SUMMARY
Nonsense-mediated mRNA decay (NMD) is a well-studied cellular quality-control pathway. It decreases the half-lives of eukaryotic mRNAs that aberrantly contain premature termination codons and additionally regulates an estimated 10–20% of normal transcripts. NMD factors play crucial roles during embryogenesis in many animals. Here, we review data indicating that NMD factors are required for proper embryogenesis by discussing the abnormal developmental phenotypes that result when the abundance of individual NMD factors is either downregulated or completely eliminated. We conclude that while NMD per se affects the embryogenesis of all animals, it is required to avoid embryonic lethality in only some animals. The critical roles of many NMD factors in other metabolic pathways undoubtedly also contribute to embryonic development if not viability.
INTRODUCTION
Nonsense-mediated mRNA decay (NMD) protects many heterozygous carriers of defective genes that encode premature termination codons (PTCs) from manifesting dominantly inherited disorders that would result if the encoded truncated proteins were expressed ([1] and references therein). It is very important for cells to remove PTC-containing mRNAs, whether they are somatically acquired or genetically inherited, because the encoded truncated proteins have the potential to manifest dominant-negative or gain-of-function activities. Thus, NMD plays a key protective role in a long list of human diseases that are due to frameshift or nonsense mutations and result in the premature termination of mRNA translation, including β0-thalassemia, cystic fibrosis, Duchenne muscular dystrophy and a number of cancers that involve PTCs within, e.g., BRCA1 or WT1 tumor suppressor mRNAs ([1] and references therein).
NMD typifies all eukaryotes that have been examined, including budding yeast, fission yeast, kinetoplastids, ciliates, nematodes, fish, flies, birds, mice, humans and plants. While NMD functions largely to protect cells from the potentially deleterious effects of routine inaccuracies in gene expression that accidentally generate PTCs, it also helps to maintain cellular homeostasis by degrading physiologic transcripts. In mammals, routine inaccuracies occur during transcription initiation, pre-mRNA splicing or the somatic-cell rearrangement and hypermutation of immunoglobulin or T-cell receptor genes that generate immune diversity, to name a few examples (reviewed in [1,2]). NMD targets in Saccharomyces cerevisiae and Paramecium tetraurelia are primarily the result of inefficient pre-mRNA splicing (see, e.g., [3,4]) and a large number of NMD targets in Caenorhabiditis elegans consist of the pseudogenes that are estimated to exist for 6–12% of functional genes [5].
In every eukaryote, NMD depends on the RNA helicase and RNA-dependent ATPase called up-frameshift suppressor (UPF)1, whose activities are activated by UPF2 and UPF3 ([6] and references therein). UPF2 binding to the cysteine-histidine-rich (CH) domain of human or S. cerevisiae UPF1 was recently shown to promote helicase activity by converting UPF1 from a closed conformation, in which the CH-domain occupies the helicase region and has a tightened hold on RNA, to an open conformation, in which the helicase domain has a looser and less extensive hold on RNA ([7], in press), presumably allowing UPF1 to slide in the 5′-to- 3′ direction on an NMD target and remove proteins as it progresses [8●]. Remarkably, overexpressing UPF1 in S. cerevisiae overrides the need for UPF2 [9], indicating that unphysiologically high levels of UPF1 assume an active conformation at a sufficient level to support NMD. Additionally, UPF2 siRNA-insensitive or UPF3 (also called UPF3a) siRNA- and/or UPF3X (also called UPF3b) siRNA-insensitive NMD targets have been reported for mammalian cells [10–13], suggesting that UPF1 can function in some instances with abnormally low levels of one or more of the other UPF proteins. The two mammalian UPF3 paralogues derive from two distinct genes, one of which is X-linked as the name UPF3X implies [14–16]. Two UPF3 paralogues also exist in Danio rerio (zebrafish) [17●●], as well as, e.g., Anolis carolinensis (Anole lizard) and Xenopus tropicalis (Ensembl Genome Browser).
The composition of an mRNP that is targeted for NMD can vary depending on the organism and, as implied above, depending on the particular mRNA. NMD in mammalian cells requires the cap-binding protein (CBP) complex that consists of CBP80 and CBP20 and typifies newly synthesized mRNAs [18]. Generally, exon-junction complexes (EJCs), which are comprised of proteins deposited upstream of the exon-exon junctions of newly synthesized mRNAs as a consequence of pre-mRNA splicing, are also critical for mammalian-cell NMD [1,18,19]. EJCs generally contain not only UPF2 and UPF3 or UPF3X but also the RNA-binding-motif protein Y14, the cancer susceptibility candidate gene 3 protein Barentsz (BTZ, also called MLN51), a homologue of Drosophila melanogaster mago nashi named MAGOH, and the eukaryotic translation initiation factor (eIF)-like protein eIF4AIII, among other proteins (reviewed in [1,18,19]).
EJC-dependent NMD generally requires that translation terminates sufficiently upstream of an EJC so that the EJC is not removed by the terminating ribosome. EJCs reside ~ 20–24-nucleotides upstream of splicing-generated exon-exon junctions, explaining the rule that in mammalian cells NMD targets transcripts containing PTCs that are situated more than ~50–55-nucleotides upstream of an exon-exon junction [24]. Notably, exceptions to the rule exist. For example, a PTC can trigger NMD if it resides closer than ~50–55-nucleotides upstream of an exon-exon junction in the case of T-cell receptor and immunoglobulin mRNAs ([1,2] and references therein; [25]). Possibly analogously, for a number of mRNAs that derive from experimenter-generated constructs in mammalian cells, NMD can occur in the absence of an EJC situated downstream of a PTC although an intron has to exist upstream of the PTC ([26] and references therein; [27,28]). NMD in S. pombe can also target naturally occurring mRNAs if a PTC exists either upstream or downstream of an intron, but the mechanism is EJC-independent [29]. As in mammals, NMD in fish appears to utilize EJCs since mRNA decay is triggered by PTCs in a zebrafish reporter transcript provided they are situated more than ~50–55-nucleotides upstream of an exon-exon junction [17●●].
It is important to point out that at least in mammals and D. melanogaster, and probably in all organisms that have them, EJCs are heterogeneous in composition and do not typify every exon-exon junction [11,30,31]. Despite their presence, EJCs are dispensable for NMD in D. melanogaster [32,33]. Consistent with this, the core EJC constituent eIF4AIII does not contribute to NMD in D. melanogaster Schneider cells [34] even though Drosophila eIF4AIII is 84% identical to human eIF4AIII. eIF4AIII also does not contribute to NMD in C. elegans [35], where it is 88% identical to human eIF4AIII. An eIF4AIII-like protein that is 78% identical to human eIF4AIII exists in S. pombe, in which EJCs do not function in NMD as noted above. eIF4AIII-like proteins that are 74.5% and 78% identical to human eIF4AIII, respectively, also typify Nicotiana benthamiana and Arabidopsis thaliana, in which EJC-dependent and EJC-independent mechanisms coexist [36,37] but will not be discussed further. Other differences among NMD pathways in different organisms pertain to their dependence on suppressor with morphological effect on genitalia (SMG) proteins. SMG proteins are conserved in many multicellular organisms but are absent from ciliates and kinetoplastids as well as single-cell organisms such as baker’s and fission yeast [38].
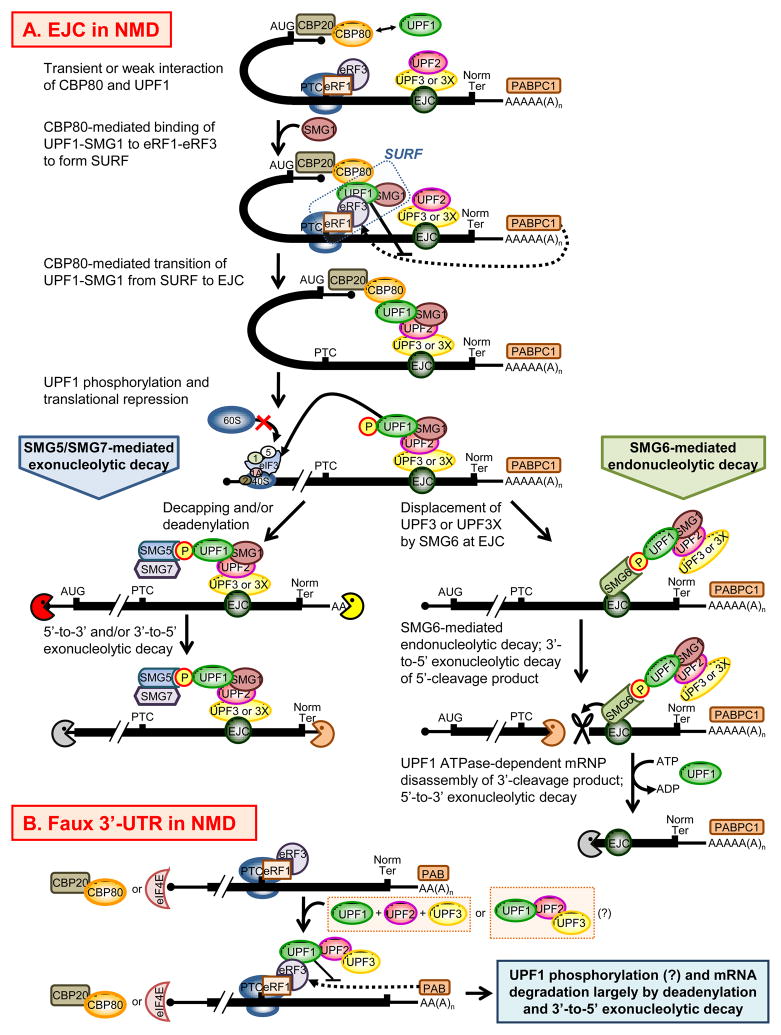
The molecular dynamics of UPF and SMG proteins on an NMD target are presently best understood for mammals (Figure 1). A transient or weak interaction of the UPF1 helicase region with mRNA cap-associated CBP80 augments NMD by promoting the association of UPF1 and its phosphatidylinositol 3-kinase-related kinase, SMG1, (i) with the translation termination factors eRF1 and eRF3 to form SURF (SMG1, UPF1, eRF1 and eRF3) at a PTC and (ii) subsequently downstream with EJC-bound UPF2 [20–22]. While the kinase activity of SMG1 in SURF is silenced by SMG8-SMG9 [39], SMG1 phosphorylates UPF1 once it associates with EJC-bound UPF2 [22]. Phosphorylated UPF1 represses further rounds of translation initiation on the NMD target by binding to the translation initiation factor eIF3 of the 43S preinitiation complex that is situated at the translation initiation codon, and it also recruits mRNA decay activities [40]. These activities consist of SMG5-and SMG7-mediated activities that degrade an NMD target from either or both ends (i.e., decapping followed by 5′-to-3′ exonucleolytic degradation and/or deadenylation followed by 3′-to-5′ exonucleolytic degradation) and also the SMG6-mediated endonucleolytic activity (reviewed in [19]). While the UPF1 ATPase, presumably together with its helicase, has been shown to promote degradation of the 3′-endonucleolytic cleavage product that results from SMG6-mediated decay [8●], it is currently uncertain how these UPF1 activities contribute to decay from either mRNA end. Data indicate that SMG5, SMG6 and SMG7 at some point recruit protein phosphatase 2A to dephosphorylate UPF1 ([41–43]; J.H. and L.E.M., unpubl. data).
Figure 1. Models for NMD.
(A) EJC function in mammalian-cell NMD. NMD is a consequence of PTC recognition during the pioneer-round of translation [18]. This round utilizes newly synthesized mRNA bound by the cap-binding protein heterodimer CBP80-CBP20 and, provided the mRNA derived from splicing, at least one exon-junction complex (EJC) situated ~ 20–24-nucleotides upstream of such a junction. The direct, but weak or transient interaction of CBP80 with the central NMD factor UPF1 promotes at least two steps during NMD [21●]. The first step is the joining of UPF1 and its kinase SMG1 to eRF1 and eRF3 at a PTC to form the SURF complex. During NMD, this step is thought to compete effectively with joining of the PABPC1 to eRF3, the latter of which is specified as a dotted line [28]. The second is the joining of UPF1 and SMG1, presumably from SURF, to a downstream EJC, which leads to UPF1 phosphorylation by SMG1 [22]. SMG5 and SMG7 form a complex with phosphorylated UPF1, as does SMG6 [42,76–79]. It is uncertain if SMG5/SMG7 and SMG6 bind multiple phosphates on the same UPF1 molecule or, as shown, different phosphorylated UPF1 molecules. In favor of the first possibility, SMG6 co-immunoprecipitates with SMG5 and SMG7 in an RNase A-resistant manner [77●]. Since SMG7-mediated mRNA decay occurs independently of SMG6 [79], it is plausible that SMG5/SMG7-mediated NMD leads to deadenylation and/or decapping followed, respectively, by exosome-mediated 3′-to-5′ and XRN1-mediated 5′-to-3′ exonucleolytic activities (reviewed in [80–82]). An alternative or additional mRNA degradation pathway involves SMG6, whose binding to hyperphosphorylated UPF1 competes with UPF3X and may replace the interaction of UPF3X with Y14-MAGOH EJC constituents [77●]. The endonuclease activity of SMG6 cleaves the NMD substrate into 5′- and 3′-cleavage products. Activation of the RNA-dependent ATPase activity of UPF1 subsequently results in the XRN1-mediated 5′-to-3′ decay of the 3′ fragment, which presumably depends on UPF1 helicase activity [8●]. PAPBC1, poly(A) binding protein C1. (B) Faux 3′-UTR function in S. cerevisiae NMD, where CBP80 and CBP20 are alternatively referred to as, respectively, CBC1 and CBC2. When a ribosome reaches a PTC that is situated abnormally upstream of the mRNA poly(A) tail, UPF1 can effectively compete with PAB for association with eRF3 so as to trigger NMD [28,44]. How and when the other UPF proteins associate with an NMD target and contribute to NMD remains uncertain, as does whether UPF1 undergoes phosphorylation during NMD. CBC, cap-binding complex constituent.
EJC-independent NMD mechanisms, like EJC-dependent mechanisms, envision the UPF proteins joining eRF1 and eRF3 at a PTC at least in part because the efficiency with which poly(A) binding protein joins the eRFs at a PTC is compromised when compared to the efficiency of joining to a termination codon that does not trigger NMD [44]. Hence, the splicing- and EJC-independent NMD mechanism that typifies S. cerevisiae has been called the ‘faux’ 3-untranslated region (3′UTR) mechanism. However, since NMD in S. cerevisiae can target mRNA in the absence of either a poly(A) tail or poly(A)-binding protein (PAB) [45], a PAB-independent mechanism that distinguishes PTCs and normal termination codons must exist.
In this review, we discuss the abnormal embryonic phenotypes and/or embryonic lethality that result when individual NMD factors are downregulated or completely eliminated in animals. Since a number of NMD factors do not function exclusively in NMD (Table 1; [1,19,46,47]), it can be difficult to attribute a developmental defect to the lack of NMD per se. Thus, we have organized our review by organism so as to point out those defects that are in common or lacking when the abundance of individual NMD factors is compromised.
Table 1.
Key NMD factors and their cellular functions in other pathways in animals
| Factor in humans | Non-NMD functions in humans | Embryonic lethality* | Other comments | References |
|---|---|---|---|---|
| UPF1 | Staufen (STAU)1- mediated mRNA decay; histone mRNA decay at the end of S phase; DNA replication; telomere maintenance; small RNA-induced mRNA downregulation | Fruitfly, zebrafish, mouse | Called SMG2 in nematode | [17●●,50●●,52,55,62,68–72] |
| UPF2 | Telomere maintenance (minor role relative to UPF1) | Fruitfly, zebrafish, mouse | Called SMG3 in nematode | [17●●,33,58,59●●,69] |
| UPF3 (UPF3a) or UPF3X (UPF3b) | None reported | None | Called SMG4 in nematode; UPF3a has peripheral role in NMD for most NMD targets in fruitful; no UPF3b orthologue in fruitfly | [14,32, 50●●] |
| SMG1 | DNA-damage response to ionizing radiation; nutrient-dependent signaling; telomere maintenance | Mouse | NMD factor for only certain NMD targets in fruitfly | ([33,51,52,57 ●●,69,73]; reviewed in [74]) |
| SMG5, SMG6 and SMG7 | Telomere maintenance | Zebrafish | No SMG7 orthologue in fruitfly | [17●●,32,69,72] |
| NAG/NBAS | Amplified in neuroblastoma | Nematode, zebrafish | Called SMGL-1 in nematode; no orthologue in fruitfly | [35,54●●] |
| DHX34 | Putative ATP-dependent RNA helicase that probably functions in non-NMD pathways | Nematode, zebrafish | Called SMGL-2 and CG32533 in nematode and fruitfly, respectively | ([35,54●●]; reviewed in [75]) |
when downregulated (nematode or zebrafish) or completely absent (fruitfly, mouse)
Nematodes
As their SMG names imply, C. elegans SMG2 (UPF1 in other organisms), SMG3 (otherwise UPF2), SMG4 (otherwise UPF3), SMG1, SMG5, SMG6 and SMG7, each of which was identified in genetic screens, are not essential for development but result in morphogenic defects in the male bursa and the hermaphrodite vulva [48,49]. Thus, NMD is not an essential process in C. elegans development. Nevertheless, SMGL-1 and SMGL-2, which were found in a reporter-based genome-wide RNAi screen to be critical NMD factors, are required for organismal viability: when depleted, each results in early embryonic lethality prior to morphogenesis [35]. SMGL-1 and SMGL-2 are absent from yeast but have readily identifiable orthologues in, e.g., fruitfly, zebrafish, pufferfish, mouse and human, where they are called, respectively, NAG/NBAS (neuroblastoma-amplified sequence) and DHX34 [DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 34] [35]. Therefore, while NMD per se is not necessary for embryonic viability in C. elegans, the NMD factors SMGL-1 and SMGL-2 are. SMGL-1 and SMGL-2 functions that do not pertain to NMD but are required to preclude embryonic lethality have yet to be identified.
Fruitflies
UPF1 and UPF2 are broadly expressed during D. melanogaster development [33]. Loss-of-function mutants demonstrated that UPF1 and UPF2, unlike UPF3 or SMG1, are required for larval development since each, e.g., provides a competitive growth or survival advantage to proliferating imaginal cells [33,50●●–52]. Unlike UPF1 and UPF2, UPF3 plays only a peripheral role in the NMD of most NMD targets [50●●]. Furthermore, SMG1 appears to have a variable role in NMD depending on the particular transcript [33,51,52], from which it can be concluded that the SMG1-mediated phosphorylation is dispensable for the NMD of at least some NMD targets. In the case of these targets, it remains to be determined if another kinase functions on behalf of SMG1. However, from those targets analyzed to date, SMG6 appears to be the major nuclease [53]. Notably, no identifiable SMG7 orthologue exists in D. melanogaster [32].
Taken together, these findings indicate that the embryonic lethality resulting from the absence of UPF1 or UPF2 must be due to UPF1 or UPF2 function in the NMD of particular transcripts and/or in one or more pathways other than NMD. Both UPF1 and UPF2 are necessary for D. melanogaster Schneider-cell progression through the G2/M phase of the cell cycle [52], which may or may not be due to their roles in NMD. The finding that depletion of UPF3, SMG5 or SMG6 has a less severe but nevertheless detectable increase in the percent of G2/M phase-cells [52] suggests that each is less critical to cell-cycle progression than is either UPF1 or UPF2. It will be important to determine if SMG5 and SMG6 are critical for embryonic viability in D. melanogaster.
Zebrafish
Whole-mount in situ-hybridization of NMD factors during zebrafish development demonstrated that UPF1, UPF2, the two paralogues of UPF3, SMG1, SMG5, SMG6 and SMG7 are maternally derived and ubiquitously expressed at 2- to 8-cell stages, during gastrulation and at one-day post-fertilization [17●●,54●●]. DHX34 and NAG/NBAS, which are orthologous to C. elegans SMGL-1 and SMGL-2, respectively, are likewise expressed ubiquitously during development [54●●]. Downregulating UPF1 using one of several antisense morpholino-modified oligonucleotides (MOs) showed a range of developmental delays and morphological abnormalities that depended on the degree to which UPF1 function was impaired [17●●]. Abnormalities included aberrant eye and brain patterning, the latter especially at the midbrain-hindbrain boundary, faulty somitogenesis and mortality rates as high as 80–85% by 5- days post-fertilization [17●●,54●●]. Essentially the same results were observed using UPF2, SMG5, SMG6, DHX34 or NBAS MOs; however, downregulating SMG7 resulted primarily in patterning defects in the brain, i.e., a visible but altered midbrain-hindbrain boundary and an elongated hindbrain. In contrast, downregulating either SMG1 or UPF3b had no detectable consequence, downregulating UPF3a had only minor effects on brain development, and downregulating both UPF3a and UPF3b was less severe than downregulating either UPF1 or UPF2, causing only 19% of morphants to die at 5-days post-fertilization [17●●].
Of course, the failure to observe a phenotype when a factor is only downregulated rather than completely absent does not rule out the importance of factor function. Furthermore, it is unclear if SMG1 and UPF3a or UPF3b are critical for NMD in zebrafish, especially since neither SMG1 nor UPF3 is critical for NMD in fruitflies. Thus, while NMD undoubtedly contributes to a normal embryonic phenotype in zebrafish, whether NMD per se or NMD factor function in one or more processes other than NMD is necessary for embryonic viability remains unclear.
Mouse
Mouse embryos completely lacking UPF1, which manifest no NMD, resorb soon after uterine implantation, and pre-plantation UPF1-null blastocysts isolated at 3.5 days post-coitum ultimately undergo apoptosis in culture [55]. The abnormal developmental processes caused by the lack of UPF1 are undoubtedly exacerbated by the failure of UPF1 to participate in non-NMD pathways such as Staufen1-mediated mRNA decay(SMD), which is in competition with NMD [12], and histone mRNA decay at the end of S phase of the cell cycle ([56] and references therein). However, data demonstrating that other NMD factors are required to preclude profound developmental defects can be used to argue for the importance of NMD. For example, SMG1-null embryos lack a vascular system and die by embryonic day 8.5, undergoing massive apoptosis and resorption [57●●]. As another example, the complete knock-out of UPF2 in hematopoietic stem cells results in stem-cell death and, after uterine implantation, lethality before embryonic-day 9.5 [58]. The conditional ablation of UPF2 expression during liver development beginning around embryonic-day 10 leads to perinatal lethality that is not due to an abnormally small liver or changes in tissue morphology but to abnormal nuclei that are arrested in mitosis and are typified by an activated DNA damage pathway [59●●].
At least in humans, while UPF1 and SMG1 have major roles in telomere maintenance and genome stability, the role of UPF2 appears to be less significant [60]. Since NMD-factor function in pathways other than NMD is probably (but not certainly) conserved between human and mouse, the less significant role of UPF2 compared to UPF1 in telomere maintenance could be used to argue that the importance of all three NMD factors to embryonic viability reflects their role in NMD per se. Nevertheless, it is difficult to tease apart the relative contributions of NMD and chromatin organization to viability considering that mouse embryonic stem cells lacking either UPF1 or UPF2 fail to form Xist RNA-mediated domains and undergo X inactivation [61]. In search of possible contributions made by NMD per se either directly or indirectly to embryonic viability, microarray analyses of cellular transcripts in primary cells that were derived from SMG1-null embryos or after the liver-specific loss of UPF2 revealed, respectively, major changes in the transcriptome and the failure of the transcriptome to undergo massive reprogramming following partial hepatectomy [57●●,59●●]. Since many of the affected transcripts appear to be NMD targets, NMD per se plays a significant role in embryonic development if not viability. However, it is difficult to imagine that NMD factor function in pathways other than NMD is not also critical for embryonic development if not viability. Besides functioning in, e.g., SMD and histone mRNA decay, UPF1, unlike UPF3X or the EJC constituent Y14, associates with the small interfering RNA-induced silencing complex and appears to promote complex binding to mRNA targets and accelerate mRNA decay [62]. Furthermore, there are likely to be other functions for NMD factors in pathways other than NMD that have yet to be discovered.
Humans
All factors shown to function in NMD in other organisms also function in NMD in humans. Of the two UPF3 paralogues, UPF3X thought to play a more significant role in NMD than UPF3 since UPF3X more efficiently binds the EJC and activates NMD in assays that directly tether either protein to mRNA at a position that resides downstream of a termination codon [14]. As noted above, NMD-factor dependence can vary depending on the particular transcript, some of which are insensitive to UPF2 downregulation and others of which are insensitive to UPF3 and/or UPF3X downregulation.
To date, the only NMD-factor deficiency known to affect humans is attributable to mutations within UPF3X. These mutations cause mild to severe X-linked mental retardation in males that is often accompanied by facial dysmorphism and other physical anomalies [63]. At least some affected males survive embryogenesis without any detectable UPF3X. They have different degrees of mental retardation depending on the amount of UPF3 [63], the levels of which are tightly upregulated by a post-transcriptional switch [10]. The existence of mental retardation in males who are deficient in functional UPF3X indicates that the degree of UPF3 upregulation is insufficient to completely compensate for the lack of UPF3X. As in mouse, NMD per se undoubtedly plays a significant role in human embryonic development if not viability.
NMD vs. NMD-factor function in other pathways: Conclusions about contributions to animal embryogenesis
We conclude that NMD factors and NMD per se are not essential for C. elegans development but NMD factors, if not NMD per se, are required not only for embryonic viability in D. melanogaster and in vertebrates but also for a normal embryogenesis. It is currently difficult to attribute specific developmental defects to a deficiency in NMD due to the roles of a number of NMD factors in one or more cellular processes in addition to NMD. Furthermore, in cases where NMD is essential for proper embryonic development, if not embryonic viability, it is difficult to know which is more important – the NMD-mediated elimination of transcripts generated in error due to either gene-derived or RNA processing defects, which could result in the production of toxic truncated proteins, or the NMD-mediated regulation of physiologic transcripts. Since NMD functions with different efficiencies in different mouse- and human-cell types [64–66] – and, not surprisingly, in different HeLa-cell strains considering their long culture time under varying conditions [67] – variabilities in the efficiency of NMD among different tissues and even the same tissue of different individual organisms will further confound determining the importance of NMD to embryogenesis in animals that are significantly higher along the evolutionary path than C. elegans.
Highlights.
> We present EJC-dependent and faux 3’-UTR mechanisms of NMD in eukaryotes.
> We highlight embryogenesis in animals: particularly worm, fly, fish, mouse and man.
> Abnormal embryonic phenotypes with particular NMD factor deficiencies are noted.
> We present evidence for NMD factor function in pathways other than NMD.
> We weight NMD vs. such non-NMD pathway roles in embryogenesis/embryo viability.
Acknowledgments
We thank Reyad Elbarbary, Chenguang Gong and Tatsuaki Kurosaki for comments on the manuscript. This work was supported by National Institutes of Health (NIH) R01 GM59614 (to L.E.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
● of special interest
●● of outstanding interest
- 1.Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem J. 2010;430:365–377. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson P, Mühlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem Soc Trans. 2010;38:1615–1620. doi: 10.1042/BST0381615. [DOI] [PubMed] [Google Scholar]
- 3.Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, Nowacki M, Serrano V, Porcel BM, Segurens B, et al. Translational control of intron splicing in eukaryotes. Nature. 2008;451:359–362. doi: 10.1038/nature06495. [DOI] [PubMed] [Google Scholar]
- 4.Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell. 2008;31:360–370. doi: 10.1016/j.molcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitrovich QM, Anderson P. mRNA surveillance of expressed pseudogenes in C. elegans. Curr Biol. 2005;15:963–967. doi: 10.1016/j.cub.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 6.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regualtion by Upf2. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.02.010. in press. [DOI] [PubMed] [Google Scholar]
- 8●.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell. 2010;143:938–950. doi: 10.1016/j.cell.2010.11.043. This paper uncovers the functional importance of ATP-hydrolysis by UPF1. During NMD, the ATP-dependent RNA helicase activity of UPF1 removes UPF1, UPF2, UPF3, SMG6 and SMG7 from the 3′-cleavage product that is generated by the SMG6 endonuclease. Removal is presumably required for this product to be accessible to degradation by the 5′-to-3′ exonuclease XRN1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maderazo AB, He F, Mangus DA, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol. 2000;20:4591–4603. doi: 10.1128/mcb.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 11.Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1015–1022. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol Cell Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17●●.Wittkopp N, Huntzinger E, Weiler C, Saulière J, Schmidt S, Sonawane M, Izaurralde E. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 2009;29:3517–3528. doi: 10.1128/MCB.00177-09. This paper reveals that some NMD factors are required for proper zebrafish embryogenesis while others, assuming their levels were sufficiently depleted, are not. Individually downregulating UPF1, UPF2, SMG5 or SMG6 or SMG7 resulted in embryonic lethality. Downregulating UPF3a individually or in combination with UPF3b resulted a milder abnormal embryonic phenotype than was observed after downregulating UPF1, UPF2, SMG5, SMG6 or SMG7, whereas individually downregulating SMG1 or UPF3b had no phenotype. In addition, downregulating UPF3a individually or in combination with UPF3b resulted in death for 19% of embryos. By using an embryonic fibroblast cell line, it was determined that NMD in zebrafish occurs when a PTC resides more than ~50–55 nucleotides upstream of an exon-exon junction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Mühlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 21●.Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell. 2010;39:396–409. doi: 10.1016/j.molcel.2010.07.004. This paper shows that during NMD the direct interaction of mRNA cap-bound CBP80 and UPF1 promotes NMD at two steps: SMG1 and UPF1 binding to eRF1 and eRF3 at a PTC to form SURF and, subsequently, the transition of SMG1 and UPF1 from SURF to UPF2-bound EJC. Additionally, UPF1 co-immunoprecipitates ~10–20-fold more efficiently with PTC-containing mRNA compared to its PTC-free counterpart in a way that is promoted by the CBP80-UPF1 interaction. Enhanced binding occurs downstream of the mRNA cap, presumably at and/or downstream of the PTC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol. 2004;11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- 24.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Gudikote JP, Olivas OR, Wilkinson MF. Boundary-independent polar nonsense-mediated decay. EMBO Rep. 2002;3:274–279. doi: 10.1093/embo-reports/kvf036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda D, Hosoda N, Kim YK, Maquat LE. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat Struct Mol Biol. 2007;14:974–979. doi: 10.1038/nsmb1297. [DOI] [PubMed] [Google Scholar]
- 27.Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen J, Brogna S. Splicing-dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO J. 2010;29:1537–1551. doi: 10.1038/emboj.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saulière J, Haque N, Harms S, Barbosa I, Blanchette M, Le Hir H. The exon junction complex differentially marks spliced junctions. Nat Struct Mol Biol. 2010;17:1269–1271. doi: 10.1038/nsmb.1890. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Krainer AR. Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components. Proc Natl Acad Sci U S A. 2007;104:11574–11579. doi: 10.1073/pnas.0704946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metzstein MM, Krasnow MA. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2006;2:e180. doi: 10.1371/journal.pgen.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 35.Longman D, Plasterk RH, Johnstone IL, Cáceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21:1075–1085. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- 37.Kerényi Z, Mérai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 2008;27:1585–1595. doi: 10.1038/emboj.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch M, Hong X, Scofield DG. NMD and the Evolution of Eukaryotic Gene Structure. In: Maquat LE, editor. Nonsense-Mediated mRNA decay. LANDES BIOSCIENCE; 2006. pp. 197–211. [Google Scholar]
- 39.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders KR, Grimson A, Anderson P. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22:641–650. doi: 10.1093/emboj/cdg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 45.Meaux S, van Hoof A, Baker KE. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol Cell. 2008;29:134–140. doi: 10.1016/j.molcel.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008 doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vicente-Crespo M, Palacios IM. Nonsense-mediated mRNA decay and development: shoot the messenger to survive? Biochem Soc Trans. 2010;38:1500–1505. doi: 10.1042/BST0381500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cali BM, Kuchma SL, Latham J, Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50●●.Avery P, Vicente-Crespo M, Francis D, Nashchekina O, Alonso CR, Palacios IM. Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA. 2011 doi: 10.1261/rna.2404211. in press. In D. melanogaster embryos, the complete absence of the UPF1 or UPF2 or loss of the UPF1-UPF2 interaction causes cellular apoptosis and, ultimately, embryonic lethality. However, the complete absence of UPF3 or loss of the UPF2-UPF3 interaction is not essential for embryonic viability. Since UPF1 and UPF2 are necessary for NMD in D. melanogaster, whereas UPF3 is not always essential, the authors conclude that certainly NMD factors, and possibly NMD itself, are essential for embryogenesis in D. melanogaster. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z, Smith KR, Batterham P, Robin C. Smg1 nonsense mutations do not abolish nonsense-mediated mRNA decay in Drosophila melanogaster. Genetics. 2005;171:403–406. doi: 10.1534/genetics.105.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54●●.Anastasaki C, Longman D, Capper A, Patton EE, Cáceres JF. Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1319. in press. This paper shows that zebrafish NAG/NBAS and DHX34, which are orthologous to nematode SMGL-1 and SMGL-2, respectively, are required for embryonic viability in zebrafish as are UPF1, SMG5 and SMG6. Furthermore, downregulating each protein results in similar abnormal embryonic phenotypes. Together with findings from [17●●], the authors conclude that NMD is essential for embryonic viability in zebrafish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 56.Koseoglu MM, Dong J, Marzluff WF. Coordinate regulation of histone mRNA metabolism and DNA replication: cyclin A/cdk1 is involved in inactivation of histone mRNA metabolism and DNA replication at the end of S phase. Cell Cycle. 2010;9:3857–3863. doi: 10.4161/cc.9.19.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57●●.McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, Mak TW. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. The complete loss of functional SMG1 at 8.5-days post-fertilization is shown to be required for the proper brain and heart development during embryogenesis in the mouse. Immortalized embryonic fibroblasts from homozygous SMG1−/− embryos exhibited slowed growth and upregulation of PTC-containing transcripts that are involved many cellular pathways, including pre-mRNA splicing, regulation of transcription and DNA repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Mönch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59●●.Thoren LA, Nørgaard GA, Weischenfeldt J, Waage J, Jakobsen JS, Damgaard I, Bergström FC, Blom AM, Borup R, Bisgaard HC, et al. UPF2 is a critical regulator of liver development, function and regeneration. PLoS One. 2010;5:e11650. doi: 10.1371/journal.pone.0011650. UPF2 is shown to be functional in NMD in mouse fetal liver based on the transcripts that are upregulated upon complete UPF2 loss. When absent in fetal liver beginning 16.5 post-fertilization, fetal-liver cells cease undergoing mitosis. The authors conclude that the NMD factor UPF2, if not NMD itself, is required for liver homeostasis during embryogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- 61.Ciaudo C, Bourdet A, Cohen-Tannoudji M, Dietz HC, Rougeulle C, Avner P. Nuclear mRNA degradation pathway(s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet. 2006;2:e94. doi: 10.1371/journal.pgen.0020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin H, Suh MR, Han J, Yeom KH, Lee Y, Heo I, Ha M, Hyun S, Kim VN. Human UPF1 participates in small RNA-induced mRNA downregulation. Mol Cell Biol. 2009;29:5789–5799. doi: 10.1128/MCB.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bateman JF, Freddi S, Nattrass G, Savarirayan R. Tissue-specific RNA surveillance? Nonsense-mediated mRNA decay causes collagen X haploinsufficiency in Schmid metaphyseal chondrodysplasia cartilage. Hum Mol Genet. 2003;12:217–225. doi: 10.1093/hmg/ddg054. [DOI] [PubMed] [Google Scholar]
- 65.Resta N, Susca FC, Di Giacomo MC, Stella A, Bukvic N, Bagnulo R, Simone C, Guanti G. A homozygous frameshift mutation in the ESCO2 gene: evidence of intertissue and interindividual variation in NMD efficiency. J Cell Physiol. 2006;209:67–73. doi: 10.1002/jcp.20708. [DOI] [PubMed] [Google Scholar]
- 66.Zetoune AB, Fontanière S, Magnin D, Anczuków O, Buisson M, Zhang CX, Mazoyer S. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 2008;9:83. doi: 10.1186/1471-2156-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense mediated decay pathway. Nucleic Acids Res. 2007;35:4542–4551. doi: 10.1093/nar/gkm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 69.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 70.Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol. 2005;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim YK, Furic L, DesGroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 72.Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L. Functional conservation of the telomerase protein Est1p in humans. Curr Biol. 2003;13:698–704. doi: 10.1016/s0960-9822(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 73.Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 74.Lovejoy CA, Cortez D. Common mechanisms of PIKK regulation. DNA Repair (Amst) 2009;8:1004–1008. doi: 10.1016/j.dnarep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chiu SY, Serin G, Ohara O, Maquat LE. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77●.Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, Izaurralde E. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes Dev. 2010;24:2440–2450. doi: 10.1101/gad.604610. Two motifs within the N-terminus of the endonuclease SMG6 that manifest sequence similarities to UPF3 and UPF3X are found to directly bind to EJCs so as to displace UPF3X (UPF3 was not tested). A point mutation within either motif inhibits not only SMG6 binding but also NMD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 79.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 80.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 82.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]