Abstract
A genome-wide association screen for primary biliary cirrhosis risk alleles was performed in an Italian cohort. The results from the Italian cohort replicated IL12A and IL12RB associations, and a combined meta-analysis using a Canadian dataset identified newly associated loci at SPIB (P = 7.9 × 10–11, odds ratio (OR) = 1.46), IRF5-TNPO3 (P = 2.8 × 10–10, OR = 1.63) and 17q12-21 (P = 1.7 × 10–10, OR = 1.38).
Primary biliary cirrhosis (PBC) is an uncommon condition (with a prevalence of ~5 per 10,000 people) of unknown etiology characterized by chronic cholestatic liver disease and, frequently, the presence of serum anti-mitochondrial antibodies (AMA). PBC's association with AMA, other autoantibodies, autoreactive T cells and HLA alleles implies an autoimmune etiology for this disease1. In addition, the association of PBC with IL12A and IL12RB2 has previously been demonstrated in a genome-wide association study (GWAS) of a Canadian PBC cohort2.
To further define the genetic factors conferring risk for PBC, we performed another GWAS in a cohort of Italian individuals with PBC (cases) and unaffected controls. We analyzed the data separately using the earlier Canadian PBC GWAS as a replication dataset and then combined the data with the Canadian GWAS in a meta-analysis. We genotyped Italian PBC cases and controls using the Illumina 610K array and Illumina 1Mb arrays, respectively. After quality control filtering and combination with additional European population controls, we performed principal components analyses, which defined a homogeneous Italian subject set encompassing 453 PBC cases and 945 controls (Supplementary Methods, Supplementary Fig. 1 and Supplementary Table 1). The Canadian PBC GWAS (which used the Illumina 370K array for genotyping) included 481 Canadian PBC cases and 3,706 population controls. We applied association methods controlling for population substructure to each dataset and used Cochran-Mantel-Haenszel adjustments when combining the separately analyzed datasets.
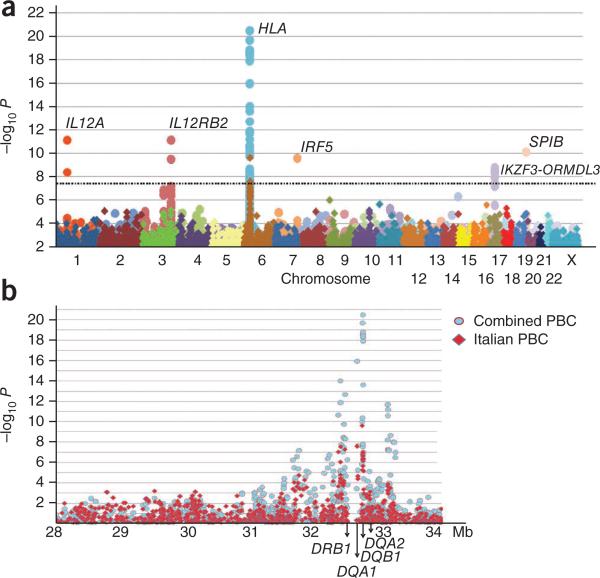
We obtained results for the homogeneous Italian subset and the meta-analysis of the entire sample set (Italian plus Canadian cohorts) (Fig. 1a). The Italian dataset showed genome-wide significant associations at the HLA region (Fig. 1a,b), with several other loci showing suggestive association signals. Analysis of the combined dataset showed multiple loci reaching conservative genome-wide P values (P < 5 × 10–8), including several newly associated loci (Table 1). For the HLA region, the SNPs showing the strongest associations nearly overlapped between the two datasets (Fig. 1b). For the Italian cases, the highest- and lowest-risk haplotypes generally corresponded with high- and low-risk HLA DRB1 alleles3 (χ2 2 × 2 contingency, P = 0.002); however, there was insufficient data for fine mapping.
Figure 1.
Results of genome-wide association tests for PBC. (a,b) The ordinate shows the level of significance for each SNP along each chromosome (a) or the HLA region (b). The Italian PBC subset (diamond symbols) and combined European dataset (circle symbols) are shown. The dashed line corresponds to P = 5 × 10–8.
Table 1.
Summary of non-MHO association tests for italian PBC and combined italian and Canadian PBC studya
| Italian |
Canadian |
Meta-analysis |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr. | Position | MAb | Gene(s) | Ctrl AFc | Case AF | P | ORd | Ctrl AF | Case AF | P | OR | P | OR |
| rs10488631 | 7 | 128,381,419 | C | IRF5-TNPO3 | 0.08 | 0.12 | 4.24 × 10-3 | 1.48 | 0.11 | 0.17 | 2.17 × 10-8 | 1.71 | 2.78 × 10-10 | 1.63 |
| rs9303277 | 17 | 35,229,995 | T | IKZF3 e | 0.45 | 0.53 | 4.01 × 10-5 | 1.41 | 0.51 | 0.58 | 2.59 × 10-5 | 1.34 | 1.69 × 10-9 | 1.38 |
| rs3745516 | 19 | 55,618,554 | A | SPIB | 0.27 | 0.36 | 1.41 × 10-5 | 1.47 | 0.23 | 0.31 | 8.88 × 10-7 | 1.45 | 7.97 × 10-11 | 1.46 |
Chr., chromosome; MA, minor allele; AF, allele frequency.
The SNP with the strongest evidence of association in the combined meta-analyses are presented for each gene and locus. Each SNP had a combined P < 10-8. See Supplementary Table 1 for IL12A and IL12RB2 data.
Minor allele base pair in forward strand.
The control minor allele frequency.
Odds ratio for the minor allele for each analysis.
SNPs for ZPBP2, GSDMB and ORMDL3 in addition to IKZF3 all showed significant association for this 17q12-21 locus.
In the combined data, IL12A and IL12RB2 were the strongest non-HLA associations, consistent with the previous Canadian GWAS (P = 3.3 × 10–10 and P = 7.5 × 10–12, respectively) (Supplementary Table 2). SNPs within SCHIP1 (located ~100 kb proximal to IL12A) also showed suggestive association with PBC (for example, rs3863075, P = 3.05 × 10–6) in the combined analysis. Independent effects in the IL12A-SCHIP1 interval were suggested by logistic regression, and risk signals from distinct haplotypes suggested that multiple and/or independently derived variations underlie the risk association in this region (Supplementary Table 3). For IL12RB2, a stronger association signal was detected in the Canadian study cohort relative to the Italian study cohort, but the P value observed for one SNP (rs3790567, P = 1.5 × 10–3) in the Italian group was among the 500 most strongly disease-associated SNPs (the most significant 0.1%) in this cohort.
Excluding the MHC region and IL12A, in the Italian cohort, 57 SNPs showed an association signal with a P < 10–5. Of these, eight SNPs reached significance (using a threshold of P < 8 × 10–4; Supplementary Methods) when the Canadian cohort was used as a replication set, and most of these SNPs had significant combined P values (Table 1). The significant SNPs included several in an ~150-kb gene-rich region on chromosome 17 containing IKZF3-ZPBP2-GSDMB-ORMDL3 that has been implicated in asthma, type 1 diabetes, Crohn's disease and ulcerative colitis4–6. A stepwise multiple logistic regression model identified a single SNP (rs907902) that could account for the observed signal. Within this region, IKZF3 (encoding IKAROS family zinc finger 3, also known as Aiolos) is a transcription factor that prevents apoptosis of IL2-deprived B cells7, regulates B-cell activation8 and has been implicated in autoimmunity; a lupus-like syndrome develops in Ikzf3-deficient mice9. However, although IKZF3 is an attractive association candidate, other genes may also account for this association signal, including ORMDL3, which has been suggested to underlie the association with asthma4.
A previously unidentified association resulting from the meta-analysis involved rs3745516 within SPIB (Table 1). A member of the ETS transcription factor family, Spi-B is an important mediator of both B-cell receptor signaling10 and early T-cell lineage decisions11. Spi-B also induces the development of plasmacytoid dendritic cells and can mediate IL7R, c-rel and IFN-γ–induced CD40 expression12. Additional studies are needed to define the causal mutation at this locus and the biological pathway connecting it to PBC predisposition.
The results from the combined data also support the association at the locus comprising IRF5 and TNPO3 (encoding interferon regulatory factor 5 and transportin 3, respectively). Although the Italian PBC cohort showed only modest association at the IRF5-TNPO3 locus (rs10488631, P = 4.2 × 10–3), the combined P value (P = 2.8 × 10–10) exceeded genome-wide significant thresholds.
Among the genes with suggestive evidence of association with PBC in the combined analysis, STAT4 (P = 1.3 × 10–5) and DENND1B (P = 1.8 × 10–4) are noteworthy because of their prior associations with autoimmune disorders. STAT4 is associated with systemic lupus erythematosus (SLE) and rheumatoid arthritis13, and its critical role in IL12-IL12R signaling makes it a strong candidate for influencing PBC risk. DENND1B has recently been identified as a susceptibility gene in pediatric asthma14. Another suggestive association signal that may merit further investigation is that for CDGAP (combined P = 1.6 × 10–7). CDGAP is a noteworthy candidate because the Cdc42 GTPase-activating protein it encodes is important in lymphocyte development and proliferation15. Other SNPs showed suggestive signals, but these signals were restricted to either the Italian or Canadian subgroup (Supplementary Table 2). Additional studies will be needed to ascertain whether these results can be replicated and whether they are restricted to a particular population subset.
The use of Illumina 610K and Illumina 1Mb arrays enabled an exploration of PBC association with mitochondrial SNPs, a potentially relevant issue in this AMA-associated disease. Although these arrays do not interrogate all mitochondrial haplogroups, the lack of disease association seen here with these SNPs (P > 0.05) suggests that variants in mitochondrial genes are unlikely to account for PBC heritability or AMA positivity. Analyses using only AMA-positive PBC cases also showed odds ratios similar to those observed with the entire set, and the odds ratios from each group showed strong correlation to each other (r2 odds ratios = 0.95), implying a similar genetic etiology regardless of AMA status (Supplementary Table 4).
Multiple genes implicated in PBC are apparently shared between different autoimmune diseases. This includes IL12A, which is shared with celiac disease; however, the SNPs and haplotypes containing the risk alleles are distinctly different for the two diseases (Supplementary Table 5). Similarly, PBC appears to be associated with at least one (the 17q12-21 locus, also shared with other autoimmune diseases) and possibly a second (DENND1B) locus implicated in pediatric asthma. For the 17q12-21 association, however, the haplotype is opposite that observed in pediatric asthma (Supplementary Fig. 2), and for DENND1B, the suggestive association also appears to be disparate from that observed with asthma. Finally, the associated SNP for IRF5-TNPO3 is within a haplotype shared with SLE and several other autoimmune diseases. In contrast, many of the other prominent associations observed in SLE, celiac disease and asthma are not apparent in PBC. Although the specific genes and variants responsible for the association signals discovered here remain to be defined, the current findings suggest specific molecular pathways as factors in the pathogenesis of PBC.
Supplementary Material
ACKNOwLEDGMENTS
We thank the subjects and physicians who contributed DNA samples and clinical data for this study. This work was supported in part by the US National Institutes of Health (R01 DK056839, R01 DK80670, K23 DK68290 and RO3 DK78527), the Canadian Institutes for Health Research (MOP 74621), the Ontario Research Fund (RE01-061), the Canadian Primary Biliary Cirrhosis Society, the American Gastroenterological Association and the A.J. and Sigismunda Palumbo Charitable Trust.
Footnotes
AUTHOR CONTRIBUTIONS
X.L., P.I., Yue Lu and R.K. contributed to initial data analyses and manuscript preparation. P.I., R.S., C.S. and A.L. managed DNA samples. A.L., F. Macciardi, A.T.L. and P.K.G. performed genotyping. M.R. provided database management. P.I., Yan Lu, I.B., M.P., C.X., G.X., A.L.M., R.P.M., K.M.P., C.N.G., F.B., M.Z., E.J.H., S.L., F.R., E.B., A.F., R.L., G.N., A.A., L. Muratori, P.M., P.L.A., P.A., M. Margotti, M.B., B.C., D.A., M.C.B., F. Marra, A. Pisano, C.R., M.C., M. Marzioni, A.B., L.F., M.S., P.P., V.O.P., C.T., L.C., S.B., S.R., M.V., C.P., A.M., P.T., A. Picciotto, A.G., C.F., S.C., G.C., L. Morini, N.C., A.C., G.S. and R.M. contributed to experimental design, subject assessment and sample collection. P.I., G.M.H., K.A.S., C.I.A., M.E.G. and M.F.S. contributed to experimental design and interpretation, statistical analyses, and initial manuscript preparation. All authors contributed to the final paper.
Note: Supplementary information is available on the Nature Genetics website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Gershwin ME, Mackay IR. Hepatology. 2008;47:737–745. doi: 10.1002/hep.22042. [DOI] [PubMed] [Google Scholar]
- 2.Hirschfield GM, et al. N. Engl. J. Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Invernizzi P, et al. Hepatology. 2008;48:1906–1912. doi: 10.1002/hep.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moffatt MF, et al. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 5.McGovern DP, et al. Nat. Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett JC, et al. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero F, Martinez AC, Camonis J, Rebollo A. EMBO J. 1999;18:3419–3430. doi: 10.1093/emboj/18.12.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JH, et al. Immunity. 1998;9:543–553. doi: 10.1016/s1074-7613(00)80637-8. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, Matthias G, Mihatsch MJ, Georgopoulos K, Matthias P. J. Immunol. 2003;170:1699–1706. doi: 10.4049/jimmunol.170.4.1699. [DOI] [PubMed] [Google Scholar]
- 10.Garrett-Sinha LA, et al. Immunity. 1999;10:399–408. doi: 10.1016/s1074-7613(00)80040-0. [DOI] [PubMed] [Google Scholar]
- 11.Dontje W, et al. Blood. 2006;107:2446–2452. doi: 10.1182/blood-2005-05-2090. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen VT, Benveniste EN. J. Biol. Chem. 2000;275:23674–23684. doi: 10.1074/jbc.M002482200. [DOI] [PubMed] [Google Scholar]
- 13.Remmers EF, et al. N. Engl. J. Med. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sleiman PM, et al. N. Engl. J. Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 15.Guo F, Velu CS, Grimes HL, Zheng Y. Blood. 2009;114:2909–2916. doi: 10.1182/blood-2009-04-214676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.