Abstract
Thrombin is a key enzyme targeted by the majority of current anticoagulants that are direct inhibitors. Allosteric inhibition of thrombin may offer a major advantage of finely tuned regulation. We present here sulfated benzofurans as the first examples of potent, small allosteric inhibitors of thrombin. A sulfated benzofuran library of 15 sulfated monomers and 13 sulfated dimers with different charged, polar and hydrophobic substituents was studied in this work. Synthesis of the sulfated benzofurans was achieved through a multiple step, highly branched strategy, which culminated with microwave-assisted chemical sulfation. Of the 28 potential inhibitors, eleven exhibited reasonable inhibition of human α-thrombin at pH 7.4. Structure activity relationship analysis indicated that sulfation at the 5-position of the benzofuran scaffold was essential for targeting thrombin. A t-butyl 5-sulfated benzofuran derivative was found to be the most potent thrombin inhibitor with an IC50 of 7.3 μM under physiologically relevant conditions. Michaelis-Menten studies showed an allosteric inhibition phenomenon. Plasma clotting assays indicate that the sulfated benzofurans prolong both the activated partial thromboplastin time and prothrombin time. Overall, this work puts forward sulfated benzofurans as the first small, synthetic molecules as powerful lead compounds for the design of a new class of allosteric inhibitors of thrombin.
Introduction
Thrombin is a key enzyme of the coagulation cascade exhibiting important roles in both pro-coagulation and anti-coagulation processes. Most clinically used anticoagulant drugs, including polymeric heparin, warfarin, hirudin, argatroban and the recently approved dabigatran, aim to reduce thrombin activity.1–3 Although these drugs have been successful in treating many thrombotic disorders, the anticoagulation therapy continues to suffer from bleeding risk, narrow therapeutic index and other drug-specific adverse effects, which highlight the critical need to develop agents with better therapeutic profile.
Structurally, thrombin is a complex serine protease of the trypsin family that displays multiple ligand-binding domains, which modulate its activity and interactions with other molecules. These domains include the active site, anion-binding exosites I and II, and the sodium binding site. The design of thrombin inhibitors has focused almost exclusively on blocking substrate’s access to the active site through a competitive process.3 This paradigm has resulted in the design of thousands of molecules distributed across a large number of scaffolds.3–6 Of these, a select group of scaffolds, including the pyrazoles, napthylamidines, indoles, biphenyls, and benzimidazoles, are preferred because of their high thrombin selectivity. Fundamentally, these designed inhibitors attempt to mimic the D-Phe-Pro-Arg recognition sequence through the incorporation of a guanidine or amidine moiety. Recently, however, newer molecules have been designed with a P-1 halophenyl substituent so as to interact with Tyr228 in the S1 pocket so as to improve the poor oral bioavailability of most P-1 guanidine-based inhibitors.3,7–9
An exciting alternative to competitive inhibition of thrombin is allosteric inhibition through either exosite I or II. It is well established that binding of ligands in these exosites can induce conformational changes in the active site of thrombin.10–13 An example of this is thrombomodulin, which interacts with exosite I so as to change the substrate specificity of thrombin from fibrinogen to protein C.12 Likewise, hirugen binding in exosite I increases or decreases the catalytic efficiency of thrombin, depending on the nature of chromogenic substrate.10,11 Similarly, exosite I – active site allosteric coupling is also the reason why peptides such as bothrojaracin inhibit thrombin.14,15 Nevertheless, no small organic molecule has been designed that utilizes exosite I to effect physiological thrombin inhibition.
Exosite II ligands include heparin, chondroitin sulfate, haemadin, and fibrinogen γ′. These can be broadly classified as either highly anionic polysaccharides or traditional peptides. Interaction of heparin with exosite II is known to induce greater reactivity with antithrombin and recent work shows that formation of antithrombin – thrombin complex disrupts exosite II.16 Also, exosite II – active site coupling is suggested by the change in fluorescence properties of para-aminobenzamidine, a non-covalent active site probe, upon heparin binding to exosite II.17 However, no exclusive exosite II ligand has been found to date that reduces the catalytic efficiency of thrombin.
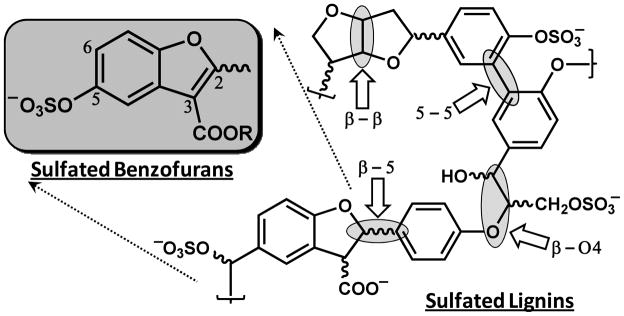
Allosteric regulation is a promising strategy for thrombin inhibition. Nature tends to utilize allosterism to confer specificity of recognition and also to effect regulation. For thrombin, the possibility of regulation, or controlled inhibition, is of considerable importance because nearly all current anticoagulants are associated with risk of bleeding. An appropriately designed allosteric regulator that does not fully nullify the procoagulant signal may maintain a finely tuned balance between procoagulant and anticoagulant signal resulting reduced bleeding complications. In pursuit of such allosteric regulators, we had earlier designed sulfated low molecular weight lignins (LMWLs) as functional mimetics of heparin targeted to exosite II-like regions of coagulation enzymes, including thrombin.18,19 Sulfated LMWLs, chemo-enzymatic variants of the naturally available lignin (Fig. 1), were found to potently inhibit thrombin with an IC50 of 18–34 nM under physiological conditions.18 More importantly, sulfated LMWLs also inhibited blood clotting under ex vivo conditions with potency comparable to the LMW heparins.20
Figure 1.
Design of sulfated benzofurans on the basis of the structure of sulfated lignins, which were found to be potent inhibitors of thrombin. Sulfated lignins are a three-dimensional network of chemo-enzymatically coupled 4-hydroxy cinnamic acid monomers. Four different inter- monomeric linkages are possible in these oligomers including β-5, β-O4, β-β and 5-5 linkages. Chiral centers are identified using curvy bonds. The chain length of sulfated lignins is typically 5–15-monomer units. Sulfated benzofurans studied in this work were designed from one specific structure generated by β-5 linked monomers, as shown by the structure in box. Positions 2, 3, 5 and 6 are numbered. See text for details.
Despite their excellent in vitro and ex vivo anticoagulant properties, sulfated LMWLs are challenging because of their polydispersity and microheterogeneity, which parallel that observed with the heparins. Sulfated LMWLs represent a large library of structures arising from multiple inter-monomeric linkages, variable sulfation and chain length, and the presence of many chiral centers (Fig. 1).21 These variables introduce considerable structural diversity to these molecules, which nevertheless bind to exosite II of thrombin and allosterically induce thrombin inhibition.18 Sulfated LMWLs represent the first examples of an exclusive exosite II-mediated inhibition of the key coagulation protease.
In this work, we present a study of 28 monomeric and dimeric sulfated benzofurans, molecules derived from the sulfated LMWL structure, as inhibitors of thrombin. Enzyme inhibition studies show that the library of molecules display a range of inhibition potential, while Michaelis-Menten kinetic studies show the inhibition to arise from an allosteric process. Collectively, this work establishes the potential of allosteric inhibition of thrombin through the interesting and unusual features of sulfated benzofurans that direct the interaction with thrombin.
Experimental Procedures
Chemicals, Reagents and Analytical Chemistry
Microwave-based sulfation reactions were performed using a CEM-Discover synthesizer (Matthews, NC) in sealed reaction vessels (7 ml) at 50 W. Temperature of the reaction was maintained at the desired setting using cooled nitrogen (45 psi) flow. Reagents and chemicals used in reactions were purchased from either Sigma-Aldrich (Milwaukee, WI) or Fisher (Pittsburgh, PA) and used as supplied. Analytical thin layer chromatography (TLC) was performed using UNIPLATE™ silica gel GHLF 250 μm pre-coated plates (ANALTECH, Newark, DE) that were analyzed by fluorescence (254 nm). Compound purity was evaluated independently through gradient reverse-phase HPLC. The purity of each compound synthesized in this work was greater than 95% and is listed in the Supplementary Material.
Column chromatography was performed using Teledyne ISCO (Lincoln, NE) Comb flash RF system and disposable normal silica cartridges of 30 – 50 μ particle size, 230 – 400 mesh size and 60 Å pore size. Sodium exchange chromatography was performed using SP Sephadex C-25 sodium cation exchange columns from GE Healthcare Life Sciences (Piscataway, NJ). Approximately 100 mg sulfated sample was loaded onto 10 g of the cation exchanger in 10 × 460 mm column and eluted with either water or 20% EtOH/H2O at 0.5 mL/min. Anhydrous reactions were carried out under nitrogen atmosphere in glassware dried at high temperature. Reagent solutions and solvents were handled under nitrogen atmosphere using syringe techniques. HPLC purification of sulfated benzofurans was carried out on a Jasco chromatography system (Easton, MD) using a 250×10 mm Varian’s Dynamax Microsorb C4 column. An acetonitrile – water gradient (5 to 100% acetonitrile in 20 min) at 3 mL/min was used to purify the compounds on milligram scale. The eluents were detected at 300 nm.
1H and 13C NMR spectra were recorded at either 300 or 400 MHz (Varian Mercury or Bruker Ultrashield™Plus) in appropriate deuterated solvents including CDCl3, DMSO-d6, CD3OD, acetone-d6, or D2O. All signals are reported in ppm with the internal chloroform, DMSO, acetone, and D2O signals at 7.26, 2.50, 3.31, 2.05, and 4.79, respectively, as standards. The data is reported as: chemical shifts (ppm), multiplicity (s = singlet, d = doublet, t = triplet, m = multiplet), coupling constant(s) (Hz), and integration. Mass spectrometry was performed on all synthesized molecules using a Micromass ZMD4000 single quadrupole mass spectrometer with ESI ionization probe operating in negative ion mode (Waters Corp., Milford, MA). The samples, dissolved in acetonitrile containing formic acid (5% v/v), were infused at 10 μL/min. The source block temperature and the probe temperature were typically held at 100 and 120 °C, respectively, while corona and cone voltages were selected through manual optimization. The desolvation nitrogen flow was 500 L/hour. Mass spectra were acquired in the mass range from 110 to 1000 daltons at 400 amu/sec.
Synthesis of Benzofuran Monomer and Dimer Precursors for Subsequent Sulfation at 5-, 5′- and Other Positions
The detailed protocol for the synthesis of each benzofuran precursor is provided in Supplementary Information. The reactions used in the synthesis are fairly common and hence not described here.
Sulfation of Appropriate Precursors to Yield Sulfated Benzofuran Monomers and Dimers
The synthesis of sulfated benzofurans was developed following our earlier report on the synthesis of sulfated flavonoids.22 Briefly, to a solution of unsulfated benzofuran monomer or dimer (typically 50 mg) in acetonitrile: DMF (4:1, 0.45 mL) in a microwave tube were added triethylamine (10 equiv) and trimethylamine-sulfur trioxide complex (15 equiv). The reaction mixture was exposed to microwaves (50 W) for 40 min at 100 °C followed by vacuum concentration to remove all solvent. The solid so obtained was directly loaded on a Sephadex C-25 cation exchange resin and eluted with water. Fractions containing the sulfated product were lyophilized to obtain a solid residue, which was further purified on reverse-phase gradient HPLC to remove residual unsulfated contaminants.
5a: 1H NMR (D2O, 400 MHz) δ 1.45 (t, J=6Hz, 3H), 2.51 (s, 3H), 3.76 (s, 3H), 4.36 (q, J =6 Hz, 2H), 5.38 (s, 1H), 7.15 (s, 1H), 7.57 (s, 1H).
5b: 1H NMR (D2O, 400 MHz) δ 2.48 (s, 3H), 3.89 (s, 3H), 4.21 (s, 3H), 6.98 (s, 1H), 6.61 (s, 1H). ESI MS (-ve) m/z calcd. for C12H11O8SNa [(M-Na)−)] 315.02, found 315.01(M-Na) −.
5c: 1H NMR (D2O, 400 MHz) δ 1.62 (s, 9H), 2.49 (s, 3H), 3.89 (s, 3H), 6.98 (s, 1H), 6.61 (s, 1H). 13C NMR (D2O) δ 13.40, 21.22, 61.33, 73.87, 99.93, 107.79, 114.00, 118.54, 139.23, 147.28, 150.45, 164.37, 165.49. ESI MS (-ve) m/z calcd for C13H17O8SNa [(M-Na) −)] 357.05, found 357.08 (M-Na) −.
5d: 1H NMR (D2O, 400 MHz) δ 2.48 (s, 3H), 3.21 (s, 3H), 3.62 (t, j=12 Hz, 2H), 3.72 (s, 3H), 4.26 (t, j=12 Hz, 2H), 6.91 (s, 1H), 7.59 (s, 1H). ESI MS (-ve) m/z calcd. for C13H13O9SNa [(M-Na) −)] 345.02, found 345.01 (M-Na) −.
5e: 1H NMR (D2O, 400 MHz) δ 1.70 (m, 6H), 2.99 (s, 3H), 4.41 (q, j= 17 Hz, 2H), 4.66 (q, j= 17 Hz, 2H), 7.55 (s, 1H), 8.21 (s, 1H). 13C NMR (D2O) δ 16.36, 16.39, 16.76, 63.15, 67.53, 99.69, 110.66, 116.09, 119.96, 141.73, 146.05, 151.33, 152.58, 165.90, 167.00. ESI HRMS (-ve) m/z calcd. for C14H16O8SNa [(M-Na) −)] 343.0512, found 343.0357 (M-Na) −.
5f: 1H NMR (D2O, 400 MHz) δ 1.37 (m, 9H), 2.36 (s, 3H), 4.23 (q, j=18 Hz, 2H), 4.54 (m, 1H), 6.96 (s, 1H), 7.60 (s, 1H). 13C NMR (D2O) δ 13.40, 13.48, 21.22, 61.33, 73.87, 99.93, 107.79, 114.00, 118.54, 139.23, 147.28, 150.45, 164.37, 165.49. ESI HRMS (-ve) m/z calcd. for C14H15O8SNa [(M-Na) −)] 357.0532, found 357.0600 (M-Na) −.
5g: 1H NMR (D2O, 400 MHz) δ 1.45 (t, j=18 Hz, 3H), 2.50 (s, 3H), 3.64 (s, 3H), 4.31 (q, j= 18 Hz, 2H), 5.33 (s, 2H), 7.15 (s, 1H), 7.64 (s, 1H). ESI HRMS (-ve) m/z calcd. for C14H15O9SNa [(M-Na) −)] 359.0420, found 359.0395 (M-Na) −.
5h: 1H NMR (D2O, 400 MHz) δ 1.39 (t, j=18 Hz, 3H), 2.57 (s, 3H), 3.87 (m, 8H), 4.29 (q, j=18 Hz, 2H), 7.45 (s, 1H), 7.57 (s, 1H). 13C NMR (D2O) δ 13.48, 13.72, 44.11, 44.94, 61.53, 61.58, 66.25, 105.77, 106.60, 108.11, 114.53, 123.18, 139.99, 140.26, 149.66, 154.36, 165.33, 166.19. ESI HRMS (-ve) m/z calcd for C17H18O10SNa [(M-Na) −)] 428.0332, found 428.0889 (M-Na) −.
5i: 1H NMR (D2O, 400 MHz) δ 2.51 (s, 3H), 3.63 (s, 3H), 4.65 (d, 2H), 5.02 (d, 1H), 5.16 (d, 2H), 5.91 (m, 1H), 6.81 (s. 1H), 7.56 (s, 1H). 13C NMR (D2O) δ 13.40, 55.05, 61.19, 70.68, 97.79, 107.91, 113.75, 114.30, 118.36, 128.61, 128.99, 138.55, 148.09, 150.16, 158.52, 163.59, 165.06. ESI HRMS (+ve) m/z calcd. for C20H19O9SNa [(M+Na)+)] 481.0632, found 481.1400 (M+Na)+.
5j: 1H NMR (D2O, 400 MHz) δ 1.28 (t, j=18 Hz, 3H), 2.09 (s, 3H), 3.54 (s, 3H), 4.12 (q, j=18 Hz, 2H), 4.78 (s, 2H), 6.57 (s, 1H), 6.68 (d, j=21 Hz, 2H), 7.17 (d, j= 21 Hz, 2H), 7.67 (s,1H). 13C NMR (D2O) δ 13.40, 55.05, 61.19, 70.68, 97.79, 107.91, 113.75, 114.30, 118.36, 128.61, 128.99, 138.55, 148.09, 150.16, 158.23, 163.59, 165.06. ESI MS (-ve) m/z calcd. for C14H13O8SNa [(M-Na) −)] 325.03, found 325.04 (M-Na) −.
6: 1H NMR (CD3OD, 300 MHz) δ 2.61 (s, 3H), 3.86 (s, 3H), 6.88 (s, 2H), 7.15 (s, 1H), 7.86 (s, 1H). 13C NMR (CD3OD) δ 10.85, 55.78, 59.93, 95.76, 110.69, 112.67, 120.71, 138.55, 150.48, 151.65, 154.22.
7: 1H NMR (CD3OD, 300 MHz) δ 0.44 (s, 6H), 1.02 (s, 9H), 2.75 (s, 3H), 3.96 (s, 3H), 6.68 (s, 2H), 6.98 (s, 1H), 7.36 (s, 1H). 13C NMR (CD3OD) δ 4.48, 12.29, 18.70, 26.02, 55.88, 56.04, 95.47, 109.94, 114.31, 120.84, 142.03, 149.32, 149.40, 151.81.
9a: 1H NMR (D2O, 400 MHz) δ 2.47 (S, 3H), 3.55–3.90 (m, 8H), 3.91 (s, 3H), 7.25 ( s, 1H), 7.41 (s, 1H). 13C NMR (D2O, 400 MHz) δ 12.48, 42.76, 47.82, 56.43, 66.41, 66.79, 96.68, 110.52, 112.70, 118.31, 137.35, 149.74, 151.36, 156.79, 166.23. ESI HRMS (-ve) m/z calcd. for C15H16NO8SNa [(M-Na) −)] 370.0544, found 370.0652 (M-Na) −.
9b: 1H NMR (D2O, 400 MHz) δ 0.89 (d, j= 17 Hz, 3H), 1.01 (m, 1H), 1.61–1.81 (m, 4H), 2.39 (s, 3H), 2.92 (m, 1H), 3.08 (m, 1H), 3.56 (m, 1H), 3.92 (s, 3H), 4.45 (m, 1H), 7.27 (s, 1H), 7.64 (s, 1H). 13C NMR (D2O) δ 12.47, 12.55, 14.88, 20.89, 30.17, 55.67, 56.59, 96.67, 111.46, 112.63, 118.67, 137.67, 149.82, 151.26, 155.45, 165.32. ESI HRMS (-ve) m/z calcd. for C17H20NO7SNa [(M-Na) −)] 482.1022, found 382.0847 (M-Na) −.
10: 1H NMR (D2O, 400 MHz) δ 1.47 (t, j=18 Hz, 3H), 2.71 (m, 4H), 3.81 (m, 4H), 3.95 (s, 3H), 4.02 (s, 2H), 4.39 (q, j= 18 Hz, 2H), 7.25 (s, 1H), 7.74 (s, 1H). 13C NMR (D2O) δ 13.45, 52.47, 52.64, 56.34, 61.82, 66.07, 96.24, 111.83, 114.82, 117.14, 137.95, 150.45, 151.64, 159.30, 164.77. ESI HRMS (-ve) m/z calcd. for C17H20NO9SNa [(M-Na) −)] 414.0933, found 414.0847 (M-Na) −.
12a: 1H NMR (CD3OD, 300 MHz) δ 1.32 ( J=6 Hz, t, 3H), 2.65 (s, 3H), 3.82 (s, 3H), 3.86 (s, 3H), 4.29 (j=6 Hz, q, 2H), 5.52 (s, 2H), 7.12 (s, 1H, Ar-H), 7.22 (s, 1H, Ar-H), 7.43 (s, 1H, Ar-H), and 8.04 ( s, 1H, Ar-H). 13C NMR (CD3OD) δ 13.12, 13.53, 13.54, 55.65, 60.20, 60.67, 63.93, 95.54, 95.63, 109.19, 112.32, 115.57, 117.40, 118.31, 139.89, 145.50, 149.74, 152.08, 152.20, 158.70, 162.67, 163.51, 164.48. ESI HRMS (-ve) m/z calcd. for C26H25O13SNa [(M-Na) −)] 577.1011, found 577.1146 (M-Na) −.
12b: 1H NMR (CD3OD, 300 MHz) δ 2.67 (s, 3H), 3.83 (s, 3H), 3.84 (s, 3H), 3.85 (s, 3H), 3.86 (s, 3H), 5.55 (s, 2H), 7.13 (s, 1H), 7.24 (s, 1H), 7.47 9s, 1H), 8.03 (s, 1H). ESI HRMS (-ve) m/z calcd. for C24H21O13SNa [(M-Na) −)] 549.0723, found 549.0795 (M-Na) −.
12c: 1H NMR (CD3OD, 300 MHz) δ 1.49 (s, 9H), 1.51 (s, 9H), 2.65 (s, 3H), 3.83 (s, 3H), 3.88 (s, 3H), 5.47 (s, 2H), 7.12 (s, 1H), 7.22 (s, 1H), 7.37 (s, 1H), 8.09 (s, 1H).13C NMR (CD3OD) δ 13.09, 27.30, 27.45, 55.59, 55.66, 64.26, 81.00, 81.62, 95.44, 95.56, 109.89, 110.21, 113.55, 115.59, 117.70, 118.42, 139.79, 145.31, 149.83, 151.87, 152.05, 158.16, 162.26, 162.63, 163.80. ESI MS (-ve) m/z calcd. for C30H33O13SNa [(M-Na) −)] 633.16, found 633.19 (M-Na) −.
12d: 1H NMR (D2O, 400 MHz) δ 2.30 (s, 3H), 3.26 (s, 3H), 3.32 (s, 3H), 3.57 (m, 2H), 3.63 (m, 5H), 3.77 (s, 3H), 4.18 (m, 2H), 4.27 (m, 2H), 5.17 (s, 2H), 6.49 (s, 1H), 6.89 (s, 1H), 6.93 (s, 1H), 7.67 (s, 1H). 13C NMR (D2O) δ 13.49, 55.40, 56.07, 58.13, 58.24, 62.74, 62.96, 63.79, 69.76, 69.89, 93.22, 95.76, 105.85, 107.40, 111.06, 114.98, 116.73, 117.17, 138.28, 144.55, 147.64, 148.10, 150.78, 151.62, 158.55, 162.96, 163.49, 164.40. ESI HRMS (-ve) m/z calcd. for C28H29O115SNa [(M-Na) −)] 637.1223, found 637.1161 (M-Na) −.
12e: 1H NMR (D2O, 400 MHz) δ 1.14 (m, 12H), 2.32 (s, 3H), 3.72 (m, 4H), 4.06 (m, 4H), 5.15 (s, 2H), 6.51 9s, 1H), 6.71 9s, 1H), 6.98 (s, 1H), 7.56 (s, 1H). ESI HRMS (-ve) m/z calcd. for C28H29O13SNa [(M-Na) −)] 605.1312, found 605.0708 (M-Na) −.
12f: 1H NMR (D2O, 400 MHz) δ 1.07 (m, 18H), 2.27 (s, 3H), 3.99 (m, 4H), 4.20 (m, 2H), 5.05 (s, 2H), 6.64 (s, 1H), 6.67 (s, 1H), 6.99 (s, 1H), 7.81 (s, 1H). 13C NMR (D2O) δ 13.29, 13.50, 13.69, 13.78, 20.94, 21.19, 21.40, 60.34, 61.09, 63.81, 71.86, 72.97, 98.90, 108.13, 111.04, 114.71, 117.74, 118.33, 139.92, 146.62, 146.72, 148.33, 148.60, 151.24, 158.69, 162.57, 163.34, 164.08. ESI HRMS (-ve) m/z calcd. for C30H33O13SNa [(M-Na) −)] 633.1236, found 633.1494 (M-Na) −.
13: 1H NMR (CD3OD, 400 MHz) δ 1.38 (m, 6H,), 2.60 (m, 3H,), 3.71 (m, 4H,), 3.91 (s, 3H,), 3.93 (s, 3H), 4.37 (m, 6H,), 5.62 (s, 2H,), 7.02 (s, 1H), 7.05 (s, 1H), 7.49 (s, 1H), 8.01 (s, 1H). ESI MS (-ve) m/z calcd. for C30H32NO14SNa [(M-Na) −)] 659.14, found 659.15 (M-Na) −.
14: 1H NMR (D2O, 400 MHz) δ 2.40 (s, 3H,), 3.24 (m, 8H), 3.67 (m, 8H), 3.81 (s, 3H,), 3.92 (s, 3H), 5.30 (s, 2H,), 6.85 (s, 1H,), 7.14 (s, 1H), 7.33 (s.1H,), 7.43 (s, 1H). 13C NMR (D2O) δ 12.36, 42.55, 56.00, 56.40, 63.52, 66.17, 66.36, 96.23, 96.73, 107.16, 110.44, 113.33, 115.29, 116.94, 117.92, 138.10, 143.04, 148.81, 149.45, 151.21, 151.81, 152.46, 156.38, 164.20, 165.93. ESI HRMS (-ve) m/z calcd. for C30H31N2O13SNa [(M-Na) −)] 659.1422, found 659.1531 (M-Na) −.
17: 1H NMR (D2O, 400 MHz) δ 1.23 (m, 6H), 2.36 (s, 3H), 3.70 (s, 3H), 4.18 (m, 4H), 5.23 (s, 2H), 6.96 (s, 1H), 7.03 (s, 1H), 7.53 (s, 1H), 7.62 (s, 1H). 13C NMR (D2O) δ 13.4, 13.51, 13.59, 56.30, 61.53, 62.08, 63.51, 96.28, 99.06, 108.05, 111.73, 114.48, 115.06, 117.13, 119.54, 138.31, 138.82, 147.96, 150.23, 150.94, 152.02, 158.07, 164.50, 164.87, 165.60. ESI HRMS (-ve) m/z calcd. for C25H22O14S2Na [(M-Na) −)] 665.0121, found 665.0366 (M-Na) −.
18: 1H NMR (D2O, 400 MHz) δ 1.55 (t, j=18 Hz, 3H),1.59 (t, j=18 Hz, 3H), 2.41 (s, 3H), 3.81 (s, 3H), 4.18 (q, j=18 Hz, 2H), 4.23 (q, j=18 Hz, 2H), 5.18 (s, 2H), 6.71 (s, 1H), 6.89 (s, 1H), 7.65 (s, 1H), 7.91 (s, 1H). 13C NMR (D2O) δ 13.47, 13.61, 13.63, 56.32, 61.53, 62.18, 63.61, 96.28, 99.16, 108.01, 111.53, 114.48, 115.06, 117.13, 119.54, 138.31, 138.82, 147.96, 150.23, 150.84, 152.12, 158.17, 164.53, 164.77, 165.60. ESI HRMS (-ve) m/z calcd. for C25H22O14S2Na [(M-Na) −)] 665.0121, found 665.0277 (M-Na) −.
19: 1H NMR (CDCl3, 300 MHz) δ 1.39 (m, 6H), 2.67 (s, 3H), 3.85 (s, 3H), 3.88 (s, 3H), 4.36 (m, 4H), 5.55 (s, 2H), 7.22 (s, 1H), 7.23 (s, 1H), 7.43 (s, 1H), 8.07 (s, 1H). 13C NMR (CD3OD) δ 13.25, 13.38, 13.52, 55.66, 55.77, 60.25, 60.79, 63.16, 95.63, 99.17, 103.95, 108.76, 112.48, 115.58, 117.27, 120.11, 139.90, 146.40, 148.00, 148.45, 152.15, 152.23, 158.27, 163.04, 163.59, 164.61. ESI HRMS (-ve) m/z calcd. for C26H25O13SNa [(M-Na) −)] 577.1011, found 577.1100 (M-Na) −.
Synthesis of Benzofuran Dimer 15 From 12a
To the solution of 12a (100 mg, 0.16 mmol) in 2 ml of methanol was added LiOH (20 mg, 0.8 mmol) and stirred for 5 h at RT. After completion, the reaction mixture was concentrated and purified using HPLC. 1H NMR (D2O, 400 MHz) δ 1H NMR (CD3OD, 300 MHz) δ 2.66 (s, 3H), 3.84 (s, 3H), 3.89 (s, 3H), 5.52 (s, 2H), 7.11 (s, 1H, Ar-H), 7.27 (s, 1H, Ar-H), 7.41 (s, 1H, Ar-H), and 8.01 ( s, 1H, Ar-H). 13C NMR (CD3OD) δ 13.43, 61.67, 63.73, 95.54, 95.63, 109.19, 112.32, 115.57, 117.40, 118.31, 139.89, 145.50, 149.74, 152.08, 152.20, 158.54, 162.57, 164.51, 165.48. ESI HRMS (-ve) m/z calcd. for C17H20NO9SNa [(M-Na) −)] 521.0322, found 521.1496 (M-Na) −.
Synthesis of Benzofuran Dimer 16 From 11a
To the solution of 11a (200 mg, 0.32 mmol) in 10 ml of anhydrous THF was added LiAlH4 (65 mg, 1.6 mmol) and stirred for 6 hours under nitrogen. After completion, reaction was quenched by adding 30 ml of water and extracted with 100 ml of diethyl ether twice. The organic layer was concentrated and purified using gradient flash chromatography (0 to 50% ethyl acetate in hexane). The reduced form of 11a was sulfated using the general sulfation procedure described above. 1H NMR (CD3OD, 300 MHz) δ 0.17 ( s, 6H), 1.01 (s, 9H), 2.71 (s, 3H), 3.87 (s, 3H), 3.91 (s, 3H) ), 5.51 (2, 2H), 6.65 (s, 2H), 6.69 (s, 2H), 7.01 (s, 1H, Ar-H), 7.04 (s, 1H,), 7.37 (s, 1H), 7.45 (s, 1H). 13C NMR (CDCl3) δ −4.47, 12.31, 18.70, 26.00, 55.45, 55.81, 55.94, 56.58, 94.60, 95.48, 103.35, 114.35, 118.34, 120.39, 121.24, 142.32, 142.65, 144.82, 148.21, 149.73, 152.05.
Thrombin and Chemicals for Biological Experiments
Human α-thrombin was from Haematologic Technologies (Essex Junction, VT). Stock solutions of thrombin were prepared in 20 mM sodium phosphate buffer, pH 7.4, containing 100 mM NaCl. Chromogenic substrate Spectrozyme TH was from American Diagnostica (Greenwich, CT). Citrated human plasma for coagulation time assays was purchased from Valley Biomedical (Winchester, VA). Thromboplastin and ellagic acid were obtained from Fisher Diagnostics (Middletown, VA).
Inhibition of Thrombin
Direct inhibition of thrombin by sulfated benzofuran derivatives was measured through a chromogenic substrate hydrolysis assay.18,19 The buffer used in these experiments was 20 mM Tris-HCl buffer, pH 7.4, containing 100 mM NaCl, 2.5 mM CaCl2, and 0.1% polyethylene glycol (PEG) 8000. Benzofuran derivatives (2 to 30 μL) at concentrations ranging from 4 μg/ml to 5 mg/ml were diluted with appropriate volume of assay buffer in PEG 20,000-coated acrylic cuvettes at 25 °C. To this solution was added 5 μL of thrombin solution to give approximately 5 nM initial thrombin concentration. After 10 min of incubation, 20 μL of 1 mM Spectrozyme TH was rapidly added and the residual thrombin activity was measured from the initial rate of increase in absorbance at 405 nm. Relative residual thrombin activity at each concentration of the inhibitor was calculated from the ratio of thrombin activity in the presence and absence of inhibitor. Logistic equation 1 was used to fit the dose-dependence of residual proteinase activity to obtain the IC50 and the efficacy ΔY (= YM – Y0) of inhibition.
| Eq. 1 |
In this equation, Y is the ratio of residual thrombin activity in the presence of inhibitor to that in its absence (fractional residual activity), YM and YO are the maximum and minimum possible values of the fractional residual proteinase activity, IC50 is the concentration of the inhibitor that results in 50% inhibition of enzyme activity, and HS is the Hill Slope, which was set constant at 1. A current version of SigmaPlot (SPSS, Inc. Chicago, IL) was used to perform non-linear curve fitting in which YM, YO and IC50 were allowed to float.
Michaelis-Menten Kinetics of Spectrozyme TH Hydrolysis
The initial rate of Spectrozyme TH hydrolysis by 5 – 10 nM thrombin was monitored from the linear increase in absorbance at 405 nm corresponding to less than 10% consumption of the substrate, as described earlier.18,19 The initial rate was measured as a function of various concentrations of the substrate (10 to 600 μM) in the presence of fixed concentration of dimer 12a (0–30 μM) in 20 mM Tris-HCl buffer, pH 7.4, at 37 °C. The data were fitted by Michaelis-Menten equation 2 to determine KM and VMAX.
| Eq. 2 |
Prothrombin Time and Activated Partial Thromboplastin Time
Clotting time was measured in a standard 1-stage recalcification assay with a BBL Fibrosystem fibrometer (Becton-Dickinson, Sparles, MD), as described previously.20,21 For PT assays, thromboplastin was reconstituted according to manufacturer’s directions and warmed to 37°C. A 10 μL sample of the sulfated benzofuran, to give the desired concentration, was brought up to 100 μL with citrated human plasma, incubated for 30 s at 37 °C followed by addition of 200 μL pre-warmed thromboplastin. Clotting time in the absence of an anticoagulant was determined using 10 μL deionized water. For the APTT assay, 10 μL inhibitor was mixed with 90 μL citrated human plasma and 100 μL of pre-warmed APTT reagent (0.2% ellagic acid). After incubation for 4 minutes, clotting was initiated by adding 100 μL of 25 mM CaCl2 (37 °C) and time to clot noted. Each clotting assay was performed in duplicate. The data were fit to a quadratic trendline, which was used to determine the concentration of the inhibitor necessary to double the clotting time, 2×APTT or 2×PT.
Results and Discussion
Design of the Sulfated Benzofuran Scaffold
Sulfated LMWLs, which recognize exosite II of thrombin, are based on an aromatic scaffold, a scaffold dramatically different from the highly anionic polysaccharide scaffold of heparin.21 Nevertheless, sulfated LMWLs retain one characteristic of heparin – structural complexity arising from the presence of multiple inter-monomeric linkages, such as β-5, β-β, β-O4, and 5-5 (Fig. 1). A simple calculation suggests that LMWL molecules as small as a tetramer can exhibit nearly 1400 distinct structures even if sulfation and inter-residue linkage are the only variables considered. To uncover distinct structures that may be the seat of anticoagulant activity, we visualized lignin structure as concatenated, aromatic units, instead of repetitions of 4-hydroxy-cinnamic acid monomers joined together through different inter-residue linkages. In this alternative analysis, oligomeric lignin can be considered as a combinatorial library of dihydro-benzofuran, phenoxy-propanoic acid, biphenyl and fused furan monomers (Fig. 1). We reasoned that one or more of these aromatic units with appropriate sulfate groups could be the seat of thrombin recognition and inhibition.
To begin assessing the contribution of these individual aromatic units, we focused on the benzofuran unit, which is a common scaffold used in many synthetic studies. In a preliminary investigation, we had prepared a library of sulfated and carboxylated benzofuran monomers, of which a small group of molecules were found to inhibit thrombin, albeit with IC50 in the millimolar range. Interestingly, mechanistic studies had suggested allosteric inhibition by these molecules.23 This promise led us to explore a more diverse set of sulfated benzofuran monomers and dimers as potential inhibitors of thrombin (Fig. 2).
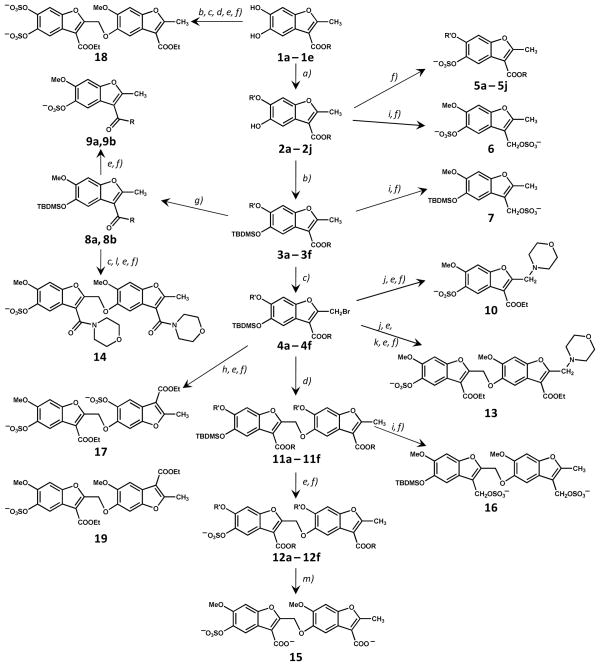
Figure 2.
Synthesis of a library of sulfated benzofurans as potential allosteric regulators of thrombin. The library consists of 15 monomers (5a – 5j, 6, 7, 9a, 9b, and 10) and 13 dimers (12a – 12f, 13–19). R and R′ groups are defined in Table 1. Synthesis of 19 is described in text and Supplementary Material. Reactions: a) Cs2CO3, alkyl or acyl halide, an. DMF; b) TBDMSCl, Imidazole, DMF; c) NBS, CCl4; d) Cs2CO3, 2; DMF; e) KF, CH3COOH, DMF; f) Et3N:SO3, CH3CN:DMF (4:1), MW; g) Me3Al, R-H, toluene; h) Cs2CO3, 1a, DMF; i) LAH, an. THF; j) Cs2CO3, morpholine, DMF; k) Cs2CO3, 4a, DMF; l) Cs2CO3, DMF, desilylated 8a; m) alc. LiOH. See text for details.
Overall Description of the Sulfated Benzofuran Library
The benzofuran unit of sulfated LMWLs contains substitutions at the 2,3,5 positions (Fig. 1). Of these, the 2 and 5 positions are utilized for inter-monomeric linkage and the 3 position may bear a carboxylate or an alkylsulfate group. Considering these variations, charged, polar and hydrophobic substitutions were engineered on this scaffold to obtain a structurally diverse library. Figure 2 describes the multi-modal approach developed to synthesize 15 sulfated monomers and 13 sulfated dimers. This library reflects significant variations in: 1) the number of negatively charged groups (1 to 3 anions); 2) the position of anionic and polar substitutions from the terminal to the center of the scaffold; 3) the size and nature of hydrophobic groups; and 4) inter-residue linkage geometry. A characteristic feature is that each member of the library is nearly fully water soluble because of the presence of one or more sulfate groups, despite the primarily hydrophobic benzofuran backbone. This combination of water solubility and hydrophobicity has been known to introduce novel physicochemical characteristics in the sulfated lignin-based scaffold, e.g., higher than expected partition co-efficient,24 and greater proportion of non-ionic forces in binding to proteins.25
Synthesis of Sulfated Benzofuran Monomers
The first step in the synthesis of the library was the generation of base structures, 5,6-dihydroxy-benzofuran-3-carboxylic acid esters (1a – 1e, Figure 2), from catechol in one step using oxidative Michael addition conditions following the report of Pei et al.26 The reaction was strongly susceptible to substituent effects, which required extensive optimization of reaction conditions, especially the organic base, reaction time and solvent. Eventually, products 1a – 1e were obtained in reasonably good yields (50 – 65%). Monoalkylation or acylation of dihydroxybenzofurans 1 proceeded smoothly to selective 6-substituted derivatives 2a – 2j. That the substitution occurred at the 6-position of the benzofuran ring was confirmed through two dimensional nuclear Overhauser spectroscopy experiment (ROESY), which showed a correlation between the protons of the 3-alkyl ester and 6-alkoxy groups (not shown). The monomeric benzofurans 2a – 2j so synthesized in two steps formed one major group of precursors for library construction. Another set of monomer precursors 3a – 3g and 4a – 4g were synthesized from 2a – 2j through silyl protection and bromination, respectively (Fig. 2).
Mono-sulfated benzofurans 5a – 5j were prepared by direct sulfation of precursors 2a – 2j. Phenolic molecules are typically more challenging to sulfate than alcoholic molecules.27 We had previously developed a microwave-based sulfation protocol for introducing multiple sulfate groups in a crowded environment using triethylamine-sulfur trioxide complex at 100 °C.22,23 This method gave good yields of mono-sulfated benzofurans 5a – 5j in their ammonium salt forms, which were exchanged for sodium using a Sephadex C-25 exchange column. To test whether the number and location of sulfate groups on the benzofuran scaffold play a significant role in recognizing thrombin, we introduced a sulfate group at the 3-position also. Thus, benzofurans 2a and 3a were reduced with LAH and sulfated to yield 6 and 7, respectively. However, both 6 and 7 were found to degrade within a couple of hours after dissolution in water at room temperature. The reason for the reduced aqueous stability of these 3-methylene sulfated benzofurans is not clear, although the dual combination of electron pull by sulfate groups and electron push by ring oxygen may be cause of more rapid de-sulfation.
To assess the importance of the nature of the carbonyl group at the 3-position, silyl-protected benzofuran ester 3 was directly converted to morpholinyl and 4-methylpiperidinyl amides 8a and 8b, respectively, using a procedure reported by Weinreb and coworkers in the late 1970s.28 Desilylation and sulfation gave mono-sulfated benzofuran amides 9a and 9b in excellent yields. Finally, 2-morpholino monomer 10 was prepared from 4a to study the effect of varying the 2-substituent.
Synthesis of Sulfated Benzofuran Dimers
The construction of the library of dimers relied on the nucleophilic displacement of the allylic bromide at the 2 position by free phenolic group at the 5-position. The apparent simplicity of this reaction29 and the availability of ten nucleophilic monomers 2a – 2j and seven allylic bromides 4a – 4g enticed us to construct a combinatorial library (Fig. 2). However, the coupling was challenging on both synthesis and purification fronts. Steric crowding around both the nucleophile and the electrophile resulted in less than optimal yields (<50%) of 11a – 11f. In addition, the synthesized dimers typically co-eluted with monomers in a wide range of multi-solvent system creating significant purification issues. We suspected that co-elution may be occurring because of pi-stacking at high concentrations. Thus, a higher yielding dimerization reaction was desirable, which was made possible through implementation of microwave coupling. Using this strategy silylated dimers 11a – 11f were synthesized in excellent yields (>90%). These were then de-protected and sulfated to yield mono-sulfated benzofuran dimers 12a – 12f. Likewise, mono-sulfated benzofuran dimers 13 and 14, which contain a morpholine ring at 2 and 3 positions, respectively, were synthesized starting from monomers 4a and 8a, respectively, through a series of silyl protection, allylic bromination, deprotection, nucleophilic coupling and sulfation steps (Fig. 2).
To evaluate the role of multiple anionic groups on the small hydrophobic scaffold, 12a was hydrolyzed in alcoholic LiOH30 to yield 15, a mono-sulfated, dicarboxylated dimer. Likewise, 11a was reduced with LAH and sulfated to yield 16, which is a di-sulfated dimer, but as with monomers 6 and 7, 16 was found to degrade rapidly in water. Finally, coupling of 4a with 1a followed by desilylation and sulfation gave 17, which is also a di-sulfated dimer, but with a structure completely different from 16. Likewise, another variant of di-sulfated dimer, i.e., 18, was synthesized by coupling 1a and 2a in five steps (Fig. 2). In combination, dimers 15 – 18 display anionic groups around three key positions (3, 5, and 6) of the benzofuran scaffold. These contain either two or three negative charges, which approximate the number of charges typically found on a heparin disaccharide.
Thrombin Inhibition Potential of Designed Sulfated Benzofuran Monomers and Dimers
Inhibition of thrombin by the designed benzofurans was measured through Spectrozyme TH hydrolysis assay, as described earlier.18,19 This assay relies on the decrease in the initial rate of substrate hydrolysis in the presence of the sulfated benzofuran derivative as a quantitative measure of the inhibition. The fractional decrease in initial rate of hydrolysis typically shows a sigmoidal profile on a semi-log plot, which is fitted by a dose – response equation 1 to derive IC50, HS, Y0 and YM parameters for each potential inhibitor.
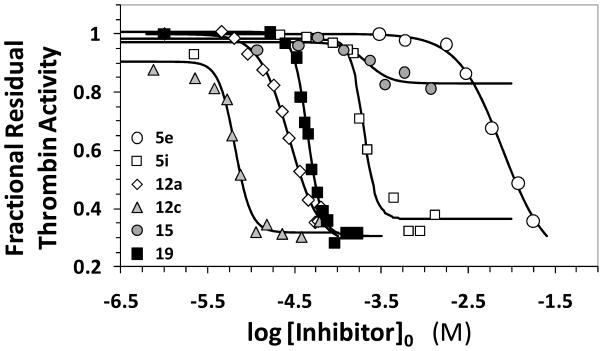
Figure 3 shows the semi-log inhibition curves observed for selected benzofuran monomers and dimers. Of the 28 potential inhibitors studied in this work, only eleven molecules exhibited reasonable inhibition of human α-thrombin at pH 7.4 and 25 °C. These included sulfated monomer 5i and dimers 12a – 12f, 13, and 17 – 19 (Tables 1 and 2). Within this group, inhibitors 12b, 12c, and 12f were the only molecules exhibiting IC50 less than 50 μM. The t-butyl derivative 12c was found to be the most potent thrombin inhibitor with an IC50 of 7.3 μM under physiologically relevant conditions.
Figure 3.
Direct inhibition of thrombin by designed sulfated benzofuran monomers and dimers. The inhibition of human thrombin was determined spectrophotometrically through a Spectrozyme TH hydrolysis assay at pH 7.4 and 25 °C. Solid lines represent sigmoidal fits to the data to obtain IC50, YM, and YO (equation 1), as described in the experimental section. See text for details.
Table 1.
Inhibition of Human α-Thrombin by Sulfated Benzofuran Monomers.a
| R | R′ | log IC50 (M) | IC50 (μM) | ΔY | |
|---|---|---|---|---|---|
| 5a | —OCH2CH3 | —CH3 | ndb | >2500c | - |
| 5b | —OCH3 | —CH3 | nd | >3000c | - |
| 5c | —OC(CH3)3 | —CH3 | nd | >3000c | - |
| 5d | —OCH2 CH2OCH3 | —CH3 | nd | >2000c | - |
| 5e | —OCH2CH3 | —CH2CH3 | −1.7±0.2d | 20560±8500 | 0.60 |
| 5f | —OCH2CH3 | —CH(CH3)2 | −1.9±0.3 | 13650±10400 | 0.75 |
| 5g | —OCH2CH3 | —CH2OCH3 | nd | >16000c | - |
| 5h | —OCH2CH3 | Morpholine-4-carbonyl | nd | >25,000c | - |
| 5i | —OCH2CH3 | Benzoyl-p-methoxy | −3.3±0.2 | 520±280 | 0.54 |
| 5j | —OCH2CH=CH2 | —CH3 | nd | >3000 | - |
| 9a | N-morpholino | —CH3 | nd | >2000c | - |
| 9b | N-piperidino | —CH3 | nd | >2000c | - |
| 10e | —CH2CH3 | —CH3 | nd | >7000c | - |
Thrombin inhibition was measured in spectrophotometric assay through initial rate of Spectrozyme TH hydrolysis as described in Experimental section.
Not determined due to incomplete and/or poor inhibition.
Estimates on the basis of the highest concentration used in experiment.
standard error shown is ±1 S.E.
10 contains a morpholine group at the allylic position (instead of a –CH3 group). See Figure 2 for the full structure.
Table 2.
Inhibition of Human α-Thrombin by Sulfated Benzofuran Dimers.a
| R | R′ | log IC50 (M) | IC50 (μM) | ΔY | |
|---|---|---|---|---|---|
| 12a | —OCH2CH3 | —CH3 | −4.22±0.16b | 60±22 | 0.75 |
| 12b | —OCH3 | —CH3 | −4.43±0.12 | 37±11 | 0.68 |
| 12c | —OC(CH3)3 | —CH3 | −5.14±0.18 | 7.3±3.1 | 0.84 |
| 12d | —OCH2 CH2OCH3 | —CH3 | −3.42±0.13 | 375±109 | 0.80 |
| 12e | —OCH2CH3 | —CH2CH3 | −3.92±0.05 | 121±14 | 0.95 |
| 12f | —OCH2CH3 | —CH(CH3)2 | −4.52±0.19 | 30±13 | 0.72 |
| 13 | nac | na | −3.89±0.20 | 129±60 | 0.62 |
| 14 | N-morpholino | —CH3 | Incompleted | >7100 | nac |
| 15 | —OH | —CH3 | −3.43±0.37 | ~374e | 0.14 |
| 17 | nac | na | −3.12±0.35 | ~764e | 0.95 |
| 18 | nac | na | −3.44±0.06 | 365±57 | 0.79 |
| 19 | nac | na | −4.05±0.26 | 89±54 | 0.70 |
Thrombin inhibition was measured in spectrophotometric assay through initial rate of Spectrozyme TH hydrolysis as described in Experimental section.
standard error shown is ±1 S.E.
Not applicable. See complete structures of 13, 14, 15, 17, 18 and 19 in Figure 2.
Full inhibition profile was not measured.
The error in IC50 could not be accurately defined because of insufficient number of data points.
The range of inhibition efficacy was found to vary considerably with the structure of the inhibitor. For example, sulfated dimer 12e displayed essentially complete inhibition of thrombin (95%, Table 2), while dimer 15 with two carboxylic acid groups inhibited to the extent of only 14% (Table 2). Nearly 85% of the inhibitors studied inhibited thrombin to more than 60% suggesting good overall efficacy in this series of sulfated benzofurans.
Comparison of the two series of benzofuran molecules shows that dimers tend to inhibit thrombin several fold better than monomers. Of the 15 sulfated monomers studied in this work and 17 sulfated and unsulfated monomers studied earlier,23 only 5i inhibited thrombin with measurable potency (~500 μM, Table 1). By contrast, most sulfated dimers displayed thrombin inhibition (Table 2). This suggested that an oligomeric scaffold may exhibit higher potency and rationalizes the nanomolar potency observed for sulfated LMWL inhibition of thrombin.18
The distinct groups at the 2, 3, 5 and 6 positions on the benzofuran ring results in interesting structure–activity relationships. The sulfate group at the 5 position was found to be the key basis for thrombin inhibition, since all unsulfated precursors studied in this work, monomers as well as dimers, did not inhibit thrombin. Such high level of dependence on one sulfate group is striking. It also appears to be unusual because thrombin is known to considerably forgiving in recognizing ligands. For example, ligands as different as heparin and RNA are known to bind in exosite II.31,32 Similarly, hirudin, which contains a sulfated tyrosine, as well as de-sulfated hirudin, i.e., desirudin, bind in exosite I with excellent affinities.2,10,13,33
The stringent requirement for 5-sulfate led to the expectation that multiple sulfate groups on the benzofuran scaffold may induce higher potency. Yet, dimers 17 and 18, which bear two sulfate groups (5 and 5′, and 5 and 6 positions, respectively) inhibited thrombin approximately 6–17-fold weaker (Table 2). Furthermore, the IC50 of 15, which contains the 5-sulfate as well as a carboxylate at each of the 3-positions of the benzofuran rings, also decreases 6.2-fold from its parent 12a. This implies that any additional anionic group on the benzofuran scaffold is counter-productive, which is contrary to what one would predict on the basis of the thrombin – heparin system.17,31,32
Comparison of IC50s of 12a, 12b and 12c shows that as lipophilicity of the 3-ester increases, the inhibition potency improves 8.2-fold (Table 2). Likewise, change in substitution from ethyl to isopropyl at the 6-position, as in 12e and 12f, also increases the potency nearly 4-fold. This implies that hydrophobicity of the scaffold plays an important role in thrombin recognition. In support of this, increase in hydrophilicity of groups markedly decreases potency. For example, the 3-methoxyethyl ester containing 12d is a 6.2-fold weaker inhibitor of thrombin than 3-ethyl containing 12a (Table 2). More importantly, introduction of an N-morpholinyl ring (dimer 14) decreased IC50 to >2000 μM, which represents a more than 270-fold loss of affinity from a comparable t-butyl ester containing dimer 2c.
While the above discussion suggests fairly stringent structural requirements on the benzofuran scaffold, two interesting results highlight the possible variances. Dimer 13 containing a morpholine ring at the 2-allylic position displays IC50 of 129 μM, which is only 2.2-fold higher than that of 12a. This suggests that introduction of a hydrophilic and sterically bulkier group at the terminus opposite the 5-sulfate is not detrimental. Likewise, dimer 19 displays no change in IC50 from its comparable parent 12a (Table 2). Dimer 19 contains a 2-CH2O-6 inter-monomeric linkage as compared to a 2-CH2O-5 linkage present in dimer 12a. An essentially identical inhibition potency of 19 as compared to 12a indicates that a change as drastic as a ring flip, or regioisomerism, is well tolerated.
Mechanism of Thrombin Inhibition By Sulfated Benzofuran Dimer 12a
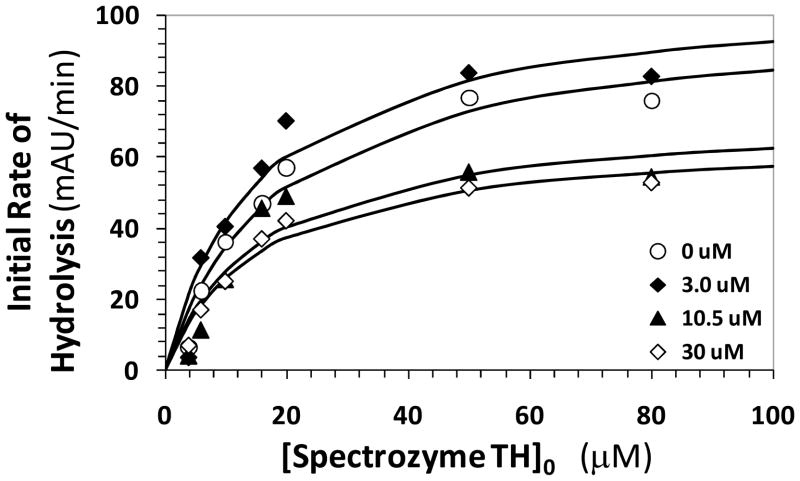
To elucidate the nature of thrombin inhibition by the designed sulfated benzofurans, the kinetics of Spectrozyme TH hydrolysis by thrombin at pH 7.4 in the presence and absence of 12a was studied. Spectrozyme TH has been routinely used as a chromogenic substrate of thrombin.18 The initial rate of amidolysis as a function of the substrate concentration displayed a hyperbolic profile, as expected (Fig. 4), from which the Michaelis constant (KM) and maximal velocity of the reaction (VMAX) were derived. The KM and VMAX for Spectrozyme TH in the absence of 12a was found to be 18.9 μM and 100 mAUmin−1μM−1, respectively. In the presence of 1.5, 3.0, 10.5 and 30 μM 12a, the KM of substrate was found to be 12.6 ± 4.7, 15.5 ± 5.5, 15.9 ± 7.4, and 15.5 ± 3.7 μM. In contrast, the VMAX was measured to be 97 ± 13, 107 ± 14, 73 ± 13 and 66 ± 6 mAUmin−1μM−1 in these experiments. This corresponds to ~34% decrease in maximal velocity of reaction upon approximately 50% saturation of the enzyme. This suggests that the presence of 12a does not significantly affect the binding of the chromogenic substrate to the active site of the enzyme, while significantly reducing its catalytic efficiency.
Figure 4.
Michaelis-Menten kinetics of Spectrozyme TH hydrolysis by human thrombin in the presence of dimer 12a. The initial rate of hydrolysis at various substrate concentrations was measured spectrophotometrically in pH 7.4 buffer as described in ‘Experimental Procedures’. The concentrations of 12a chosen for study include 0 (○), 3.0 (◆), 10.5 (▴), and 30 μM (⋄). Solid lines represent non-linear regressional fits to the data by the standard Michaelis-Menten equation to yield KM and VMAX.
The above Michaelis – Menten kinetics result indicates a non-competitive, allosteric mechanism of thrombin inhibition by 12a and, by analogy, other sulfated benzofuran molecules. The reduction in catalytic efficiency by these novel inhibitors is primarily brought about by the slow conversion of the thrombin–Spectrozyme TH Michaelis complex, which may arise from the intrinsic change in the rate of chemical reaction or by a change in the structure of the catalytic triad induced by the allosteric inhibitors.
Prolongation of Plasma Clotting Time by Sulfated Benzofurans
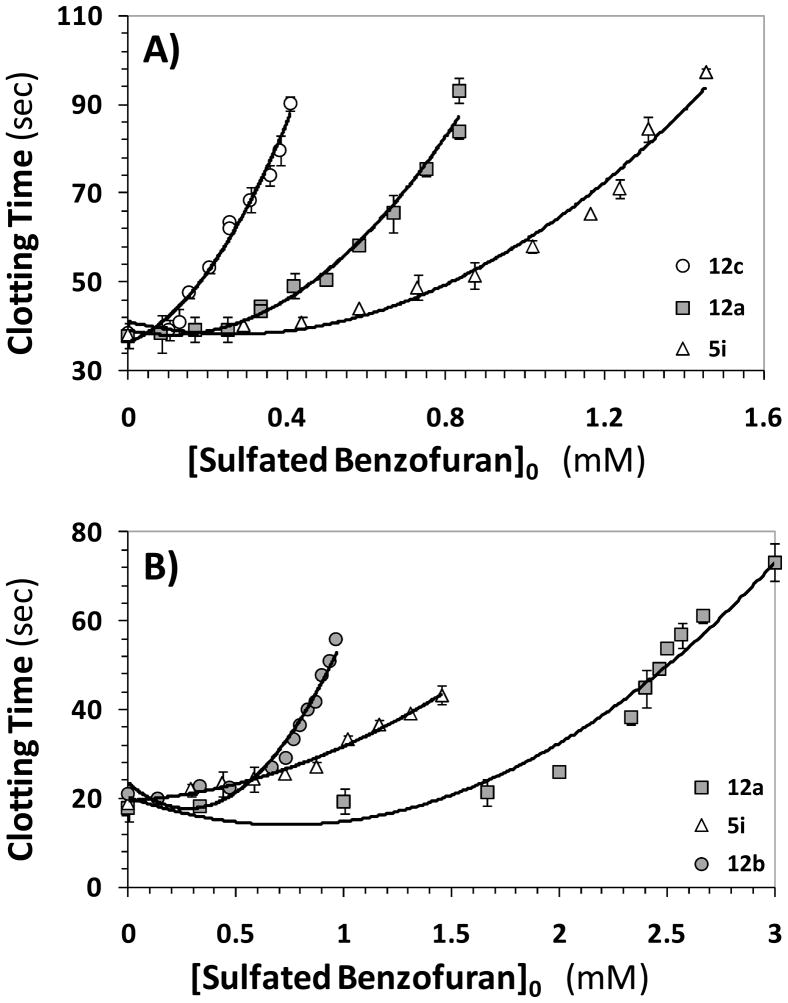
Two plasma clotting assays are typically used to assess the anticoagulant potential of new coagulation inhibitors. These are the prothrombin and activated partial thromboplastin time assays (PT and APTT, respectively), which attempt to measure the effect of an inhibitor on the extrinsic and intrinsic flux of coagulation. The concentrations of the inhibitors required to double the PT and APTT were measured, as described earlier for sulfated LMWLs.20,21 The prolongation of plasma clotting time as a function of the concentration of the sulfated benzofurans followed a pattern typical of other well-studied anticoagulants (Figure 5), except for the range of active concentrations. A 2-fold increase in PT required 850 – 2100 μM concentration of 5i, 12a or 12b (Table 3). In a similar manner, the doubling of APTT required 355 – 1250 μM of 5i, 12a, 12c, or 12e (Table 3). Most other monomers or dimers were found to be not effective. These results suggest that the designed small, sulfated benzofurans are about 100 – 250-fold less potent in human plasma than the oligomeric sulfated LMWLs from which they were designed.21 This is not too unexpected considering that thrombin inhibition by the smaller synthetic molecules is nearly 400-fold weaker than that of the parent oligomers. Yet, interesting similarities and differences are discernible. For example, dimers are better anticoagulants in plasma than monomers. Within the group of dimers, 12a, 12c and 12e, which inhibit thrombin well, are also the best in terms of prolonging APTT. Interestingly, 12b, which is comparable to 12a in thrombin inhibition, does not affect APTT at all (Table 3) and the reason for this behavior is not clear. Overall, the plasma studies suggest the feasibility of designing sulfated benzofurans that will function as anticoagulants in vivo.
Figure 5.
Prolongation of clotting time as a function of concentration of sulfated benzofuran monomers and dimers in either activated partial thromboplastin time assay (A) or prothrombin time assay (B). Solid lines are trend lines, and not non-linear regressional fits. Error bars in the range of symbol size have been omitted. APTT and PT assays were performed as described in the experimental section.
Table 3.
Effect of Designed Sulfated Benzofurans on Human Plasma Clotting Time.a
| APPT | PT | |
|---|---|---|
| (μM) | (μM) | |
| 5i | 1,250 | 1,200 |
| 5g | >1,200 | neb |
| 5c | >2,000 | ne |
| 12a | 740 | 2,100 |
| 12b | >1,000 | 850 |
| 12c | 355 | ne |
| 12d | >1,000 | ne |
| 12e | 530 | ne |
| 14 | >1,000 | ndc |
| 15 | >1,000 | ne |
PT and APTT values were deduced in in vitro human plasma experiments where the clot initiator is either thromboplastin or ellagic acid, respectively. See Methods for details.
no effect.
not determined.
Significance
The range of thrombin inhibition potency and efficacy discovered for sulfated benzofurans suggest that the designed sulfated benzofuran scaffold may be an excellent lead in developing anticoagulants radically different from those studied to-date. The β-5 inter-monomer linkage is only one of the many units present in sulfated LMWLs,18,21 which implies that computational structure-based design may be necessary to identify the most appropriate ‘small’ structure from the large number of potential molecules. One such strategy, e.g., combinatorial virtual library screening, has been put forward for highly sulfated molecules34 and also been found useful for sulfated lignins binding to antithrombin.25
This work presents small, synthetic, sulfated aromatic structures as novel allosteric modulators of thrombin function. Although it is too early to speculate on the site of binding of sulfated benzofurans, the parent oligomeric sulfated LMWLs are known to engage exosite 2 of thrombin.18 Allosteric, exosite 2-based modulators of thrombin are of special interest because they afford the possibility of controlled inhibition of this key protease that takes part in a number of physiological processes.13 Excess inhibition could, in principle, be reversed by highly sulfated small molecules, e.g., sucrose octasulfate, that are also expected to bind in this region without inducing significant reduction in thrombin’s catalytic activity. Allosteric, exosite 2-mediated inhibitors with nanomolar potency, other than sulfated LMWLs,18 have not been designed as yet and thus, sulfated benzofurans represent first molecules in a new class of thrombin modulators.
Although the potency of our first allosteric inhibitors is not high, it is not a major concern because rational design of these molecules from the parent polymers invoked a rather drastic reduction in molecular weight. Whereas sulfated LMWLs are oligomeric chains with an average molecular weight of 5000, sulfated benzofurans range from 500 to 1000.21 Thus, a relatively easy approach to higher potency would be to prepare appropriate tri- and tetramers of sulfated benzofurans.
Finally, this work also highlights the opportunity of exploring allosteric modulators of other coagulation enzymes, e.g., factors Xa, IXa and XIa, through sulfated lignin scaffolds. Considering the wealth of structural diversity accessible with the lignin scaffold, it may be possible to design allosteric inhibitors that target these enzymes in a highly specific manner.
Supplementary Material
Acknowledgments
This work was supported by grants HL099420 and HL090586 from the National Institutes of Health. We thank Dr. H. Tonie Wright for helpful discussions and reading of the manuscript.
Abbreviations
- APTT
activated partial thromboplastin time assays
- LMWL
low molecular weight lignin
- PT
prothrombin time
Footnotes
Supporting Information: Available: Synthesis and spectral properties of molecules depicted in Figure 2. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bauer KA. Hematology Am Soc Hematol Educ Program. 2006. New anticoagulants; pp. 450–456. [DOI] [PubMed] [Google Scholar]
- 2.Henry BL, Desai UR. Anticoagulants. In: Abraham DJ, Rotella DP, editors. Burger’s Medicinal Chemistry, Drug Discovery and Development. 7. John Wiley; Hoboken: 2010. pp. 365–408. [Google Scholar]
- 3.Straub A, Roehrig S, Hillisch A. Oral, direct thrombin and factor Xa inhibitors: The replacement of warfarin, leeches and pig intestines? Angew Chem Int Ed. 2011;50:4574–4590. doi: 10.1002/anie.201004575. [DOI] [PubMed] [Google Scholar]
- 4.Steinmetzer T, Sturzebecher J. Progress in the development of synthetic thrombin inhibitors as new orally active anticoagulants. Curr Med Chem. 2004;11:2297–2321. doi: 10.2174/0929867043364540. [DOI] [PubMed] [Google Scholar]
- 5.Saiah E, Soares C. Small molecule coagulation cascade inhibitors in the clinic. Curr Top Med Chem. 2005;5:1677–1695. doi: 10.2174/156802605775009702. [DOI] [PubMed] [Google Scholar]
- 6.Smallheer JM, Quan ML. Recent advances in coagulation serine protease inhibitors. Ann Rep Med Chem. 2009;44:189–208. [Google Scholar]
- 7.Tucker TJ, Brady SF, Lumma WC, Lewis SD, Gardell SJ, Naylor-Olsen AM, Yan YW, Sisko JT, Stauffer KJ, Lucas BJ, Lynch JJ, Cook JJ, Stranieri MT, Holahan MA, Lyle EA, Baskin EP, Chen IW, Dancheck KB, Krueger JA, Cooper CM, Vacca JP. Design and synthesis of a series of potent and orally bioavailable noncovalent thrombin inhibitors that utilize nonbasic groups in the P1 position. J Med Chem. 1998;41:3210–3219. doi: 10.1021/jm9801713. [DOI] [PubMed] [Google Scholar]
- 8.Burgey CS, Robinson KA, Lyle TA, Nantermet PG, Selnick HG, Isaacs RCA, Lewis SD, Lucas BJ, Krueger JA, Singh R, Miller-Stein C, White RB, Wong B, Lyle EA, Stranieri MT, Cook JJ, McMasters DR, Pellicore JM, Pal S, Wallace AA, Clayton FC, Bohn D, Welsh DC, Lynch JJ, Yan YW, Chen ZG, Kuo L, Gardell SJ, Shafer JA, Vacca JP. Pharmacokinetic optimization of 3-amino-6-chloropyrazinone acetamide thrombin inhibitors. Implementation of P3 pyridine N-oxides to deliver an orally bioavailable series containing P1N-benzylamides. Bioorg Med Chem Lett. 2003;13:1353–1357. doi: 10.1016/s0960-894x(03)00099-4. [DOI] [PubMed] [Google Scholar]
- 9.Brånalt J, Gustafsson D, Nilsson I, Polla M. New heterocyclic carboxamides for use as thrombin inhibitors. 2009/157860. International patent application. WO. :A1. AstraZeneca AB.
- 10.Liu LW, Vu TK, Esmon CT, Coughlin SR. The region of the thrombin receptor resembling hirudin binds to thrombin and alters enzyme specificity. J Biol Chem. 1991;266:16977–16980. [PubMed] [Google Scholar]
- 11.Hortin GL, Trimpe BL. Allosteric changes in thrombin’s activity produced by peptides corresponding to segments of natural inhibitors and substrates. J Biol Chem. 1991;266:6866–6871. [PubMed] [Google Scholar]
- 12.Ye J, Liu LW, Esmon CT, Johnson AE. The fifth and sixth growth factor-like domains of thrombomodulin bind to the anion-binding exosite of thrombin and alter its specificity. J Biol Chem. 1992;267:11023–11028. [PubMed] [Google Scholar]
- 13.Huntington JA. Molecular recognition mechanisms of thrombin. J Thromb Haemost. 2005;3:1861–1872. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- 14.Monteiro RQ, Rapôso JG, Wisner A, Guimarães JA, Bon C, Zingali RB. Allosteric changes of thrombin catalytic site induced by interaction of bothrojaracin with anion-binding exosites I and II. Biochem Biophys Res Commun. 1999;262:819–822. doi: 10.1006/bbrc.1999.1297. [DOI] [PubMed] [Google Scholar]
- 15.Monteiro RQ. Targeting exosites on blood coagulation proteases. An Acad Bras Cienc. 2005;77:275–280. doi: 10.1590/s0001-37652005000200007. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Johnson DJ, Adams TE, Pozzi N, De Filippis V, Huntington JA. Thrombin inhibition by serpins disrupts exosite II. J Biol Chem. 2010;285:38621–38629. doi: 10.1074/jbc.M110.144964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson ST, Halvorson HR, Bjork I. Quantitative characterization of the thrombin-heparin interaction. Discrimination between specific and nonspecific binding models. J Biol Chem. 1991;266:6342–6352. [PubMed] [Google Scholar]
- 18.Henry BL, Monien BH, Bock PE, Desai UR. A novel allosteric pathway of thrombin inhibition. Exosite II mediated potent inhibition of thrombin by chemo-enzymatic, sulfated dehydropolymers of 4-hydroxycinnamic acids. J Biol Chem. 2007;282:31891–31899. doi: 10.1074/jbc.M704257200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry BL, Abdel Aziz M, Zhou Q, Desai UR. Sulfated, low molecular weight lignins are potent inhibitors of plasmin, in addition to thrombin and factor Xa: Novel opportunity for controlling complex pathologies. Thromb Haemost. 2010;103:507–515. doi: 10.1160/TH09-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry BL, Thakkar JN, Martin EJ, Brophy DF, Desai UR. Characterization of the plasma and blood anticoagulant potential of structurally and mechanistically novel oligomers of 4-hydroxycinnamic acids. Blood Coag Fibrinol. 2009;20:27–34. doi: 10.1097/MBC.0b013e328304e077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monien BH, Henry BL, Raghuraman A, Hindle M, Desai UR. Novel chemo-enzymatic oligomers of cinnamic acids as direct and indirect inhibitors of coagulation proteinases. Bioorg Med Chem. 2006;14:7988–7998. doi: 10.1016/j.bmc.2006.07.066. [DOI] [PubMed] [Google Scholar]
- 22.Raghuraman A, Riaz M, Hindle M, Desai UR. Rapid, high-yielding microwave-assisted per-sulfation of organic scaffolds. Tetrahedron Lett. 2007;48:6754–6758. doi: 10.1016/j.tetlet.2007.07.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verghese J, Liang A, Sidhu PS, Hindle M, Zhou Q, Desai UR. First steps in the direction of synthetic, allosteric, direct inhibitors of thrombin and factor Xa. Bioorg Med Chem Lett. 2009;19:4126–4129. doi: 10.1016/j.bmcl.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang A, Thakkar JN, Desai UR. Study of physico-chemical properties of novel highly sulfated, aromatic, mimetics of heparin and heparan sulfate. J Pharm Sci. 2010;99:1207–1216. doi: 10.1002/jps.21908. [DOI] [PubMed] [Google Scholar]
- 25.Henry BL, Connell J, Liang A, Krishnasamy C, Desai UR. Interaction of antithrombin with sulfated, low molecular weight lignins. Opportunities for potent, selective modulation of antithrombin function. J Biol Chem. 2009;284:20897–20908. doi: 10.1074/jbc.M109.013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei L, Li Y, Bu X, Gu L, Chan ASC. One-pot synthesis of 5,6-dihydroxylated benzo[b]furan derivatives. Tetrahedron Lett. 2006;47:2615–2618. [Google Scholar]
- 27.Al-Horani RA, Desai UR. Chemical sulfation of small molecules – Advances and challenges. Tetrahedron. 2010;66:2907–2918. doi: 10.1016/j.tet.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basha A, Lipton M, Weinreb S. A mild, general method for conversion of esters to amides. Tetrahedron Lett. 1977;48:4171–4174. [Google Scholar]
- 29.Flessner T, Doye S. Cesium carbonates: A powerful inorganic base in organic synthesis. J Prakt Chem. 1999;341:186–190. [Google Scholar]
- 30.Dayal B, Salen G, Toome B, Tint GS, Shefer S, Padia J. Lithium hydroxide/aq. methanol: A mild reagent for hydrolysis of bile acids methyl esters. Steriods. 1990;55:233–237. doi: 10.1016/0039-128x(90)90021-3. [DOI] [PubMed] [Google Scholar]
- 31.Carter WJ, Cama E, Huntington JA. Crystal structure of thrombin bound to heparin. J Biol Chem. 2005;280:2745–2749. doi: 10.1074/jbc.M411606200. [DOI] [PubMed] [Google Scholar]
- 32.Jeter ML, Ly LV, Fortenberry YM, Whinna HC, White RR, Rusconi CP, Sullenger BA, Church FC. RNA aptamer to thrombin binds anion-binding exosite-2 and alters protease inhibition by heparin-binding serpins. FEBS Lett. 2004;568:10–14. doi: 10.1016/j.febslet.2004.04.087. [DOI] [PubMed] [Google Scholar]
- 33.Liu CC, Brustad E, Liu W, Schultz PG. Crystal structure of a biosynthetic sulfo-hirudin complexed to thrombin. J Am Chem Soc. 2007;129:10648–10649. doi: 10.1021/ja0735002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raghuraman A, Mosier PD, Desai UR. Finding needle in a haystack. Development of a combinatorial virtual screening approach for identifying high specificity heparin/heparan sulfate sequence(s) J Med Chem. 2006;49:3553–3562. doi: 10.1021/jm060092o. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.