media After 48h, the cells were washed with phosphate buffered saline (PBS) then resuspended into Iron Uptake Buffer (25 mM Tris, 25 mM MES, 140 mM NaCl, 5.4 mM KCl, 5 mM glucose, 1.8 mM CaCl2, pH 5.5) and transferred to glass test tubes. Ascorbic acid was added to 1 mM FeSO4 at a 44:1 ratio. 55FeCl3 (PerkinElmer Life Science) was added to the iron/ascorbic acid mixture, which was then added to the cells in Iron Uptake Buffer to a final concentration of 20 µM. Cells were incubated at 37°C with shaking for 30 minutes. The cells were washed twice with cold PBS + 0.5 mM EDTA and harvested. An aliquot of resuspended cells was taken for protein assay using the Bio-Rad Protein Assay Reagent; the protein concentrations of individual samples were used to quantitate 55Fe incorporation (cpm/µg protein). Samples were normalized to control.
In primary cortical neurons
Cells were dissected out of E16-E18 wild-type or nNOS knockout mice and plated in 6 well plates at 3 × 106 cells per well. Cells were maintained in Primary Neuron Media (Neurobasal media supplemented with B27 serum, 2 mM L-glutamine and 100U/ml PS) at 37°C with 5% CO2 atmosphere in a humidified incubator. Using this media, the growth of the glia is suppressed and the amount of glia is less than 0.5% of the total culture, giving us an essentially pure neuronal culture. Because Mg2+ can block NMDA receptors, we measured the free Mg2+ levels of the Primary Neuron Media using a fluorescent Mg2+ indicator (Magnesium Green, Invitrogen) incorporating 0.2 mM EGTA to chelate free calcium. The Mg2+ level in the media is 88 µM, much lower than concentrations of Mg2+ that provide extensive blockade of NMDA receptors.
Neurons were aged 14–20 days after plating before being used for iron uptake assays. Cells were treated as follows: 100µM NMDA for 30 min, 300µM NMDA for 10 minutes or 5 µM ionomycin for 10 min. For samples with MK801, cells were pre-treated with 10 µM MK801 for 10 min, then 100 µM NMDA was added for 30 min. Cells were then washed once with warm PBS. For samples with MK801, the drug was added back after NMDA treatment.
For NTBI uptake assays, 1 ml of warm Iron Uptake Buffer containing 20 µM iron prepared as above was added to each well and incubated at 37°C with 5% CO2 atmosphere in a humidified incubator for 30 min. Cells were harvested, washed twice in cold PBS + 0.5 mM EDTA and processed as above.
For Tf-iron uptake pathway, assays were performed as previously described (Kim, and Ponka, 2002). In brief, apo-transferrin was loaded with 55Fe2+ and the concentration measured. After treatment, cells were incubated in a final concentration of 10 µM transferrin (1 µM 55Fe-transferrin and 9 µM holotransferrin) in 1 ml of Primary Neuron Media for 2h. Cells were harvested, washed twice in cold PBS + 0.5 mM EDTA and processed as above.
RNA Interference in PC12 cells
The Dexras1 RNA interference (RNAi) insert was designed using the Genescript siRNA design center and cloned into pRNAT-U6.1/Neo (Genescript). To obtain knockdown in PC12 cells, 24 µg of Dexras1 RNAi were transfected into cells using Lipofectamine 2000 (Invitrogen) in Optimem overnight. Control transfection used the pRNAT-U6.1/Neo empty vector. The next morning, cells were fed with full media and allowed to recover for 24h. They were transfected again overnight, and then allowed to recover for 2 days before treatment with GSNO for 3h. The transfection efficiency after the double transfection was greater than 90%. To monitor knockdown, lysate (100 µg) was run on a Western blot (mock transfected cells had no DNA), and Dexras1 protein levels were monitored using a rabbit anti-Dexras1 antibody.
Heme Incorporation Assay
Primary rat cortical neurons were treated with 100 µM NMDA, then incubated in 55Fe-transferrin as described above. Cells were washed in PBS + 0.5 mM EDTA, then resuspended in 1 ml of 0.2N HCl. Using an aliquot of cell lysate, the protein concentration of individual samples was determined using the Biorad Protein Assay. The resuspended cells were heated at 100°C for 10 minutes, cooled on ice, and then transferred to glass test tubes. 10% TCA (3 ml) was added to each sample, vortexed, and incubated on ice for 15 minutes. Protein precipitate was pelleted by centrifugation, and the pellet was washed in 3 ml of 10% TCA, and resuspended in 500 µL of 0.2N HCl. Scintillation fluid was added and 55Fe2+ incorporated into heme counted in a scintillation counter. Samples were normalized to control.
Measurement of Reactive Oxygen Species
To measure highly reactive oxygen species (hROS), such as hydroxyl free radicals generated by the Fenton reaction, we employed 2-[6-(4'-hydroxy)phenoxy-3H-xanthen-3-on-9-yl]benzoic acid (HPF) (Cell Technology, Inc, Mountain View, CA). This scarcely fluorescent compound developed by T. Nagano (Setsukinai, and Urano, et al, 2003) binds specifically to hydroxyl free radicals, and less specifically to peroxynitrate, and converts to a highly fluorescent molecule.
Rat primary cortical neurons were prepared as above and aged for 14 days after plating. Measurement of hROS was conducted according to the manufacturer’s protocol. Briefly, neurons were pretreated with either 0 µM or 100 µM SIH for 3 hours in full media, which was then removed, and the cells were washed once with Hanks Balanced Salt Solution (HBSS). 5 µM HPF in HBSS was added to the cells and incubated at 37°C with 5% CO2 atmosphere in a humidified incubator for 30 min. 0 µM or 300 µM NMDA was then added to the cells and incubated for another 30 minutes. Cells were harvested in HBSS and fluorescence was measured in a fluorimeter utilizing excitation 488 nm and emission 515 nm.
Measurement of NMDA-induced Neurotoxicity
Rat primary cortical neurons were prepared as above and aged for 14 days after plating. Briefly, neurons were pretreated with either water (vehicle), 10 µM MK801 or 100 µM SIH for 3 hours in full media, which was then removed. The cells were then treated with 300 µM NMDA for 10 minutes in full media, then replaced with full media and incubated for 24 hours at 37°C with 5% CO2 atmosphere in a humidified incubator. Cells were washed with PBS and incubated in 1 µg/ml propidium iodide for 10 minutes to stain for dead cells. The neurons were then fixed in 4% paraformaldehyde in PBS and stained with Hoechst stain to visualize total cells using an Olympus IX81 fluorescent microscope. Cell death was assessed by the ratio of propidium iodide stained cells to total cells and expressed as % cell survival.
Acknowledgements
We thank Mahil Rao for technical assistance and Barbara Zeigler for editorial assistance. This work was supported by US Public Health Service grant DA000266, Research Scientist Award DA00074 (to S.H.S.) and Canadian Institute of Health Research fellowship (to S.F.K).
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests.
REFERENCES
- Blumer JB, Cismowski MJ, Sato M, Lanier SM. AGS proteins: receptor-independent activators of G-protein signaling. Trends Pharmacol. Sci. 2005;26:470–476. doi: 10.1016/j.tips.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Bredt DS, Snyder SH. Nitric oxide: a physiologic messenger molecule. Annu. Rev. Biochem. 1994;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- Brenman JE, Christopherson KS, Craven SE, McGee AW, Bredt DS. Cloning and characterization of postsynaptic density 93, a nitric oxide synthase interacting protein. J. Neurosci. 1996;16:7407–7415. doi: 10.1523/JNEUROSCI.16-23-07407.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P. Nramp defines a family of membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 1995;92:10089–10093. doi: 10.1073/pnas.92.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Obrietan K, Cain SW, Lee BY, Agostino PV, Joza NA, Harrington ME, Ralph MR, Penninger JM. Dexras1 potentiates photic and suppresses nonphotic responses of the circadian clock. Neuron. 2004;43:715–728. doi: 10.1016/j.neuron.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Choi DW. Glutamate receptors and the induction of excitotoxic neuronal death. Prog. Brain Res. 1994;100:47–51. doi: 10.1016/s0079-6123(08)60767-0. [DOI] [PubMed] [Google Scholar]
- Cismowski MJ, Ma C, Ribas C, Xie X, Spruyt M, Lizano JS, Lanier SM, Duzic E. Activation of heterotrimeric G-protein signaling by a ras-related protein. Implications for signal integration. J. Biol. Chem. 2000;275:23421–23424. doi: 10.1074/jbc.C000322200. [DOI] [PubMed] [Google Scholar]
- Cismowski MJ, Takesono A, Bernard ML, Duzic E, Lanier SM. Receptor-independent activators of heterotrimeric G-proteins. Life Sci. 2001;68:2301–2308. doi: 10.1016/s0024-3205(01)01019-0. [DOI] [PubMed] [Google Scholar]
- Dawson VL, Dawson TM, London ED, Bredt DS, Snyder SH. Nitric oxide mediates glutamate neurotoxicity in primary cortical cultures. Proc. Natl. Acad. Sci. U. S. A. 1991;88:6368–6371. doi: 10.1073/pnas.88.14.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk JD, Vargiu P, Foye PE, Usui H, Perez J, Danielson PE, Lerner DL, Bernal J, Sutcliffe JG. Rhes: A striatal-specific Ras homolog related to Dexras1. J. Neurosci. Res. 1999;57:782–788. [PubMed] [Google Scholar]
- Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- Farcich EA, Morgan EH. Uptake of transferrin-bound and nontransferrin-bound iron by reticulocytes from the Belgrade laboratory rat: comparison with Wistar rat transferrin and reticulocytes. Am. J. Hematol. 1992;39:9–14. doi: 10.1002/ajh.2830390104. [DOI] [PubMed] [Google Scholar]
- Fleming MD, Trenor CC, 3rd, Su MA, Foernzler D, Beier DR, Dietrich WF, Andrews NC. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat. Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- Fleming MD, Romano MA, Su MA, Garrick LM, Garrick MD, Andrews NC. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick LM, Dolan KG, Romano MA, Garrick MD. Nontransferrin-bound iron uptake in Belgrade and normal rat erythroid cells. J. Cell. Physiol. 1999;178:349–358. doi: 10.1002/(SICI)1097-4652(199903)178:3<349::AID-JCP9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Graham TE, Key TA, Kilpatrick K, Dorin RI. Dexras1/AGS-1, a steroid hormone-induced guanosine triphosphate-binding protein, inhibits 3',5'-cyclic adenosine monophosphate-stimulated secretion in AtT-20 corticotroph cells. Endocrinology. 2001;142:2631–2640. doi: 10.1210/endo.142.6.8209. [DOI] [PubMed] [Google Scholar]
- Graham TE, Prossnitz ER, Dorin RI. Dexras1/AGS-1 inhibits signal transduction from the Gi-coupled formyl peptide receptor to Erk-1/2 MAP kinases. J. Biol. Chem. 2002;277:10876–10882. doi: 10.1074/jbc.M110397200. [DOI] [PubMed] [Google Scholar]
- Graham TE, Qiao Z, Dorin RI. Dexras1 inhibits adenylyl cyclase. Biochem. Biophys. Res. Commun. 2004;316:307–312. doi: 10.1016/j.bbrc.2004.02.049. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Huang Z, Huang PL, Panahian N, Dalkara T, Fishman MC, Moskowitz MA. Effects of cerebral ischemia in mice deficient in neuronal nitric oxide synthase. Science. 1994;265:1883–1885. doi: 10.1126/science.7522345. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Fang M, Snyder SH. Nitrosopeptide mapping: a novel methodology reveals s-nitrosylation of dexras1 on a single cysteine residue. Chem. Biol. 2002;9:1329–1335. doi: 10.1016/s1074-5521(02)00293-4. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snowman AM, Eliasson MJ, Cohen NA, Snyder SH. CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron. 1998;20:115–124. doi: 10.1016/s0896-6273(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001;2001 doi: 10.1126/stke.2001.86.pl1. PL1. [DOI] [PubMed] [Google Scholar]
- Kemppainen RJ, Behrend EN. Dexamethasone rapidly induces a novel ras superfamily member-related gene in AtT-20 cells. J. Biol. Chem. 1998;273:3129–3131. doi: 10.1074/jbc.273.6.3129. [DOI] [PubMed] [Google Scholar]
- Kim S, Ponka P. Nitrogen monoxide-mediated control of ferritin synthesis: implications for macrophage iron homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12214–12219. doi: 10.1073/pnas.192316099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh JY, Choi DW. Vulnerability of cultured cortical neurons to damage by excitotoxins: differential susceptibility of neurons containing NADPH-diaphorase. J. Neurosci. 1988;8:2153–2163. doi: 10.1523/JNEUROSCI.08-06-02153.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Degenhardt B, Tobin D, Yao ZX, Tasken K, Papadopoulos V. Identification, localization, and function in steroidogenesis of PAP7: a peripheral-type benzodiazepine receptor- and PKA (RIalpha)-associated protein. Mol. Endocrinol. 2001;15:2211–2228. doi: 10.1210/mend.15.12.0736. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;12:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- Moos T, Morgan EH. Evidence for low molecular weight, nontransferrin-bound iron in rat brain and cerebrospinal fluid. J. Neurosci. Res. 1998;54:486–494. doi: 10.1002/(SICI)1097-4547(19981115)54:4<486::AID-JNR6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Nguyen CH, Watts VJ. Dexras1 blocks receptor-mediated heterologous sensitization of adenylyl cyclase 1. Biochem. Biophys. Res. Commun. 2005;332:913–920. doi: 10.1016/j.bbrc.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Picard V, Govoni G, Jabado N, Gros P. Nramp 2 (DCT1/DMT1) expressed at the plasma membrane transports iron and other divalent cations into a calcein-accessible cytoplasmic pool. J. Biol. Chem. 2000;275:35738–35745. doi: 10.1074/jbc.M005387200. [DOI] [PubMed] [Google Scholar]
- Ponka P. Hereditary causes of disturbed iron homeostasis in the central nervous system. Ann. N. Y. Acad. Sci. 2004;1012:267–281. doi: 10.1196/annals.1306.022. [DOI] [PubMed] [Google Scholar]
- Richardson DR, Ponka P. The molecular mechanisms of the metabolism and transport of iron in normal and neoplastic cells. Biochim. Biophys. Acta. 1997;1331:1–40. doi: 10.1016/s0304-4157(96)00014-7. [DOI] [PubMed] [Google Scholar]
- Rouault TA. Systemic iron metabolism: a review and implications for brain iron metabolism. Pediatr. Neurol. 2001;25:130–137. doi: 10.1016/s0887-8994(01)00260-0. [DOI] [PubMed] [Google Scholar]
- Setsukinai K, Urano Y, Kakinuma K, Majima HJ, Nagano T. Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J. Biol. Chem. 2003;278:3170–3175. doi: 10.1074/jbc.M209264200. [DOI] [PubMed] [Google Scholar]
- Shoham S, Youdim MB. Iron involvement in neural damage and microgliosis in models of neurodegenerative diseases. Cell. Mol. Biol. 2000;46:743–760. [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yamamoto A, Yano A, Nakamura N, Ikehara Y. Identification and characterization of a novel Golgi protein, GCP60, that interacts with the integral membrane protein giantin. J. Biol. Chem. 2001;276:45298–45306. doi: 10.1074/jbc.M108961200. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Umeda N, Tsutsumi Y, Fukumura R, Ohkaze H, Sujino M, van der Horst G, Yasui A, Inouye ST, Fujimori A, et al. Mouse dexamethasone-induced RAS protein 1 gene is expressed in a circadian rhythmic manner in the suprachiasmatic nucleus. Brain Res. Mol. Brain Res. 2003;110:1–6. doi: 10.1016/s0169-328x(02)00543-0. [DOI] [PubMed] [Google Scholar]
- Vargiu P, Morte B, Manzano J, Perez J, de Abajo R, Sutcliffe JG, Bernal J. Thyroid hormone regulation of rhes, a novel Ras homolog gene expressed in the striatum. Brain Res. Mol. Brain Res. 2001;94:1–8. doi: 10.1016/s0169-328x(01)00140-1. [DOI] [PubMed] [Google Scholar]
- Xu H, Jin J, DeFelice LJ, Andrews NC, Clapham DE. A spontaneous, recurrent mutation in divalent metal transporter-1 exposes a calcium entry pathway. PLoS Biol. 2004;2:378–386. doi: 10.1371/journal.pbio.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca L, Youdim MB, Riederer P, Connor JR, Crichton RR. Iron, brain ageing and neurodegenerative disorders. Nat. Rev. Neurosci. 2004;5:863–873. doi: 10.1038/nrn1537. [DOI] [PubMed] [Google Scholar]