Abstract
DNA double stranded breaks (DSBs) are generated by the RAG endonuclease in all developing lymphocytes as they assemble antigen receptor genes1. DNA cleavage by RAG occurs only at the G1-phase of the cell cycle and generates two hairpin-sealed DNA (coding) ends that require nucleolytic opening prior to their repair by classical non-homologous end-joining (NHEJ)1–3. Although there are several cellular nucleases that could perform this function, only the Artemis nuclease is able to do so efficiently2, 3. Here we show, in vivo, that the histone protein H2AX prevents nucleases other than Artemis from processing hairpin-sealed coding ends; in the absence of H2AX, CtIP can efficiently promote the hairpin opening and resection of DNA ends generated by RAG cleavage. This CtIP-mediated resection is inhibited by γ-H2AX and by MDC-1, which binds to γ-H2AX in chromatin flanking DNA DSBs. Moreover, the ATM kinase activates antagonistic pathways that modulate this resection. CtIP DNA end resection activity is normally limited to cells at post-replicative stages of the cell cycle where it is essential for homology-mediated repair4, 5. In G1-phase lymphocytes, DNA ends that are processed by CtIP are not efficiently joined by classical NHEJ and the joints that do form frequently use micro-homologies and exhibit significant chromosomal deletions. Thus, H2AX preserves the structural integrity of broken DNA ends in G1-phase lymphocytes thereby preventing these DNA ends from accessing repair pathways that promote genomic instability.
V(D)J recombination, the reaction that assembles the second exon of all antigen receptor genes, requires the generation and repair of DNA DSBs and occurs exclusively during the G1-phase of the cell cycle1. The Rag-1 and Rag-2 proteins, which together form an endonuclease referred to as RAG, initiate V(D)J recombination by introducing DNA DSBs at the border of two gene segments and their associated RAG recognition sequences (recombination signals, RSs)1. Cleavage by RAG generates a pair of hairpin-sealed coding ends and a pair of blunt signal ends1. These distinct pairs of DNA ends are processed and joined by NHEJ to form a coding joint and signal joint, respectively2, 3. Signal ends undergo minimal nucleolytic processing prior to joining. In contrast, hairpin-sealed coding ends must first be opened by an endonculease and are also frequently processed by exonucleases prior to joining2, 3. This nucleolytic processing results in antigen receptor gene sequence diversification that is essential for adaptive immunity.
Efficient opening of the hairpin-sealed coding ends generated by RAG cleavage is dependent on the Artemis nuclease2, 3. Although other cellular nucleases have enzymatic activity that could open and resect hairpin-sealed coding ends, such functional redundancy is not evident in Artemis-deficient cells; as a result, Artemis-deficient mice and humans are severely lymphopenic3. Thus, the nucleolytic processing of broken DNA ends in G1-phase lymphocytes must be tightly regulated; however, the cellular components that mediate this regulation are not known. Unrepaired coding ends are resolved as chromosomal translocations at a higher frequency in lymphocytes deficient in both Artemis and H2AX as compared to those with an isolated deficiency of Artemis6. As the formation of these translocations requires the Artemis-independent opening of these hairpin-sealed DNA ends, we considered that H2AX may function to restrict the ability of nucleases to act on broken DNA ends in G1-phase lymphocytes.
To test this notion, we generated pre-B cell lines transformed with the v-abl kinase (hereafter referred to as abl pre-B cells), each of which contain a single integrant of the pMX-DELCJ retroviral recombination substrate and are wild-type (WT:DELCJ), deficient in Artemis (Artemis−/−:DELCJ) or deficient in both Artemis and H2AX (Artemis−/−:H2AX−/−:DELCJ)6–8. pMX-DELCJ has a single pair of RSs and undergoes rearrangement by deletion resulting in a coding joint that remains within the chromosomal context (Fig. 1a)7. Treatment of abl pre-B cells with the v-abl kinase inhibitor, STI571, leads to G1 cell cycle arrest and induction of RAG, which in NHEJ-deficient cells results in the accumulation of unrepaired coding ends6–9. In G1-arrested Artemis−/−:DELCJ abl pre-B cells, these unrepaired coding ends are homogenous in size as expected given their hairpin-sealed structure (Fig. 1 b and Supplementary Figs. 1 and 2). In contrast, coding ends in G1-arrested Artemis−/−:H2AX−/−:DELCJ abl pre-B cells are heterogeneous in size and significantly smaller (up to 1kb) suggesting that the hairpin-sealed coding ends have been opened and resected in an Artemis-independent fashion (Fig. 1b and Supplementary Fig. 1 and 2). Importantly, H2AX also regulates coding end resection in DNA ligase IV-deficient (Lig IV−/−) abl pre-B cells where hairpins can be efficiently opened by Artemis but cannot be ligated (Supplementary Fig. 3). Moreover, blunt chromosomal signal ends generated by RAG cleavage at the pMX-DELSJ retroviral substrate also exhibit significant resection in Lig IV−/−:H2AX−/−:DELSJ abl pre-B cells (Supplementary Fig. 4)7.
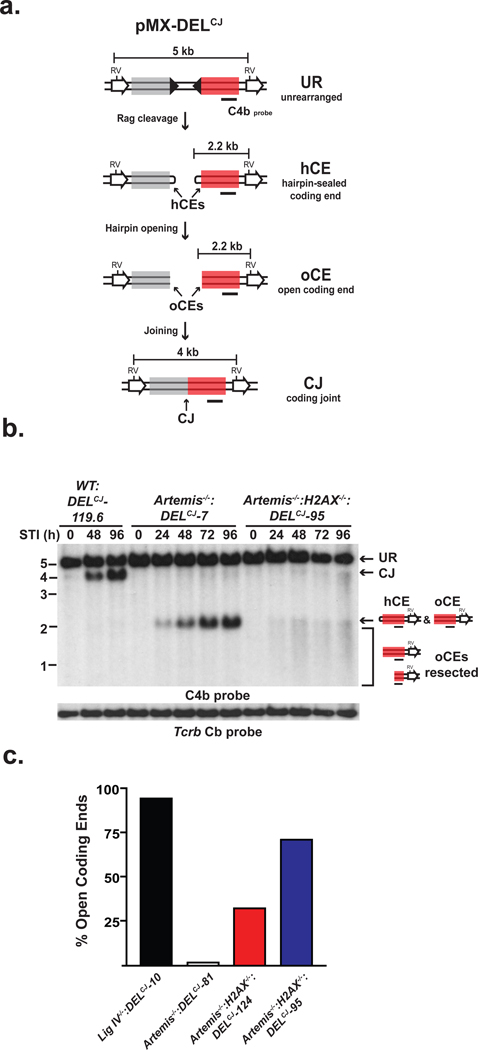
Figure 1. H2AX inhibits DNA end resection.
(a) pMX-DELCJ retroviral recombination substrate un-rearranged (UR), coding ends (CEs) that are hairpin-sealed (hCE) or open (oCE), and coding joint (CJ). RSs (filled triangles), EcoRV sites (RV), the C4b probe (black bar) and fragment sizes are shown. (b) Southern blot analysis of EcoRV-digested genomic DNA from wild-type (WT:DELCJ), Artemis−/−:DELCJ and Artemis−/−:H2AX−/−:DELCJ abl pre-B cells treated with STI571 (STI, hours). C4b hybridizing UR, CJ and hCEs are indicated as are oCEs that have been resected (bracket). Tcrb Cb hybridization is a loading control. (c) Quantification of denaturing Southern blot analyses for open CEs.
To demonstrate that hairpin-sealed coding ends have been opened, DNA end structure was assayed by TdT-assisted PCR, which can detect hairpin-opened but not hairpin-sealed coding ends10 (Supplementary Fig. 5a). In this regard, analysis of Artemis−/−:H2AX−/−:DELCJ abl pre-B cells revealed robust TdT-assisted PCR products indicating that many of the hairpin-sealed coding ends have been opened (Supplementary Fig. 5c). To quantify the fraction of open coding ends in Artemis−/−:H2AX−/−:DELCJ abl pre-B cells, we carried out Southern blot analyses of native and denatured genomic DNA. Denaturing hairpin-opened coding ends dissociates the complementary DNA strands into single-stranded fragments that migrate differently than hairpin-sealed coding ends where the complementary strands are covalently linked and, thus, do not dissociate (Supplementary Fig. 6). Analyses of denatured coding ends from Artemis−/−:H2AX−/−:DELCJ abl pre-B cells reveal that up to 70% of the hairpin-sealed coding ends have been opened (Fig. 1c and Supplementary Fig. 6).
The requirement for H2AX in regulating coding end processing was also observed at the immunoglobulin light chain (Igl) k locus in primary G1-phase bone marrow derived pre-B cell cultures from Artemis−/− and Artemis−/−:H2AX−/− mice expressing an immunoglobulin heavy chain (Igh) transgene (Ightg) (Supplementary Fig. 7)8. Southern blotting and TdT-assisted PCR revealed that the Jk coding ends in Artemis−/−:H2AX−/−:Ightg primary pre-B cells have open hairpins and are resected whereas those from Artemis−/−:Ightg pre-B cells remain hairpin-sealed (Supplementary Fig. 8). Together, these data demonstrate that H2AX restricts the activity of nucleolytic pathways that would otherwise aberrantly resect unrepaired hairpin-sealed or open coding ends in lymphocytes at the G1-phase of the cell cycle.
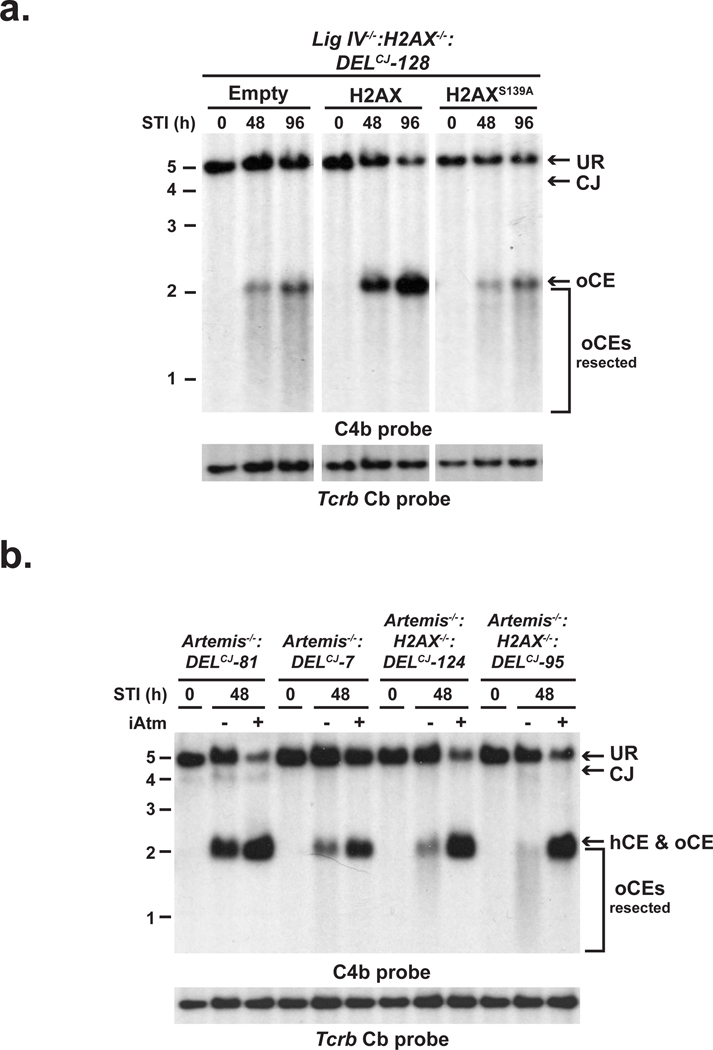
H2AX-dependent DNA damage responses generally depend on the phosphorylation of serine 139 by ATM or DNAPKcs to form γ-H2AX in chromatin flanking DNA DSBs including those generated by RAG11, 12. Reconstitution of Lig IV−/−:H2AX−/−:DELCJ and Artemis−/−:H2AX−/−:DELCJ abl pre-B cells with wild-type H2AX but not a serine 139 to alanine mutant (H2AXS139A), inhibits coding end resection in these cells (Fig. 2a and Supplementary Fig. 9). These findings implicate γ-H2AX formation at broken DNA ends in maintaining the structure of these ends in G1-phase lymphocytes but do note exclude the possibility that H2AX also inhibits resection through additional pathways that are independent of γ-H2AX formation. Unexpectedly, although ATM is required for the optimal formation of γ-H2AX in chromatin flanking RAG DSBs, treatment of Artemis−/−:DELCJ abl pre-B cells with the ATM kinase inhibitor, KU-55933, did not lead to the opening and resection of hairpin-sealed coding ends (Fig. 2b)12. Rather, treatment of Artemis−/−:H2AX−/−:DELCJ abl pre-B cells with KU-55933 resulted in a near complete block in end resection with a large fraction of DNA ends in these cells being hairpin-sealed (Fig. 2b and data not shown). We conclude that ATM functions to inhibit coding end resection through the formation of γ-H2AX; however, ATM activity is also required to promote this nucleolytic resection. Thus, ATM modulates the function of antagonistic pathways that both positively and negatively regulate DNA end resection in G1-phase lymphocytes.
Figure 2. ATM and γ-H2AX regulate DNA end resection.
pMX-DELCJ Southern blot analysis (as described in Fig. 1 a and b) of STI571-treated (a) Lig IV−/−:H2AX−/−:DELCJ abl pre-B cells reconstituted with an empty retroviral vector (empty) or vectors encoding wild-type H2AX (H2AX) or H2AXS139A and (b) Artemis−/−:DELCJ and Artemis−/−:H2AX−/−:DELCJ abl pre-B cell lines treated with either the ATM inhibitor KU55933 (+) or a DMSO vehicle control (−).
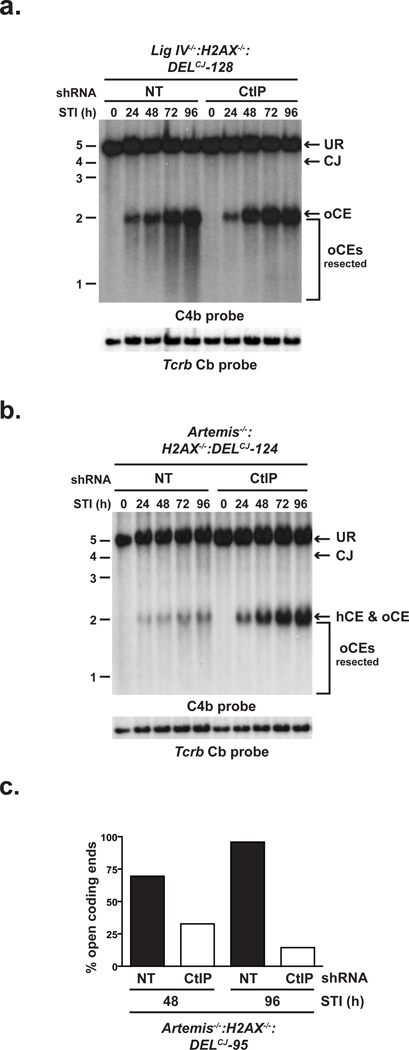
We show that, in the absence of H2AX, nucleases other than Artemis can efficiently open and resect hairpin sealed coding ends in a manner that is dependent on ATM. In this regard, ATM positively regulates CtIP activity in promoting DNA end resection5. CtIP binds directly to Nbs1, a component of the Mre11-Rad50-Nbs1 (MRN) complex that associates with RAG DSBs and functions in their repair5, 11, 13–15. Furthermore, Sae2, the S. cerevisiae ortholog of CtIP, functions with Mre11 to promote the opening and resection of hairpin-sealed DNA ends in vitro16. Together, these data suggest that H2AX could regulate the ability of CtIP to mediate hairpin opening and DNA end resection in G1-phase lymphocytes. Indeed, knockdown of CtIP in both Artemis−/−:H2AX−/− and Lig IV−/−:H2AX−/−:DELCJ abl pre-B cells largely blocked the aberrant coding end resection observed in these cells (Fig. 3a and b and Supplementary Figs. 10 and 11). Moreover, upon CtIP depletion, the majority of unrepaired coding ends in Artemis−/−:H2AX−/− abl pre-B cells are hairpin-sealed (Fig. 3c). Importantly, although our data demonstrate that the hairpin coding ends in Artemis−/−:H2AX−/−:DELCJ abl pre-B cells have been opened, we cannot determine the position of opening nor the extent of resection after opening. However, it is notable that Sae2 can mediate hairpin opening at significant distances from the hairpin tip in vitro16.
Figure 3. H2AX prevents CtIP-mediated DNA end resection.
(a–b) pMX-DELCJ Southern blot analysis (as described in Fig. 1 a and b) of STI571-treated (a) Lig IV−/−:H2AX−/−:DELCJ and (b) Artemis−/−:H2AX−/−:DELCJ abl pre-B cells expressing either non-targeting (NT) or CtIP-specific (CtIP) shRNAs. (c) Quantification of denaturing Southern blot analysis for open CEs from STI571-treated Artemis−/−:H2AX−/−:DELCJ abl pre-B cells expressing either NT or CtIP-specific shRNAs.
The DNA damage response protein MDC-1 is recruited by γ-H2AX to chromatin flanking DNA DSBs17, 18. We find that, like γ-H2AX, MDC-1 is also required to inhibit the ATM-dependent resection of coding ends in G1-phase lymphocytes (Supplementary Fig. 12). As CtIP binds to the FHA domain of Nbs1 that also binds MDC-1, this raises the possibility that upon recruitment to DSBs by γ-H2AX, MDC-1 may disrupt CtIP-Nbs1 interactions19–23. 53BP1, which is also retained at DSB sites in a γ-H2AX-dependent manner, regulates DNA end resection during V(D)J recombination in thymocytes and during immunoglobulin class switch recombination10, 24. Additionally, 53BP1 inhibits CtIP-dependent DNA end resection in Brca-1-deficient cells at post-replicative stages of the cell cycle; thus, γ-H2AX may inhibit DNA end resection in these cells by recruiting 53BP125. Notably, at post-replicative stages of the cell cycle, H2AX also functions to promote DNA DSB repair by homologous recombination (HR), which requires CtIP-mediated DNA end resection5, 26. Thus, H2AX likely modulates the activity of diverse cell cycle-specific DNA end resection pathways.
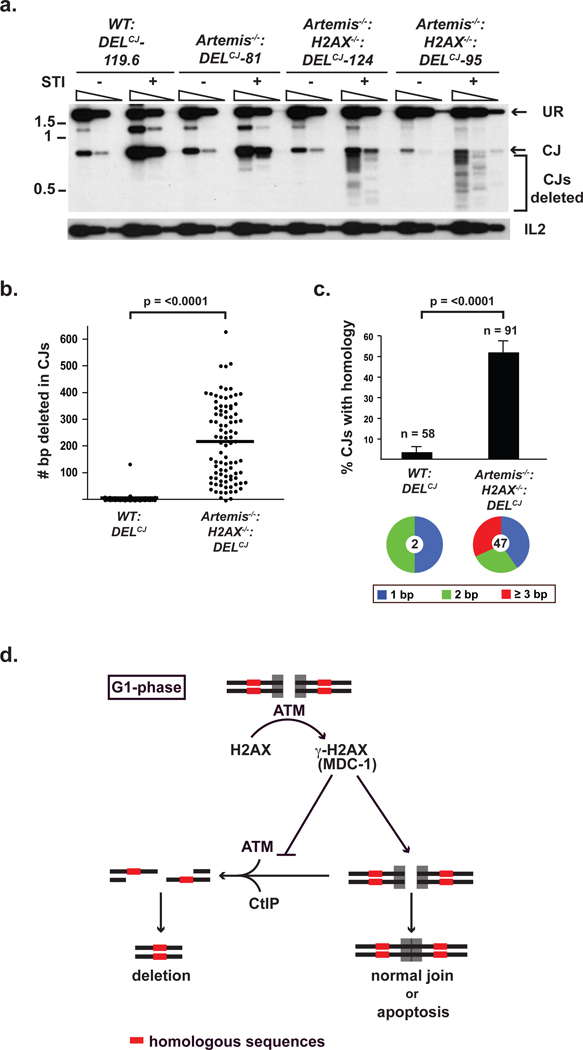
What is the fate of broken DNA ends processed by CtIP in G1-phase lymphocytes? During V(D)J recombination, Artemis functions primarily to open hairpin-sealed coding ends after which core NHEJ factors join these DNA ends3. However, hairpin-sealed coding ends opened in a CtIP-dependent manner persist unrepaired at high levels in Artemis−/−:H2AX−/− abl pre-B cells (Fig. 1b and Supplementary Fig. 2). In this regard, the single strand overhangs generated by CtIP-mediated resection during HR would likely be poor substrates for NHEJ in G1-phase cells4. However, this resection could expose regions of homology flanking the DSB, which, if used to mediate DSB repair by homology-driven repair pathways, would form joints with chromosomal deletions4. Indeed, PCR and sequence analyses of coding joints formed in Artemis−/−:H2AX−/−:DELCJ abl pre-B cells revealed that they are heterogeneous in size as compared to those that form in WT:DELCJ or Artemis−/−:DELCJ abl pre-B cells (Figs. 4a and b, and Supplementary Figs. 13 and 14). These joints have significant deletions extending up to 700 base pairs, which is the maximum size deletion that would be amplified by the PCR approach used (Figs. 4a and b, and Supplementary Fig. 14). Moreover, the coding joints formed in Artemis−/−:H2AX−/−:DELCJ abl pre-B cells use microhomologies at a higher frequency than those formed in WT:DELCJ abl pre-B cells (50% vs. 5%) (Fig. 4c and Supplementary Fig. 14). Analysis of T cell receptor β (Tcrb) Db1 to Jb1.1/Jb1.2 joints in Artemis−/−:H2AX−/− thymocytes revealed that these joints similarly exhibit significant deletions as compared to those formed in either Artemis−/− or wild type thymocytes (Supplementary Fig. 15). We conclude that RAG-mediated DNA breaks generated in H2AX-deficient lymphocytes are processed in a CtIP-dependent manner and are resistant to repair by classical NHEJ in Artemis−/−:H2AX−/−:DELCJ abl pre-B cells. However, these DNA ends can be channeled into homology-driven repair pathways resulting in joints that form significant chromosomal deletions.
Figure 4. Aberrant joining in H2AX-deficient cells.
(a) PCR products representing normal and deleted (bracket) pMX-DELCJ CJs from WT:DELCJ, Artemis−/−:DELCJ and Artemis−/−:H2AX−/−:DELCJ abl pre-B cells treated with STI571. Serial five-fold dilutions of genomic DNA were amplified. IL2 gene PCR is a loading control. (b–c) (b) Base-pairs deleted and (c) microhomology utilization in pMX-DELCJ CJs sequenced from WT:DELCJ and Artemis−/−:H2AX−/−:DELCJ abl pre-B cells (Supplementary Fig. 14). The total number of sequenced coding joints (n) is indicated. Pie chart shows fraction of joints with 1, 2 or ≥ 3 microhomologies and total number (center) of joints using microhomologies. (d) Model for H2AX function in DNA end processing in G1-phase lymphocytes. Error bars represent standard error of the mean (SEM). p-values were determined using Student’s t-test with Welch’s correction for unequal variances.
The requirement for H2AX to prevent CtIP-dependent resection and the resolution of RAG DSBs as chromosomal deletions is congruent with the phenotype of H2AX-deficient mice which are predisposed to lymphoid tumors that can harbor chromosomal lesions indicative of aberrantly resolved RAG DSBs27, 28. However, chromosomal V(D)J recombination proceeds efficiently in H2AX-deficient abl pre-B cells6,29. Moreover, DbJb joints formed in H2AX−/− thymocytes do not exhibit significant deletions or an increase in micro-homology usage as compared to wild type thymocytes (Supplementary Figs. 16 and 17). Thus, H2AX may be required for the repair of a limited subset of RAG DSBs. Alternatively, other proteins may compensate for a more general requirement for H2AX in DSB repair during V(D)J recombination. In agreement with this notion, while isolated deficiencies in H2AX or XLF have a minimal effect on chromosomal V(D)J recombination, the combined deficiency of H2AX and XLF in abl pre-B cells results in a significant accumulation of unrepaired coding ends29. Notably, the coding ends in H2AX−/−:XLF−/− abl pre-B cells appear to be more extensively resected than those in either Artemis−/−:H2AX−/− or Lig IV−/−:H2AX−/− abl pre-B cells raising the possibility that H2AX and XLF have overlapping activities in modulating DNA end resection29.
We demonstrate that H2AX maintains the structural integrity of broken DNA ends generated during V(D)J recombination in G1-phase lymphocytes. This protective function of H2AX would ensure that unrepaired DNA ends are either joined via classical NHEJ or signal for elimination of the cell by apoptosis (Fig. 4d). In the absence of H2AX, ATM/CtIP-dependent resection creates genomic instability by allowing DNA ends to be shuttled into homology-driven repair pathways that can form potentially dangerous chromosomal deletions (Fig. 4d). The chromosomal translocations observed in H2AX-deficient mice may rely on these defects in DNA end processing coupled with the diminished retention of coding ends in post-cleavage complexes in H2AX-deficient cells6. Important parallels can be drawn between mechanisms that protect, process and repair RAG DSBs and those incurred by environmental genotoxins. In this regard, a subset of genotoxic DSBs requires Artemis for their repair by NHEJ, suggesting that these broken DNA ends must undergo nucleolytic processing30. Furthermore, the repair of these genotoxic DSB may also depend on H2AX30. Thus, our finding that H2AX regulates the processing of unrepaired DNA ends during V(D)J recombination likely reflects a broader function of H2AX in regulating the nucleolytic processing of DNA DSBs generated during other physiologic processes and by harmful genotoxic agents.
Methods Summary
Abl pre-B cell lines containing the pMX-DELCJ retroviral recombination substrate were generated and maintained, and IL7-dependent pre-B cells were cultured as described previously7, 8. Standard protocols for Southern blotting (native and denaturing), Western blotting, flow cytometry, retroviral-mediated protein expression and shRNA-mediated knockdown were followed and are detailed in the Methods. TdT-assisted PCR for coding ends generated during rearrangement of pMX-DELCJ and the Iglk locus was performed as described in the Methods10.
Methods
Mice
All animals were housed in a specific pathogen-free facility at Washington University School of Medicine, and all animal protocols were approved by the Washington University Institutional Animal Care and Use Committee.
Cell line generation and cell culture
Rag−/−, Artemis−/−, and WT abl pre-B cells were described previously7, 8. Artemis−/−:H2AX−/− v-abl transformed pre-B cells were generated as previously described6. Ligase IV−/− and Ligase IV−/−:H2AX−/− abl preB cells were generated by treating Ligase IVloxp/loxp and Ligase IVloxp/loxp:H2AX−/− abl preB cells with a Tat-Cre fusion protein6, 31. Cells were incubated in media containing Tat-Cre at 50 µg/mL for 1 hour and allowed to recover for 48 hours prior to subcloning. Cre-mediated deletion of the Ligase IV gene was confirmed by PCR and Southern blotting. All lines were transduced with pMX-DELCJ of pMX-DELSJ by co-centrifugation and clonal populations each containing a single integrant of the retroviral substrate were isolated by limiting dilution as described previously8. Cells were treated with 3 µM STI571 (Novartis) for the indicated amount of time at a concentration of 106 cells/mL. KU-55933 (Tocris) or DMSO (vehicle control) was used at a concentration of 15 µM. Primary bone marrow pre-B cell cultures were generated by harvesting bone marrow from Rag−/−:Ightg, Artemis−/−:Ightg or Artemis−/−:H2AX−/−:Ightg mice followed by culture for 6–10 days in media containing IL-7 at 5 ng/mL maintained at a concentration of approximately 2 × 106 cells/mL prior to withdrawal of IL-77.
Retroviral reconstitution and lentiviral knock-down
Reconstitution of Artemis−/−:H2AX−/− and Ligase IV−/−:H2AX−/− abl preB cells was performed by retroviral transduction with either empty retrovirus or retrovirus containing cDNAs encoding H2AX or H2AXS139A. A cDNA encoding H2AXS139A (serine 139 changed to alanine) was generated by PCR-based site-directed mutagenesis of WT H2AX cDNA. cDNAs encoding WT H2AX and H2AXS139A were cloned into the pMX-PIE retroviral vector and cells were transduced by co-centrifugation as described previously8. Cells expressing the retroviral construct were obtained by flow cytometric cell sorting of cells expressing GFP using a FACSVantage (BD Biosciences). Generation of lentiviral shRNAs vectors was carried using the previously described pFLRU:YFP lentiviral vector32, 33. CtIP-specific and non-targeting (NT) shRNAs were cloned into the pFLRU:YFP lentiviral vector. Sequences targeted by the shRNA are as follows: CtIP (GAGCAGACCTTTCTCAGTA) and NT (GGTTCGATGTCCCAATTCTG). 4 µg of these pFLRU:shRNA:YFP vectors were individually co-transfected with 4 µg pHR’Δ8.2R packaging vector and 1 µg of pCMV-VSVg envelope plasmid into HEK293T cells plated at approximately 70% confluency in 6 cm2 plates using Lipofectamine 2000 (Invitrogren). Media was replaced at 12 hours post-transfection. Supernatants were harvested 24 hours later. Transduction of abl pre-B cells was performed by co-centrifugation with viral supernatant at 1800 rpm for 90 min with polybrene added to 5 µg/mL. Cells expessing the pFLRU-shRNA vectors were obtained by flow cytometric cell sorting of cells expressing YFP using a FACSVantage (BD Biosciences).
Southern blotting analyses
Standard Southern blot analysis of pMX-DELCJ and pMX-DELSJ was performed utilizing EcoRV digested genomic DNA and the C4b probe as described previously8. Southern blot analyses of coding ends generated during rearrangement at the Iglk locus was performed as described previously on genomic DNA digested with SacI and EcoRI using the JκIII probe7. Denaturing agarose electrophoresis gels was carried out as described previously with modifications34. Briefly, 40µg of genomic DNA was digested overnight with EcoRV in a 400 µl volume and concentrated to 30µl The DNA was then re-suspended with the addition of 5 volumes of a solution containing 8M urea, 1% NP-40, 1mM Tris pH 8.0 and 0.5 mg/ml bromophenol blue. This DNA solution was divided in two, with one half heated at 90°C for 8 min to denature the genomic DNA while the other half was incubated on ice. After 8 min, the heated DNA samples were placed on ice prior to electrophoresis on a 1.2% agarose TAE gel with 1M urea at 50 V for ~24hrs at 4°C in TAE buffer also containing 1M urea. Quantification was carried out using ImageJ software. To calculate the percent of hairpin-sealed coding ends, we measured the integrated density (ID) of the band representing open coding ends (oCEs) and that representing hairpin-sealed coding ends (hCEs) and subtracted background ID levels to obtain the corrected ID levels for each (corr. IDoCE and corr. IDhCE, respectively). The ID for the closed ends was divided by the sum of the IDs of closed and open ends and that was multiplied by 100. % hCE = (corr. IDhCE)/(corr. IDhCE + corr. IDoCE) * 100
TdT-assisted PCR
TdT-assisted PCR analysis of pMX-DELCJ coding ends was performed as described previously using the IRES REV5 oligo (CTCGACTAAACACATGTAAAGC) for the primary PCR reaction, the IRES REV4 oligo (CCCTTGTTGAATACGCTTG) for the secondary PCR reaction and the I4 oligo (TAAGATACACCTGCAAAGGCG) as a probe10. Similar conditions were used for PCR analyses of coding ends generated during rearrangement of the endogenous Iglk locus, using the Jk2 ds oligo (CCACAAGAGGTTGGAATGATTTTC) for amplification and the Jκ oligo (GTAGTCTTCTCAACTCTTGTTCACT) as a probe. IL2 gene PCR, which is provided as a DNA loading control, was performed as described previously10.
Analysis of pMX-DELCJ coding joints
pMX-DELCJ coding joints were amplified using oligos pC (GCACGAAGTCTTGAGACCT) and IRES REV5 (CTCGACTAAACACATGTAAAGC). 300 ng of genomic DNA was used in the original amplification with serial 5-fold dilutions. Oligo IR4 (CCCTTGTTGAATACGCTTG) was used as a probe. Cloning and sequencing of pMX-DELCJ coding joints was performed as described previously15. P-values for both Figure 4c and 4d were calculated by Student’s t-test with Welsh's correction for unequal variances.
Analysis of Tcrb coding joints
Db1-Jb1.1 and Db1-Jb1.2 CJs were amplified using oligos Db1 us (CCTTCCTTATCTTCAACTC) and Jb1.2 ds (CCTGACTTCCACCCGAGGTT) with the following PCR parameters: 92°C for 1:30, 55°C for 2:30, 72°C for 1:00. 300 ng of thymic DNA was used in the original amplification (13 cycles). PCR products were digested with BglII for 3 hours prior to a second round of amplification (30 cycles) to permit the isolation of DJb CJs without the competing amplification from the germline Tcrb locus. Southern blot analyses of the PCR products were done using Oligo Jb (GTAATCAGAGGAAGGATG) as a probe. Thymic DNA isolated from 3 mice for each genotype (WT, Artemis−/− and Artemis−/−:H2AX−/−) was analyzed. Db1-Jb1.1 CJs from WT and H2AX−/− thymic DNA were cloned and sequenced.
Flow Cytometric Analyses
Flow cytometric analyses were performed on a FACSCaliber (BD Biosciences) using fluorescein isothocyanate (FITC)-conjugated anti-CD45R/B220 and allophycocyanin (APC)-conjugated IgM and the appropriate isotype control. All antibodies were from BD Biosciences. Cell cycle analyses were carried out by assessing DNA content by incubating with Hoechst 33342 dye (Invitrogen) for one hour at 37°C prior to flow cytometric analysis. Percent of cells in G1 and S-G2-M were approximated using the Dean-Jett-Fox Method in the FLOW-JO 8.8.6 software.
Supplementary Material
Acknowledgements
We thank Drs. Eugene Oltz and Frederick Alt for critical review of the manuscript, Dr. Richard Baer for providing us with the anti-CtIP antibodies and Drs. Greg Longmore and Yunfeng Feng for pFLRU, pHR’Δ8.2R and pCMV-VSVg. This work is supported by the National Institutes of Health grants AI074953 (B.P.S.), AI47829 (B.P.S.), CA136470 (B.P.S and C.H.B), CA125195 (C.H.B.) and CA096832 (P.J.M.). J.J.B. is supported by an NIH Ruth L. Kirschstein National Research Service Award (T32 HD007499) and a Children's Discovery Institute Fellows Award. C.H.B. was a Pew Scholar in the Biomedical Sciences program and is a Leukemia and Lymphoma Society Scholar.
Footnotes
Author Contributions
B.P.S. and B.A.H. conceived the study and wrote the paper. B.P.S. and C.H.B. designed experiments and interpreted data. B.A.H., A.T., Y.D., G.S., J.Z. and J.J.B. designed and performed experiments and interpreted data. Z.F. and L.M.W. performed experiments. P.J.M. provided important reagents.
References
- 1.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 4.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.You Z, Bailis JM. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010;20:402–409. doi: 10.1016/j.tcb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin B, et al. Histone H2AX stabilizes broken DNA strands to suppress chromosome breaks and translocations during V(D)J recombination. J Exp Med. 2009;206:2625–2639. doi: 10.1084/jem.20091320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bredemeyer AL, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 8.Bredemeyer AL, et al. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–823. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol. 2003;4:31–37. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 10.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HT, et al. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savic V, et al. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 14.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helmink BA, et al. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med. 2009;206:669–679. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 18.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 19.Melander F, et al. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spycher C, et al. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lloyd J, et al. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams RS, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bothmer A, et al. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie A, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassing CH, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 28.Celeste A, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zha S, et al. XLF Has Redundant Functions with ATM and H2AX in V(D)J Recombination and non-homologous DNA End-joining. Nature. 2010 In Press. [Google Scholar]
- 30.Riballo E, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 31.Shull ER, et al. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 2009;23:171–180. doi: 10.1101/gad.1746609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu L, et al. Actin stress fiber pre-extension in human aortic endothelial cells. Cell Motil Cytoskeleton. 2008;65:281–294. doi: 10.1002/cm.20260. [DOI] [PubMed] [Google Scholar]
- 33.Langer EM, et al. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14:424–436. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hegedus E, Kokai E, Kotlyar A, Dombradi V, Szabo G. Separation of 1-23-kb complementary DNA strands by urea-agarose gel electrophoresis. Nucleic Acids Res. 2009;37:e112. doi: 10.1093/nar/gkp539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.