Abstract
Notch signaling plays a key role in osteoblast differentiation. A major transcriptional downstream regulator of this pathway is the Helix-Loop-Helix (HLH) transcription factor Hairy/Enhancer of Split 1 (Hes-1). Here we investigated the function of Hes-1 in osteoblastic cells. Endogenous Hes-1 gene expression decreases during progression of bone cell phenotype development in MC3T3-E1 osteoblasts suggesting that it is a negative regulator of osteoblast differentiation. Forced expression of Hes-1 inhibits osteocalcin (OC) mRNA levels, and luciferase assays indicate that Hes-1 directly represses OC promoter activity. In vitro and in vivo protein/DNA interaction assays reveal that recombinant Hes-1 binds specifically to an E-box in the proximal promoter of the OC gene. Deletion of the Hes-1 WRPW domain (MHes-1) that recruits the co-repressor Groucho abrogates repression of OC promoter activity by Hes-1, but also blocks Hes-1 binding to the promoter. The latter result suggests that exogenous Hes-1 may be recruited to the OC promoter by both protein/DNA and protein/protein interactions. We conclude that the Notch-responsive Hes-1 protein is capable of repressing OC gene transcription in osteoblastic cells through an E-box in the proximal promoter. Hes-1 may contribute to osteoblast growth and differentiation by controlling basal bone-specific transcription directly through interactions with transcriptional regulators that are known to bind to the OC gene promoter.
Keywords: Osteoblast, Bone, Osteogenesis, HLH protein, E-box, Notch signaling, Promoter
The Notch signaling cascade mediates cell fate determination during development by translating cell-cell interactions into specific transcriptional programs [Artavanis-Tsakonas et al., 1999; Bray, 2006]. This pathway is intricately modulated to control proliferation, differentiation and apoptotic events [Weinmaster, 1997; Wang et al., 2008]. In mammals, activation of Notch receptors by membrane-bound ligands initiates the release of the Notch intracellular domain (NICD), which then enters the nucleus to regulate gene expression.
Cell culture studies have indicated that the Notch pathway can stimulate or inhibit osteoblast differentiation depending on timing and context [Deregowski et al., 2006; Sciaudone et al., 2003; Tezuka et al., 2002]. Recent in vivo studies are consistent with the biological role of Notch signaling in osteoblast function and bone homeostasis; osteoblast-specific gain-of-function mutations in the Notch pathway cause severe osteosclerosis due to increased proliferation of immature osteoblasts and upregulation of genes encoding cyclin D, cyclin E and Sp7 (Osterix) [Engin et al., 2008]. However, other reports have suggested that Notch signaling may suppress osteoblast differentiation to maintain a pool of mesenchymal progenitors [Hilton et al., 2008].
Hairy and enhancer of split 1 (Hes-1) is a downstream negative regulator of Notch signaling [Jarriault et al., 1995]. Several studies have indicated that Hes-1 regulates neurogenesis, myogenesis, hematopoiesis, and sex determination [Jarriault et al., 1995; Ishibashi et al., 1995; Tan-Pertel et al., 2000; Parkhurst and Meneely, 1994]. Hes factors form homo- or heterodimers through their helix-loop-helix domains and each dimer binds DNA through the E box (CANNTG) or related sequences [Sasai et al., 1992]. A carboxy-terminal peptide (the WRPW domain) of Hes factors interacts with the co-repressor protein Groucho (or its mammalian homologues TLE/Grg) to repress transcription at target promoters [Fisher et al., 1996; Paroush et al., 1994].
Although in vivo and in vitro studies show that the Notch signaling pathway plays a critical role during osteogenesis, there are limited data on the molecular mechanisms by which Hes-1 functions as a downstream effector of Notch signaling during osteogenesis. Hes-1 stimulates the transcriptional activity of Runx2 by increasing Runx2 protein stability during osteoblast differentiation [Suh et al., 2008] and cooperates with pRb to regulate Runx2-dependent transcription [Lee et al., 2006]. Additionally, Hes-1 may activate or inhibit osteopontin gene expression depending on the experimental context [Shen and Christakos, 2005; Matsue et al., 1997]. The osteocalcin (OC) promoter contains an E-box sequence overlapping with a C/EBP binding site that acts as a principal positive element for OC gene transcription [Tamura and Noda, 1994]. Our previous studies revealed that E-boxes in the Runx2 and OC promoters recruit HLH factors including Twist1 and Twist2 [Zhang et al., 2008] as a component of an HLH related gene regulatory network. In this study, we investigate Hes-1 dependent transcriptional control during osteoblast differentiation. The main finding of our study is that recombinant Hes-1 represses osteocalcin gene transcription through multiple mechanisms and interacting with other functional regulators through its WRPW domain
Material and Methods
Transient Transfections and Luciferase Reporter Assays
ROS17/2.8 cells were grown in F-12 media supplemented with 5% fetal bovine serum. MC3T3-E1 cells were maintained in α-minimal essential media supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA). For differentiation studies, MC3T3-E1 cells were cultured in medium containing 10 mM β–glycerol phosphate and 50 µg/ml ascorbic acid and medium was replenished every second day at confluence.
Cells in 6-well plates at 70% confluence were treated with 6 µl of FuGENE 6 transfection reagent (Roche Diagnostics, Sommerville, NJ) and 1 µg of total DNA per well in accordance with the manufacturer’s protocol. The 1.1-kb rat osteocalcin gene promoter fused to the firefly luciferase reporter gene (‘OC-Luc’) [Ryoo et al., 1997] was used in transfection assays. Hes-1 responsiveness of the OC-promoter was examined by co-transfection of expression vectors for wild type Hes-1 (pCMV2-FLAG-Hes-1) or a truncated mutant Hes-1 protein that lacks the last C-terminal 6 amino acids (pCMV2-FLAG-MHes-1) (kindly provided by Stefano Stifani, McGill University, Montreal, Quebec) [McLarren et al., 2000].
Luciferase reporter plasmids, expression plasmids and the Renilla luciferase reference gene plasmid (pRLh-null-Renilla luciferase construct) were co-transfected in duplicate wells. The total amount of DNA was maintained at a constant level by adding empty vector. Cells were harvested after 24 h using 200 µl lysis buffer (Promega, Madison, WI) per well. Firefly luciferase activity in cell lysates (20 µl) was evaluated using the Dual-Luciferase reporter assay kit (Promega) and normalized to values for pRLh-null-Renilla luciferase. For all transcription studies, promoter activity (firefly/Renilla luciferase) is represented as fold induction compared to luciferase activity observed for a promoterless pGL2 construct. All transfection experiments were performed in duplicate and repeated at least three times, and representative results are shown.
RNA Isolation and Analysis
RNA was isolated from cells using TRIzol reagent (Invitrogen, Carlsbad, CA). After purification, 5 µg of total RNA was DNase treated using a DNA-free RNA column purification kit (Zymol Research, Orange, CA). RNA (1 µg) was then reverse transcribed using random hexamers as primers and the SuperScript 1st Strand Synthesis kit (Invitrogen) according to the manufacturer’s protocol. Reverse-transcriptase quantitative real-time PCR (qPCR) was used to assess the expression of a panel of genes including osteocalcin, alkaline phosphatase, Hes-1 and Runx2. Primer Express software was used to predict optimum reverse transcription-PCR primer sets (Fig. 1A), except for primers detecting glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA that were purchased from Applied Biosystems. Quantitative PCR was performed using SYBR Green 2 ×master mix (Applied Biosciences, Foster City, CA) and a two-step cycling protocol (anneal and elongate at 60°C, denature at 94°C). Specificity of primers was verified by dissociation of amplicons using SYBR Green as a detector. All transcript levels were normalized to that of Gapdh.
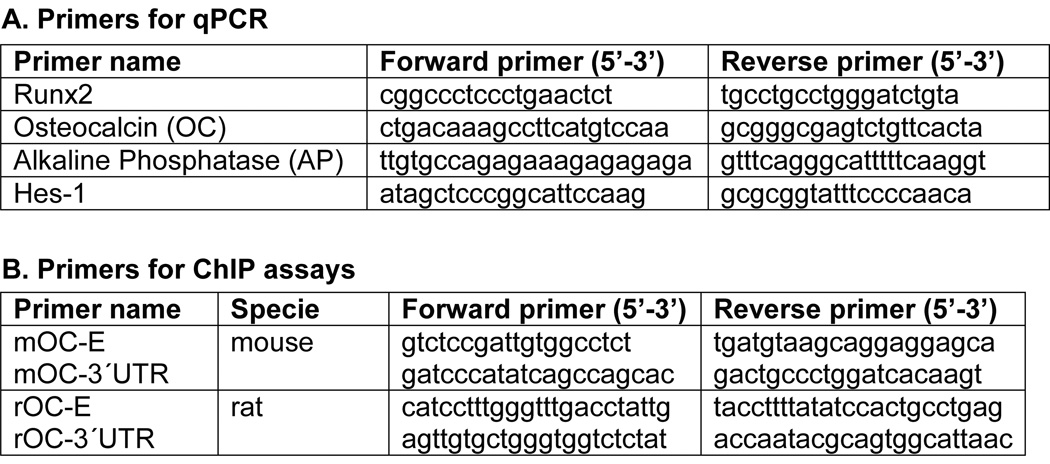
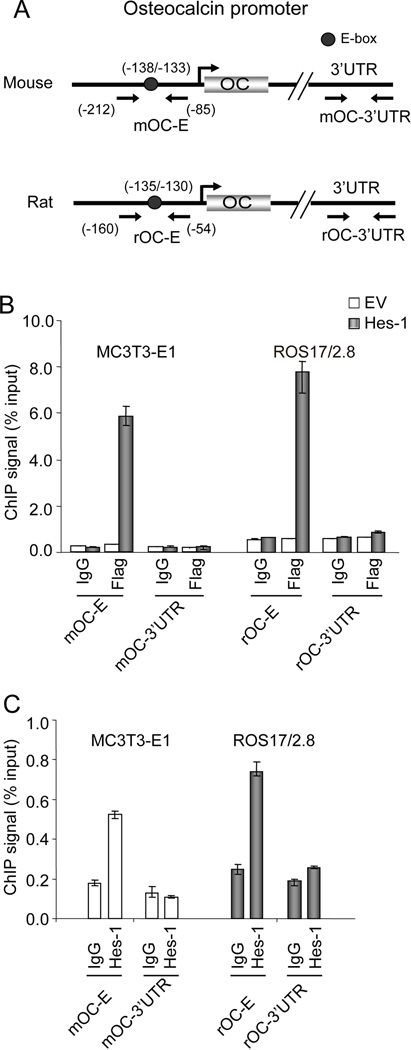
Fig. 1. Primers for quantitative PCR (qPCR) and chromatin immunoprecipitation assay (ChIP assay).
A. Primer sets used for detecting expression of bone-phenotypic genes (Runx2, osteocalcin, and alkaline phosphatase) and Hes-1 in MC3T3-E1 cells. B. Primer sets used in ChIP assays to detect transcription factor binding to mouse (mOC-E and mOC-3’UTR) and rat (rOC-E and rOC-3’UTR) osteocalcin promoters.
Chromatin Immunoprecipitation (ChIP) Assays
Chromatin Immunoprecipitation studies were performed as described previously [Zhang et al., 2009] with the following modifications. Formaldehyde cross-linking was quenched by addition of glycine to final concentration of 0.125 M at room temperature for 10 min, followed by washing with ice-cold 1×PBS before re-suspending in lysis buffer. Sonication was performed 6 times at setting 3 (model 550 sonic dismembrator, Fisher Scientific, Pittsburgh, PA) for 10 s in 1.5 ml tubes. The pre-cleared cell lysate was incubated overnight at 4°C with 3 µg of anti-flag M2 mouse antibody (Sigma), anti-Hes-1 antibodies (Santa Cruz, CA) and normal mouse or goat IgG as a negative control. After reversal of cross-links at 65°C overnight, the DNA was recovered by phenol-chloroform extraction and ethanol precipitation using 5 µg of glycogen as carrier. An aliquot of each sample was assayed using quantitative PCR.
To detect the presence of specific DNA fragments containing putative E-box motifs, primer sets that amplify the osteocalcin (OC) promoter were used (mOC-E, rOC-E; listed in Fig. 1B). Control primer pairs from 3´untranslated regions (UTR) of the OC gene were used to verify specific and non-specific binding of DNA fragments (amplified by primers including mOC-3´UTR and rOC-3´UTR) (Fig. 1B).
Electrophoretic mobility shift analysis (EMSA)
Nuclear protein preparations were prepared from MC3T3-E1 and ROS 17/2.8 cells that were cultured to 90% confluence and harvested in ice-cold PBS buffer by scraping. Cell pellets were lysed and nuclear extracts were prepared as previously described [Drissi et al., 2000]. Protein concentrations were determined by the Bradford method (Pierce Chemical Co., Rockford, IL). Wild type (WT) oligonucleotides containing the HLH motif derived in proximal osteocalcin promoter (OC-WT) (5'GGT TTG ACC TAT TGC GCA CATG ACC CCC AA3' [the E-box/HLH motif is underlined]) was end labeled with [γ-32P] ATP using T4 polynucleotide kinase (New England Biolabs, Beverly, Massachusetts) as described previously [Hassan et al., 2004]. Mutant oligonucleotides (OC-MUT, 5’GGT TTG ACC TAT TGC GCA CAg aAC CCC CAA3’ [lower case letters indicate mutant base pairs in the underlined E-box/HLH motif]) were used as competitors. Nuclear protein extracts (5 µg) were incubated for 30 min at room temperature with 1 µg of non-specific competitor DNA poly (dI·dC) (Pharmacia, Piscataway, NJ) and 80,000 cpm of labeled oligonucleotides. Competition assays were performed by mixing 100-fold molar excess unlabeled oligonucleotides with non-specific competitor DNA and probes before addition of nuclear extracts. Hes-1 antibody (2 µg; Santa Cruz, CA) was used for block shift experiments. Normal goat IgG (Santa Cruz, CA) was used as a non-specific antibody. Antibodies were mixed with nuclear protein on ice for 20 min before the addition of probe DNA. Protein–DNA complexes were resolved in a 6% acrylamide gel and visualized by autoradiography.
Immunoblotting
ROS17/2.8 cells were lysed in 50 µl lysis buffer (2% sodium dodecyl sulfate [SDS], 10 mM dithiothreitol, 10% glycerol, 12% urea, 10 mM Tris-HCl/pH 7.5, 1 mM phenylmethylsulfonyl fluoride, 1µl protease inhibitor cocktail and 25 µM MG132) and boiled for 5 min. Equal amounts of total protein were analyzed by SDS-polyacrylamide gel electrophoresis and probed with anti-flag antibody (Santa Cruz, CA). Immunocomplexes were detected using Western Lightning chemiluminescence reagent (Perkin Elmer, MA).
RESULTS
Forced expression of Hes-1 inhibits osteocalcin gene expression and transcriptional activity in a cell type-dependent manner
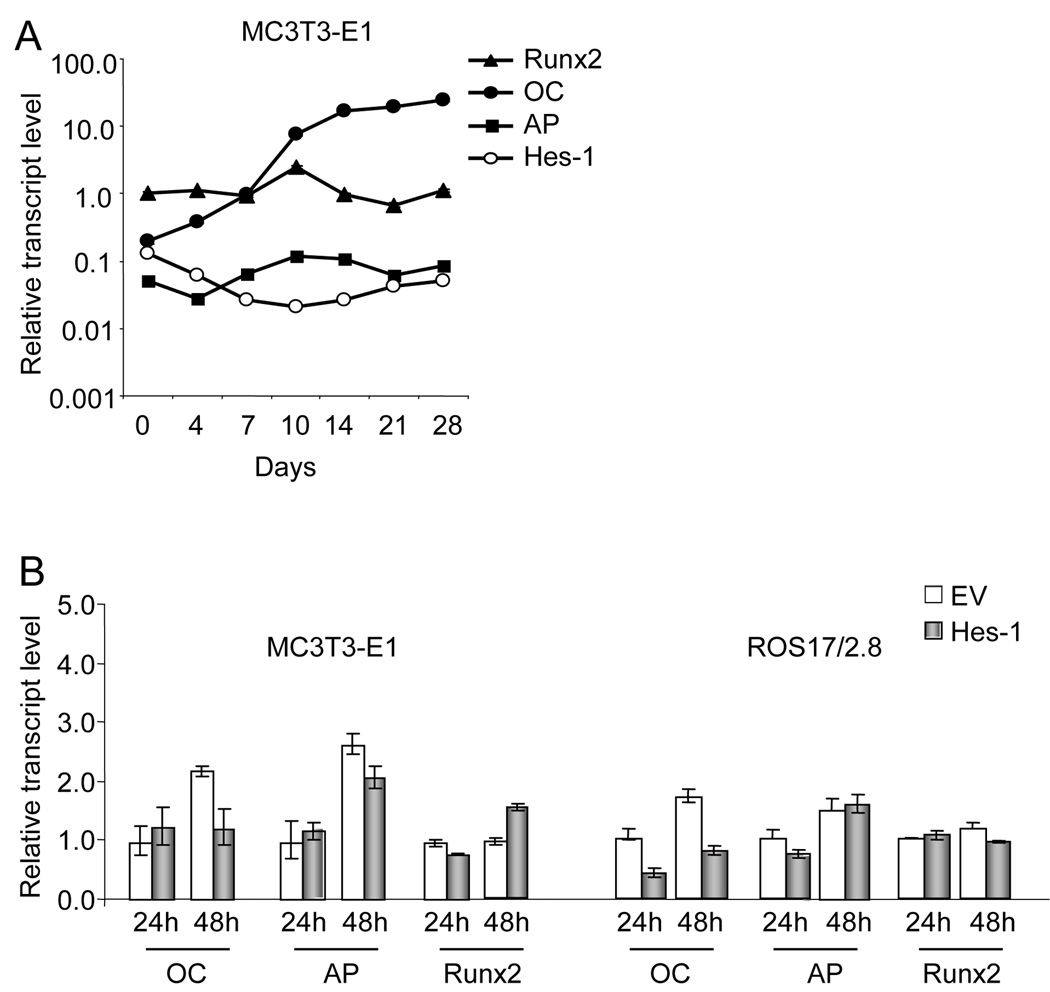
To explore the functions of Hes-1 in osteoblast differentiation, we first detected expression of Hes-1 during MC3T3-E1 differentiation. Phenotypic maturation of MC3T3-E1 osteoblasts was established by monitoring the increased expression of several osteoblast marker genes, including Runx2 (the master regulator of osteoblast maturation), alkaline phosphatase (AP; an early osteoblastic marker), and osteocalcin (OC; a late-stage marker of mature osteoblasts) (Fig. 2A). Hes-1 gene expression gradually decreases upon induction of differentiation in MC3T3-E1 (Fig. 2 A). From day 0 to day 10, Hes-1 mRNA levels are dramatically downregulated (~10 fold), while osteocalcin expression increases about 10 fold. The reduced expression of Hes-1 suggests that this transcription factor may function as a negative regulator of bone-specific gene expression during osteoblast differentiation.
Fig. 2. Regulation of Hes-1 gene expression and Hes-1 effects on bone marker gene expression in osteoblastic cells.
A. Hes-1 gene expression during MC3T3-E1 differentiation. Expression of Hes-1 and bone marker genes including Runx2, osteocalcin (OC), and alkaline phosphatase (AP) was analyzed by qPCR using RNA isolated between day 0 to day 28 of the MC3T3-E1 culture period. Expression values were normalized to values obtained for Gapdh at each time point. Runx2 expression at day 0 was arbitrarily set as 1. B. Effects of exogenous Hes-1 expression on transcript levels of OC, AP and Runx2 in MC3T3-E1 cells. MC3T3-E1 and ROS17/2.8 cells were transfected with empty vector (EV) or Hes-1 expression construct for 24 h and 48 h, and RNA was analyzed for expression of bone marker genes. Hes-1 inhibits OC transcript levels in ROS17/2.8 cells upon exogenous expression of Hes-1. Expression levels were determined by qPCR using RNA isolated 24h after transfection and expression values are the mean (± S.D.) of three independent observations after normalization based on Gapdh mRNA levels.
To investigate the functional role of Hes-1 in regulating expression of bone-phenotypic genes, Hes-1 was transiently overexpressed in MC3T3-E1 cells. Quantitative real time PCR (qPCR) analysis revealed that Hes-1 has no effect on AP mRNA levels. In immature MC3T3-E1 cells, Hes-1 does not affect OC and Runx2 gene expression at 24 h post-transfection, but Hes-1 inhibits OC gene expression 2-fold and modestly increases Runx2 gene expression by 48 h (Fig. 2B). However, in rat osteosarcoma cells (ROS17/2.8), transient transfection of Hes-1 results in a 50% decrease of OC mRNA levels after 24h transfection (Fig. 2B). The difference in results between MC3T3-E1 cells and ROS17/2.8 cells suggests that exogenous Hes-1 inhibits OC transcription through different mechanisms in distinct cell types.
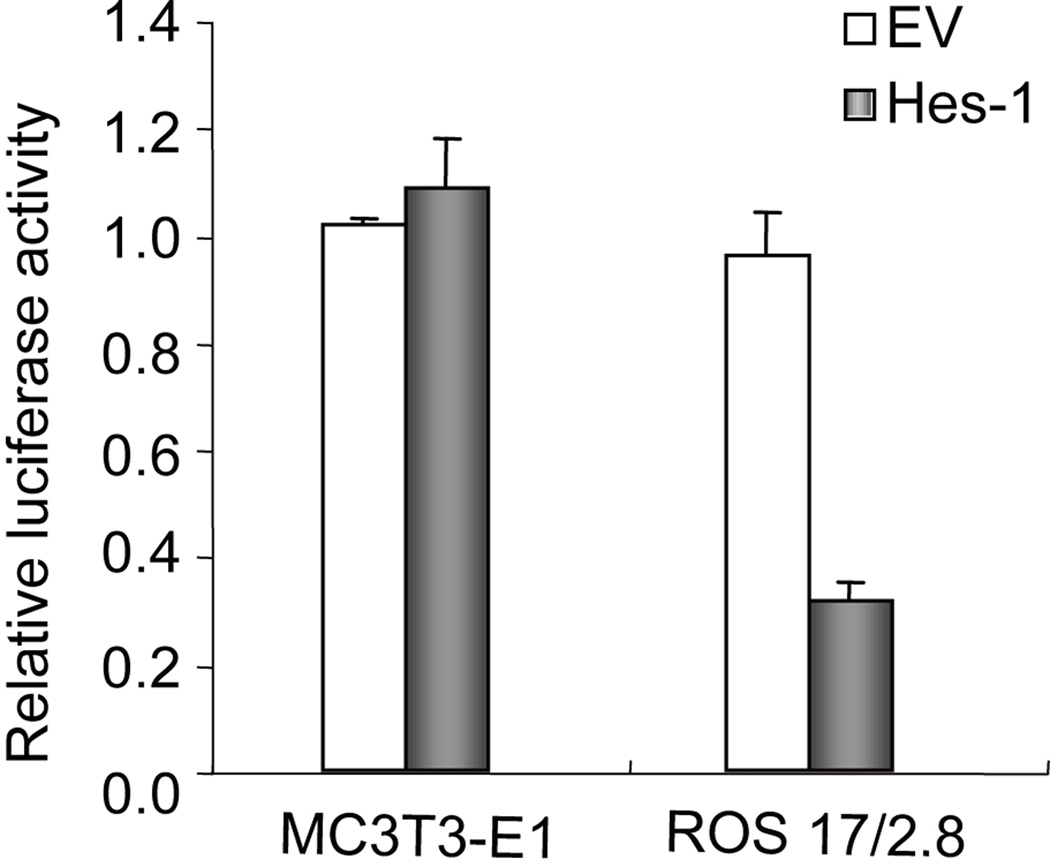
Transient reporter gene assays were performed to determine whether Hes-1 regulates OC gene expression through a promoter-dependent mechanism. MC3T3-E1 and ROS17/2.8 cells were each co-transfected for 24 h with a Hes-1 expression vector and an OC promoter/Luciferase reporter gene construct. Luciferase assays revealed that Hes-1 inhibits OC transcriptional activity by 70% in ROS17/2.8 cells (Fig. 3). However, Hes-1 does not modulate OC gene transcription in MC3T3-E1 cells at 24h post-transfection (Fig. 3), consistent with the qPCR results. Taken together, these data suggest that exogenous Hes-1 inhibits OC gene expression by both direct and indirect mechanisms.
Fig. 3. Regulation of osteocalcin (OC) promoter activity by Hes-1 in MC3T3-E1 osteoblasts and ROS17/2.8 osteosarcoma cells.
Cells were co-transfected with an OC promoter/Luciferase reporter gene construct in the presence of a Hes-1 expression vector or the corresponding empty vector. Cells were harvested with lysis buffer after 24 h and firefly luciferase activity was measured and normalized relative to Renilla luciferase activity, relative to the background expression observed for the promoter-less reporter vector (pGL2). Hes-1 inhibits OC promoter activity in ROS17/2.8 cells after 24 h transfection, but has no effect on OC promoter activity in MC3T3-E1 cells. Expression values represent the means ± S.D (n=3).
Hes-1 directly binds to the proximal E-box of the osteocalcin promoter in vitro
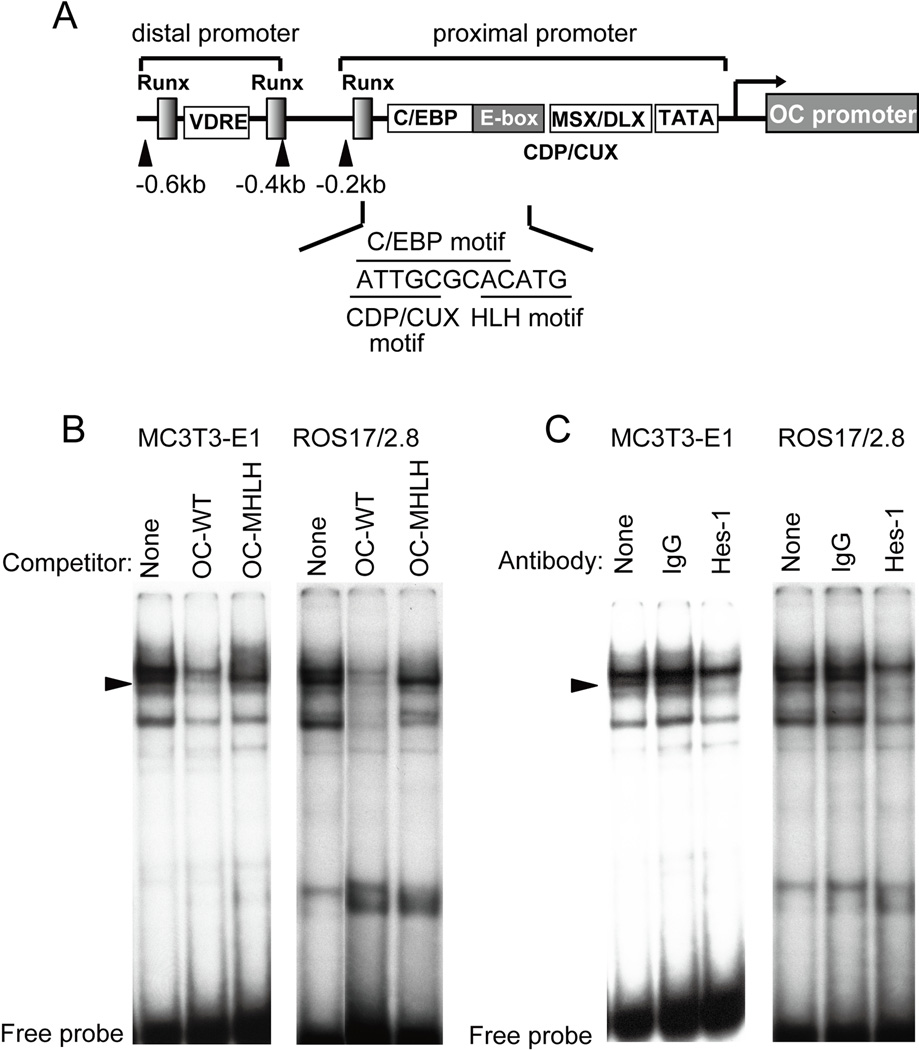
To investigate the regulatory mechanisms of OC transcription by Hes-1, we performed gel shift assays. The proximal OC promoter contains an E-box that overlaps with a C/EBP binding site (5’ATTGCGCACATG; E-box/HLH motif underlined) [Tamura and Noda, 1994] (Fig. 4A). We labeled a dsDNA oligonucleotides probe (-152/-122, OC-WT) spanning this E-box and a corresponding mutant oligonucleotide (OC-MHLH) in which the two of the six E-box nucleotides (5´ CACATG 3´) were altered (5´ CACAga 3´). Nuclear extracts were prepared from MC3T3-E1 and ROS17/2.8 cells in which Hes-1 was transiently over-expressed for 24 h. Competition assays using the wild type probe (OC-WT) show that Hes-1 related complexes (indicated by arrowheads) disappear using unlabeled wild type competitor (OC-WT), but not with corresponding mutant nucleotide (OC-MHLH) (Fig. 4B). These results indicate that the shifted complexes specifically require the integrity of the E-box motif, and thus that the factors mediating these complexes are related to helix-loop-helix factors.
Fig. 4. Hes-1 interacts with the HLH motif in the proximal OC promoter in MC3T3-E1 and ROS17/2.8 cells.
A. Diagram of the rat OC gene promoter. The OC promoter contains distal Vitamin D responsive enhancer and Runx motifs and a proximal bone-tissue specific promoter that contains interaction motifs for C/EBPβ, Msx2, Dlx5 and CDP/Cux1 [Hassan et al., 2004; Gutierrez et al., 2002; van Gurp et al., 1999] as indicated. The E-box in the OC promoter overlaps with the C/EBP binding site and is adjacent to a CAAT-like motif that interacts with CDP/Cux1. B. Gel shift competition assays were performed with labeled wild type OC probe (OC-WT), which spans the HLH motif, and unlabeled wild type or mutant oligos in which the HLH motif was mutated (OC-MHLH). The unlabeled OC-WT oligo but not the OC-MHLH oligo probe inhibited binding of E box related protein/DNA complexes (indicated by black arrows). C. Gel shift immuno-assays were performed with the same wild type probe using non-specific antibody (normal IgG) or a Hes-1 specific antibody. Immunoreactivity by the latter antibody is reflected by a block shift. Nuclear extracts used in Panels B and C were prepared from cells in which Hes-1 was over-expressed for 24 h.
To determine whether Hes-1 mediates formation of the shifted complexes, we used Hes-1 antibody in gel shift immunoassays. In lysates from cells exogenously expressing Hes-1, the Hes-1 antibody caused a clear reduction of HLH related bands (Fig. 4C). We did not observe a reduction in shifted bands using normal IgG with either MC3T3-E1 or ROS17/2.8 nuclear extracts. The in vitro gel shift results indicate that recombinant Hes-1 is capable of directly binding to the OC promoter.
Hes-1 is recruited to the OC gene promoter in vivo
Chromatin immunoprecipitation (ChIP) assays were performed to investigate whether Hes-1 directly binds to the OC promoter to regulate OC transcription in vivo. Two sets of primers were designed to determine the occupancy of Hes-1 on the OC promoter. Primers (mOC-E in MC3T3-E1; rOC-E in ROS17/2.8) were designed to amplify the proximal osteocalcin gene promoter region which contains the E-box motif (Fig. 5A), and the 3’ untranslated regions (UTR) of the OC genes to detect non-specific binding of DNA fragments (mOC-3´UTR; rOC-3´UTR). After overexpression of a Flag-tagged Hes-1 protein (for 24 h) in MC3T3-E1 and ROS17/2.8 cells, ChIP assays were performed with a Flag-antibody or normal IgG as a negative control. We observed a strong binding signal for the proximal OC promoter including the E-box in cells expressing Hes-1 but not in cells transfected with empty vector. In both MC3T3-E1 and ROS17/2.8 cells, there was no specific binding to the 3’UTR of the OC gene that served as negative controls (Fig. 5B). Having established that exogenous epitope-tagged Hes-1 is recruited to the OC promoter, we also examined interactions of endogenous Hes-1 with the OC gene (Fig. 5C). The data indicate that endogenous Hes-1 occupies the OC promoter in both MC3T3-E1 and ROS17/2.8 cells (Fig. 5C). These data suggest that Hes-1 regulates OC transcription by binding to the proximal OC promoter.
Fig. 5. In vivo interaction of Hes-1 with the genomic OC promoter.
A. Diagram of the mouse and rat OC genes and the primer pairs used to amplify the proximal promoters or 3’UTRs (negative control). The location of the E-box is indicated by a circle. B and C. Chromatin immunoprecipitation (ChIP) assays reveal that Hes-1 binds to the E-box containing proximal promoter of the OC promoter in MC3T3-E1 and ROS17/2.8 cells. In Panel B, binding of Hes-1 is examined with chromatin isolated from cells in which either empty vector (EV) or Flag-epitope tagged Hes-1 was transfected. Interactions with the OC gene promoter were examined in chromatin immunoprecipitation reactions obtained using 3 µg of Flag antibody or normal IgG (as a negative control). In Panel C, binding of endogenous Hes-1 to OC promoter was detected using 3 µg of Hes-1 antibody or the normal IgG. The ChIP data presented are representative of independent three experiments.
Transcriptional repression of osteocalcin by Hes-1 depends on its WRPW motif in ROS 17/2.8
Our data show that Hes-1 can bind to the E-box of the OC promoter in vitro (Fig. 4) and the OC gene locus in vivo (Fig. 5), and this interaction is capable of acutely repressing OC gene transcription in ROS17/2.8 cells (Fig. 2 and 3). A combination of direct and indirect mechanisms may account for the delayed responsiveness of OC gene expression to Hes-1 levels in MC3T3-E1 cells. Transcriptional repression modulated by Hes-1 is thought to occur at least partly by recruitment of co-repressor proteins, Groucho/transducin-like Enhancer of split (Gro/TLE) via the WRPW tetra-peptide motif conserved in the C terminus of all family members [Giebel and Campos-Ortega, 1997; Wainwright and Ish-Horowicz, 1992]. To investigate whether the WRPW motif of Hes-1 is involved in transcriptional repression of the osteocalcin gene, we compared the functional effects of exogenously expressing wild type Hes-1 and a mutant Hes-1 protein (MHes-1) in which the WRPW motif was deleted.
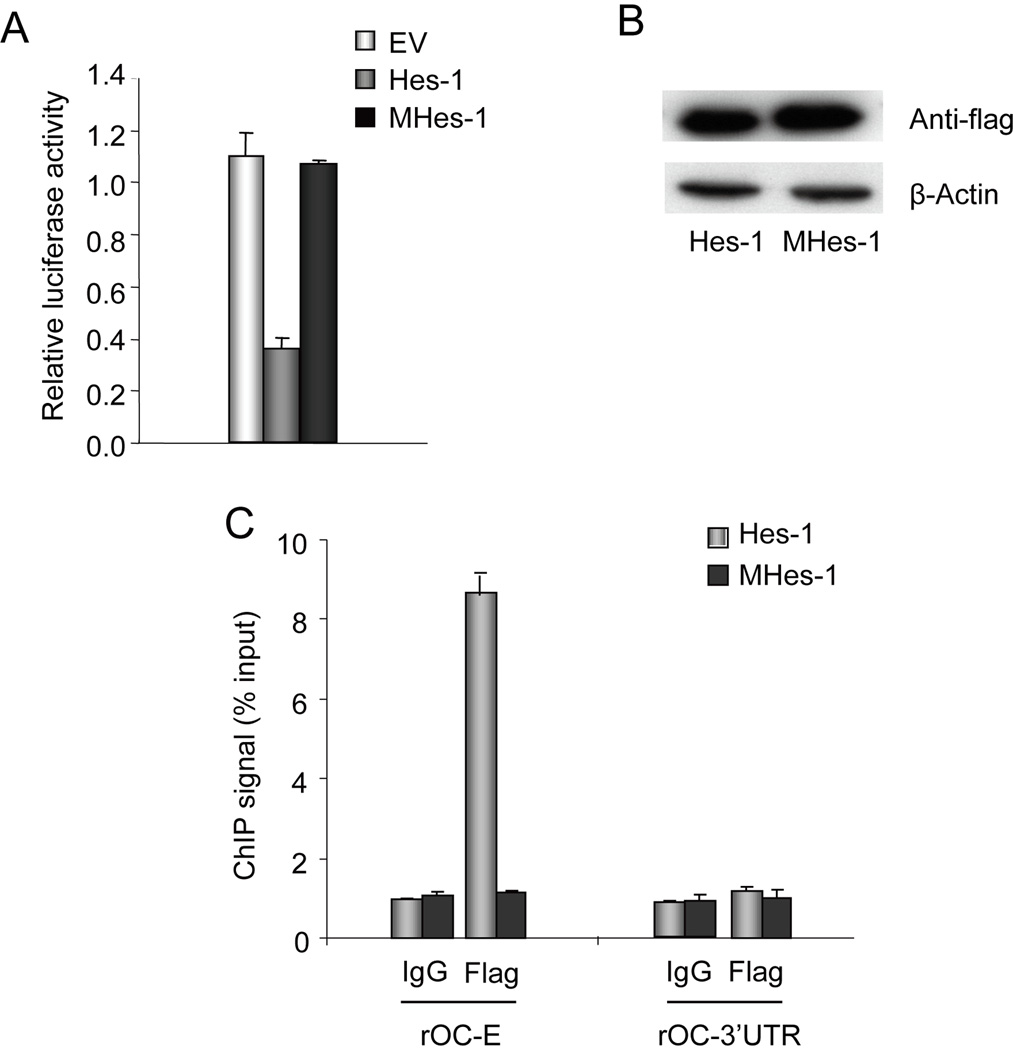
We monitored luciferase activity in ROS17/2.8 cells upon co-transfection for 24 h of the OC promoter with either wild type Hes-1 or the MHes-1 mutant protein. We determined by western blot analysis that Hes-1 and MHes-1 are expressed at similar levels (Fig. 6B). Luciferase assays reveal that expression of Hes-1 inhibits promoter activity by 60%, but expression of MHes-1 has no effect (Fig. 6A). Hence, the WRPW motif of Hes-1 is essential for repression of OC gene transcription by Hes-1. We then performed ChIP assays to assess whether transiently expressed wild type Hes-1 or mutant MHes-1 are recruited to the OC promoter. Strikingly, we detected a 90% decrease in the binding of the mutant MHes-1 to the proximal OC promoter compared to cells over-expressing wild type Hes-1 (Fig. 6C). We conclude that while Hes-1 can directly bind to the OC promoter in vitro (Fig. 4), both the repression of OC transcription by Hes-1 and its physical association in vivo with the genomic OC promoter depend on a functional WRPW motif in Hes-1. The latter finding suggests that Hes1 may interact with the OC gene promoter through both protein/protein and protein/DNA interactions.
Fig. 6. The WRPW motif of Hes-1 (MHes-1) is required for suppression of OC gene transcription.
A. Co-transfection assays were performed with the OC promoter/Luciferase reporter gene construct and either wild type Hes-1 or mutant Hes-1 (MHes-1) in which the WRPW motif is deleted. Firefly luciferase activity was measured after 24 h and normalized to Renilla luciferase activity. Wild type Hes-1 but not MHes-1 inhibits OC promoter activity in ROS17/2.8. B. Western blot analysis shows that both wild type Hes-1 and mutant Hes-1 (MHes-1) are expressed at similar levels. C. Chromatin immunoprecipitations were performed to examine whether wild type Hes-1 or mutant Hes-1 (MHes-1) are recruited to the OC promoter in ROS 17/2.8 cells transfected with expression constructs for either protein. ChIP assays were performed with anti-flag antibody or normal IgG. Primer sets to amplify the OC promoter or the 3’UTR are described in Figure 4. The results represent the means ± S.D. of triplicate samples.
DISCUSSION
In vitro and in vivo studies have demonstrated that Notch signaling is critical for bone development. In this study, we have obtained data indicating that Hes-1, a key downstream target of Notch signaling, attenuates osteocalcin (OC) gene expression in osteoblastic cells. Exogenous Hes-1 indirectly inhibits osteocalcin (OC) gene expression in MC3T3-E1 cells, while the factor may directly repress OC transcription in ROS17/2.8 osteosarcoma cells through binding to the proximal OC promoter and/or by indirect protein/protein interaction mechanisms that depends on the WRPW domain of Hes-1.
Notch intracellular domain (NICD) has been reported to bind directly to Runx2 and to repress trans-activation of type X collagen and OC promoters [Engin et al., 2008]. In the nucleus, NICD interacts with the DNA-binding protein CSL to induce the expression of Hes-1 or Hes-5 [Artavanis-Tsakonas et al., 1999; Ohtsuka et al., 1999] or Hes-3 [Iso et al., 2001]. Other studies have reported that Hes-1 may regulate osteopontin (OP) gene expression and increase Runx2 protein stability depending on the experimental context [Suh et al., 2008; Lee et al., 2006; Shen and Christakos, 2005]. Our study shows that over-expression of Hes-1 has no effects on OP expression (data not shown) in MC3T3-E1 cells. However, we observed that forced expression of Hes-1 inhibits OC transcription through both direct and indirect mechanisms. Notch signaling stimulates osteoblast proliferation, but inhibits early osteoblast differentiation by regulating bone marker genes such as Runx2 expression. Our data suggest that Notch signaling may regulate bone marker gene expression through multiple mechanisms and Hes-1 may play an important regulatory role by attenuating late osteoblast differentiation and by restricting OC gene expression as well as expression of other bone-phenotypic genes to mature osteoblasts.
Hes-1 is expressed in almost all undifferentiated cells. Elevated expression of Hes-1 blocks differentiation of many cell types, and Hes-1-deficient mice show premature differentiation and severe defects in various tissues such as the brain, eye, and pancreas [Ishibashi et al., 1995; Jensen et al., 2000; Tomita et al., 1996]. In addition, Hes-1 is important for the generation of multiple cell types from common precursors. Hes-1 may function as a cell determining factor in part by maintaining precursor pools at developmental stages when distinct differentiation cues are present or alternatively, by promoting differentiation of phenotypically distinct cell types. Our results show that Hes-1 expression decreased approximately 10 fold during osteoblast differentiation, consistent with the concept that Hes-1 negatively regulates bone phenotypic maturation by permitting the expansion of committed osteoprogenitors and/or supporting the progression of bone cell differentiation.
Because forced expression of the dominant negative HLH protein Id-1 decreases OC gene promoter activity in ROS17/2.8 cells, it has been proposed that the proximal E-box in the OC promoter is a positive element [Tamura and Noda, 1994]. Our in vivo and in vitro protein/DNA interaction data show that exogenous Hes-1 can be recruited to the OC promoter in MC3T3 presumably through the E-box and perhaps WRPW dependent protein/protein interactions. However, forced expression of Hes-1 inhibits OC gene expression after 48 h. Our previous studies revealed that the HLH proteins Twist1 and Twist2, which are also important HLH factors in osteogenesis, are also capable of repressing OC transcription. In contrast to Hes-1, the effects of Twist1 and Twist2 are indirect because these proteins do not appear to be recruited to the E-box in the OC promoter [Zhang et al., 2008]. Taken together, we propose that binding of Hes-1 to the functional E-box of the OC gene promoter may cooperate with a number of factors including other HLH factors (e.g., Twist1 and Twist2) or other transcriptional regulators (e.g., C/EBPβ, Dlx5, Dlx3, Msx2, Cux1 or Runx2), and that the combined output of these interactions determines the basal levels of OC gene transcription.
Hes-1 modulates target gene transcription not only through binding to the E-box of gene promoters, but also by interacting with the co-repressor protein TLE/Grg/Groucho through its WRPW domain [Wainwright and Ish-Horowicz, 1992]. Our data show that in ROS17/2.8 cells, forced expression of Hes-1 inhibits OC gene expression, but deletion of the WRPW domain in Hes-1 (MHes-1) abrogates repression of OC promoter activity and mutant Hes-1 cannot bind to the genomic OC promoter in the context of chromatin. Because the WRPW motif is essential for TLE/Grg/Groucho binding and is not known to affect DNA binding, it appears that the interaction of these co-repressors through the WRPW motif is required for Hes-1 binding to the OC promoter in vivo to repress its transcription. Apart from direct binding of Hes-1 to the E-box, Hes-1 may form protein/protein bridges through other transcription factors bound to the OC promoter [McLarren et al., 2000]. Specifically, the Runt-related transcription factor Runx2 has a VWRPY motif that recruits TLE/Groucho [Javed et al., 2000] and perhaps interactions of Runx2 and Hes-1 with the OC promoter may be stabilized by TLE/Groucho.
In conclusion, our study shows that the Notch-responsive Hes-1 protein can inhibit transcription of the bone-specific osteocalcin gene, consistent with the biological role of Notch signaling in controlling skeletal development and bone mass in mammalian species.
ACKNOWLEDGMENTS
Contract grant sponsor: NIH; Contract grant number: R01 AR039588 (Stein GSS) and R37 DE012528 (JBL). The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
We thank the members of our laboratories for stimulating discussions and technical advice. We thank Jonathan Gordon for critical reading of the manuscript and Judy Rask for assistance with manuscript preparation.
REFERENCES
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Deregowski V, Gazzerro E, Priest L, Rydziel S, Canalis E. Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone morphogenetic protein signaling. J Biol Chem. 2006;281:6203–6210. doi: 10.1074/jbc.M508370200. [DOI] [PubMed] [Google Scholar]
- Drissi H, Luc Q, Shakoori R, Chuva de Sousa Lopes S, Choi J-Y, Terry A, Hu M, Jones S, Neil JC, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J Cell Physiol. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, Boyce BF, Lee B. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel B, Campos-Ortega JA. Functional dissection of the Drosophila enhancer of split protein, a suppressor of neurogenesis. Proc Natl Acad Sci U S A. 1997;94:6250–6254. doi: 10.1073/pnas.94.12.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez S, Javed A, Tennant D, van Rees M, Montecino M, Stein GS, Stein JL, Lian JB. CCAAT/enhancer-binding proteins (C/EBP) β and δ Activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem. 2002;277:1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Iso T, Sartorelli V, Poizat C, Iezzi S, Wu HY, Chung G, Kedes L, Hamamori Y. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol Cell Biol. 2001;21:6080–6089. doi: 10.1128/MCB.21.17.6080-6089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Javed A, Guo B, Hiebert S, Choi J-Y, Green J, Zhao S-C, Osborne MA, Stifani S, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Groucho/TLE/R-Esp proteins associate with the nuclear matrix and repress RUNX (CBFα/AML/PEBP2α) dependent activation of tissue-specific gene transcription. J Cell Sci. 2000;113:2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Lee JS, Thomas DM, Gutierrez G, Carty SA, Yanagawa S, Hinds PW. HES1 cooperates with pRb to activate RUNX2-dependent transcription. J Bone Miner Res. 2006;21:921–933. doi: 10.1359/jbmr.060303. [DOI] [PubMed] [Google Scholar]
- Matsue M, Kageyama R, Denhardt DT, Noda M. Helix-loop-helix-type transcription factor (HES-1) is expressed in osteoblastic cells, suppressed by 1,25(OH)2 vitamin D3, and modulates 1,25(OH)2 vitamin D3 enhancement of osteopontin gene expression. Bone. 1997;20:329–334. doi: 10.1016/s8756-3282(97)00005-7. [DOI] [PubMed] [Google Scholar]
- McLarren KW, Lo R, Grbavec D, Thirunavukkarasu K, Karsenty G, Stifani S. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor cbfa1. J Biol Chem. 2000;275:530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst SM, Meneely PM. Sex determination and dosage compensation: lessons from flies and worms. Science. 1994;264:924–932. doi: 10.1126/science.8178152. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Ryoo H-M, Hoffmann HM, Beumer TL, Frenkel B, Towler DA, Stein GS, Stein JL, van Wijnen AJ, Lian JB. Stage-specific expression of Dlx-5 during osteoblast differentiation: involvement in regulation of osteocalcin gene expression. Mol Endocrinol. 1997;11:1681–1694. doi: 10.1210/mend.11.11.0011. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Sciaudone M, Gazzerro E, Priest L, Delany AM, Canalis E. Notch 1 impairs osteoblastic cell differentiation. Endocrinology. 2003;144:5631–5639. doi: 10.1210/en.2003-0463. [DOI] [PubMed] [Google Scholar]
- Shen Q, Christakos S. The vitamin D receptor, Runx2, and the Notch signaling pathway cooperate in the transcriptional regulation of osteopontin. J Biol Chem. 2005;280:40589–40598. doi: 10.1074/jbc.M504166200. [DOI] [PubMed] [Google Scholar]
- Suh JH, Lee HW, Lee JW, Kim JB. Hes1 stimulates transcriptional activity of Runx2 by increasing protein stabilization during osteoblast differentiation. Biochem Biophys Res Commun. 2008;367:97–102. doi: 10.1016/j.bbrc.2007.12.100. [DOI] [PubMed] [Google Scholar]
- Tamura M, Noda M. Identification of a DNA sequence involved in osteoblast-specific gene expression via interaction with helix-loop-helix (HLH)-type transcription factors. J Cell Biol. 1994;126:773–782. doi: 10.1083/jcb.126.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Pertel HT, Walker L, Browning D, Miyamoto A, Weinmaster G, Gasson JC. Notch signaling enhances survival and alters differentiation of 32D myeloblasts. J Immunol. 2000;165:4428–4436. doi: 10.4049/jimmunol.165.8.4428. [DOI] [PubMed] [Google Scholar]
- Tezuka K, Yasuda M, Watanabe N, Morimura N, Kuroda K, Miyatani S, Hozumi N. Stimulation of osteoblastic cell differentiation by Notch. J Bone Miner Res. 2002;17:231–239. doi: 10.1359/jbmr.2002.17.2.231. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- van Gurp MF, Pratap J, Luong M, Javed A, Hoffmann H, Giordano A, Stein JL, Neufeld EJ, Lian JB, Stein GS, van Wijnen AJ. The CCAAT displacement protein/cut homeodomain protein represses osteocalcin gene transcription and forms complexes with the retinoblastoma protein-related protein p107 and cyclin A. Cancer Res. 1999;59:5980–5988. [PubMed] [Google Scholar]
- Wainwright SM, Ish-Horowicz D. Point mutations in the Drosophila hairy gene demonstrate in vivo requirements for basic, helix-loop-helix, and WRPW domains. Mol Cell Biol. 1992;12:2475–2483. doi: 10.1128/mcb.12.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Y, Banerjee S, Sarkar FH. Exploitation of the Notch signaling pathway as a novel target for cancer therapy. Anticancer Res. 2008;28:3621–3630. [PubMed] [Google Scholar]
- Weinmaster G. The ins and outs of notch signaling. Mol Cell Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hassan MQ, Li ZY, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Intricate gene regulatory networks of helix-loop-helix (HLH) proteins support regulation of bone-tissue related genes during osteoblast differentiation. J Cell Biochem. 2008;105:487–496. doi: 10.1002/jcb.21844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hassan MQ, Xie RL, Hawse JR, Spelsberg TC, Montecino M, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Co-stimulation of the bone-related Runx2 P1 promoter in mesenchymal cells by SP1 and ETS transcription factors at polymorphic purine-rich DNA sequences (Y-repeats) J Biol Chem. 2009;284:3125–3135. doi: 10.1074/jbc.M807466200. [DOI] [PMC free article] [PubMed] [Google Scholar]