Abstract
Heterotopic ossification (HO) occurs at a high frequency in severe orthopaedic extremity injuries; however, the etiology of traumatic HO is virtually unknown. Osteogenic progenitor cells have previously been identified within traumatized muscle. Although the signaling mechanisms that lead to this dysregulated differentiation pathway have not been identified, it is assumed that inflammation and fibrosis, which contribute to an osteoinductive environment, are necessary for the development of HO. The hypothesis of this study was that cytokines related to chronic inflammation, fibrogenesis and osteogenesis become up-regulated following severe muscle trauma where HO forms. Classification of these cytokines by their differential expression relative to control muscle will provide guidance for further study of the mechanisms leading to HO. Real-time RT-PCR analysis revealed no significant up-regulation of cytokines typically associated with HO (e.g., BMP-4, as observed in the genetic form of heterotopic ossification, Fibrodysplasia Ossificans Progressiva). Instead, the cytokine gene expression profile associated with the traumatized muscle included up-regulation of cytokines associated with osteogenesis and fibrosis (i.e., BMP-1 and TGF-beta1). Using immunohistochemistry, these cytokines were localized to fibroproliferative lesions, which have previously been implicated in HO. This study identifies other cell and tissue-level interactions in traumatized muscle that should be investigated further to better define the etiology of HO.
Keywords: muscle injury, heterotopic ossification, cytokines, bone morphogenetic protein, gene expression profiling
Introduction
Heterotopic ossification (HO) is a complication characterized by the formation of mature bone in the soft tissues (1). HO occurs as an uncommon condition following general musculoskeletal trauma (2), although it has been encountered in greater than 60% of the military population that has sustained poly-traumatic injuries during Operation Iraqi Freedom (OIF) and Operation Enduring Freedom (OEF) (3). HO can result in pain and dysfunction, which result in difficult rehabilitation and revision surgeries. Little is known about the formation and growth of HO except that it typically forms out of fibrotic lesions, which provide an inductive environment for osteochondral ossification (1). It is unclear why HO is more likely to occur as the result of severe and systemic trauma characteristic of combat injuries. Current treatment options for HO, including radiation therapy, non-steroidal anti-inflammatory drugs (NSAIDs) and bisphosphonates, have significant limitations, especially in a multi-traumatized patient population (2). A more directed prophylactic strategy for prevention of HO will require investigation into the biochemical signaling that occurs during the wound healing and regeneration processes in traumatized muscle to identify mechanisms that may be pathological.
Fibrodysplasia ossificans progressiva (FOP), a rare genetic disorder, provides the best characterized model of a pathological soft tissue environment that is inductive to bone formation. FOP has been linked to a mutation in the bone morphogenetic protein (BMP) receptor ACVR-1 (activin A receptor, type 1) (4). In FOP individuals, in response to minor trauma, ACVR-1 appears to become constitutively activated, which is correlated with up-regulation of BMP-4 (5) and inappropriate tissue-level regulation of BMP antagonists, including noggin and gremlin (6). Over time, a fibroproliferative lesion forms and generates an environment where osteoinduction factors can be concentrated in the vicinity of progenitor cells that possess the potential to differentiate and form ectopic bone (7). FOP provides an example of how dysregulated cytokine signaling can result in pathological wound healing, although it is not clear whether any of these events or similar mechanisms occur in muscle tissue following high-energy, combat-related trauma.
We have previously identified a population of multipotent mesenchymal progenitor cells (MPCs) within traumatized muscle with characteristic similarities to mesenchymal stem cells, including the ability to become chondrogenic progenitors and osteoprogenitors (8–9). Dysregulation of inflammatory and wound healing factors could signal the MPCs (10) or other nearby cells (11) to initiate ectopic bone formation. Additionally, the formation of fibrotic lesions in the regenerating muscle tissue provides a hypoxic, osteoinductive environment that could encourage the cells to undergo endochondral ossification (7; 12). However, the cytokine gene expression profile of human muscle tissue following severe trauma has not been completely characterized, and thus there is no rational basis to identify cytokines that may be contributing to a pathological environment in the injured tissue. Therefore, the objective of this study was to determine the differential gene expression of traumatized muscle compared to control muscle tissue to identify signaling mechanisms that may be characteristic of pathological wound healing mechanisms. Our hypothesis was that the gene expression of cytokines that are associated with chronic inflammation, fibrosis and osteogenesis will be up-regulated during the tissue repair processes that follow traumatic muscle injury. Our specific aims were (1) to measure the gene expression of cytokines in traumatically injured muscle that are associated with a genetic forms of HO, i.e., FOP, (2) to determine the cytokine gene expression profile associated with traumatic injury in muscle tissue, and (3) to substantiate the presence of the differentially regulated factors by localizing them within the traumatized muscle samples. The results of this study thus describe a novel cytokine profile for muscle tissue following orthopaedic trauma, and classification of these cytokines based on their differential expression will provide a rational basis to prioritize further investigation into the etiology of HO following traumatic injury.
Materials and Methods
Tissue Collection
Tissue samples were collected with Institutional Review Board approval at Walter Reed Army Medical Center (WRAMC). Debrided tissues containing traumatically injured muscle were obtained during serial washouts (13.1±4.5 days after initial injury) of lower-extremity wounds sustained as a result of high-energy, penetrating trauma during OEF and OIF (n=7, age: 24.8 ± 7.8, all males), and treatment for all but one of the patients resulted in lower extremity amputation. All of these patients developed post-traumatic HO within 1 year of their injury. Control muscle samples were obtained during routine ligament reconstruction when autograft tendons were harvested (n=5, age: 34.4 ± 11.65, all males). Tissue samples were first washed in saline to remove any particulate debris, and areas of fibrous, fat or necrotic tissue were removed, leaving behind only the viable tissue that was adjacent to the new wound margin that was generated during debridement. The tissue was divided into equal portions for histology and RNA extraction.
Real-Time Reverse Transcription – Polymerase Chain Reaction (RT-PCR) Assays
A portion of each individual muscle was flash frozen in liquid nitrogen and stored at −80°C to prevent RNA degradation. Prior to harvesting the RNA, the samples were flash frozen a second time in liquid nitrogen and then homogenized to very fine powder by manual crushing in a mortar that was also cooled in liquid nitrogen. The frozen tissue powder was placed in TRIzol Reagent (Invitrogen, Carlsbad, CA), and further homogenized with an electric rotor-stator. TRIzol based RNA extraction was then performed per the manufacturer’s protocol and further purified using the RNeasy Kit mini-columns (Qiagen, Valencia, CA). RNA concentration was estimated on the basis of A260, and RNA quality was assessed to ensure A260/280>1.8 and A260/230>1.7. cDNA synthesis was performed using the Superscript III First Strand Synthesis System for RT-PCR (Invitrogen). Real-time RT-PCR was performed using an iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, CA) using SYBR Green (Bio-Rad) and primers that were either designed in Beacon Designer (Palo Alto, CA; See Table 1 for primer sequences, annealing temperatures and product sizes) or purchased from SABiosciences (Rockville, MD). Gene expression levels of mRNA were normalized against the gene for β-actin (ACTB) using the comparative cycle threshold (Ct) method.
Table 1.
Primer sequences for gene expression analysis by real-time RT-PCR.
| Gene | Primer Sequence (5’→ 3’) | TA [°C] | Product Size [bp] | |
|---|---|---|---|---|
| ACTA | forward reverse |
TGGAGTGTGATGGCAAGG GAGTGGAAGGACAGTGAGG |
55 | 174 |
| ACVR1 | forward reverse |
GGTAGGGACTGGAGGAACAC GCAATAAAGAAGAGAAGCACAGG |
55 | 163 |
| ACTB | forward reverse |
ATTGCCGACAGGATGCAGAA GCTGATCCACATCTGCTGGAA |
57 | 150 |
| BMP2 | forward reverse |
TGAGTTCACAAGTTCAAGTCC CAGCAATGTCTGGTTCTTATCC |
53 | 168 |
| BMP4 | forward reverse |
TTAACCTCAGCAGCATCC CAGTCTCGTGTCCAGTAG |
50 | 188 |
| GREM | forward reverse |
GGATAATCAGCAGCGTAACTAC TATCATTACATCATCAGTGACTCG |
54 | 175 |
Cytokine gene expression profiles were assayed using the Common Cytokine RT2 Profiler™ PCR Arrays (SA Biosciences). RNA samples were harvested as described above. Prior to reverse transcription, RNA integrity was assessed electrophoretically using a 6000 RNA Pico lab-on-a-chip in a Bioanalyzer (Agilent, Santa Clara, CA). cDNA was synthesized using the RT2 First Strand Kit (SA Biosciences), and the PCR arrays assays were performed according to the manufacturer’s instructions.
Immunohistochemistry
A portion of each tissue sample was immediately fixed in 4% phosphate-buffered paraformaldehyde at 4 °C, dehydrated through a graded series of ethanol, infiltrated with xylenes, embedded in paraffin, and sectioned at a thickness of 5 µm. Sections were deparaffinized in xylenes, rehydrated using a graded series of ethanol and stained with hematoxylin and eosin (H&E) or Mallory’s trichrome stain. H&E staining was used to reveal the local tissue architecture, and the fibrous tissue components were identified using Mallory’s trichrome stain.
Immunohistochemistry was performed by first heating the sections in an Antigen Unmasking Solution (VectorLabs, Burlingame, CA) according to the manufacturer’s instructions. The sections were stained using the Vectastain ABC peroxidase system (VectorLabs) and with mouse anti-CD105 IgG (clone 4G11; VectorLabs), mouse anti-TGF-β1 IgG (clone TFGB17; VectorLabs), rabbit anti-BMP-1 (clone B784; LifeSpan Biosciences, Seattle, WA) and mouse anti-GDF-11 (clone 3g6; Novus Biologicals, Littleton, CO), all of which were reactive against human antigens. Antigen localization was visualized with the 3,3’-diaminobenzidine (DAB) substrate for the horseradish peroxidase and the tissue was counterstained with hematoxylin QS (VectorLabs). The tissue sections were imaged using a Leica FireCam (Wetzlar, Germany).
Statistical Analysis
Relative gene expression levels measured by real-time RT-PCR and PCR arrays were analyzed using Student’s t-tests, with N=7 for the real-time RT-PCR arrays (4 traumatized samples and 3 controls) and N=12 for the PCR array assays (7 traumatized samples and 5 controls). Statistical significance was assigned to p<0.05 for the RT-PCR assays and p<0.015 for the PCR arrays to limit the false discovery rate. By applying the Simes procedure to the PCR array data, we determined the false discovery rate was controlled when p<0.0036 (13).
Results
Gene Expression in Traumatized Muscle
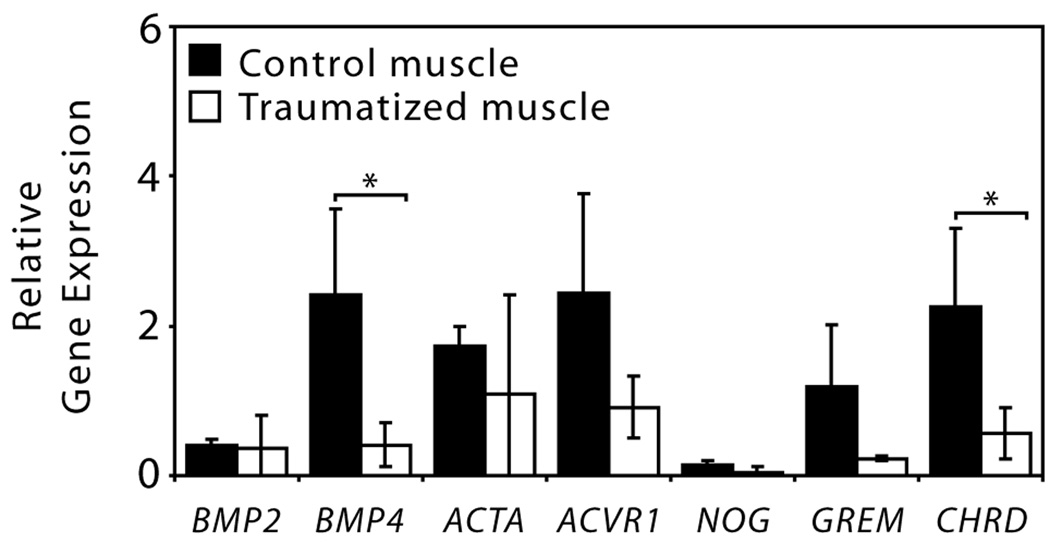
The tissue-level gene expression of cytokines that have been associated with the genetic disorder FOP was evaluated using real-time RT-PCR (Figure 1). BMP4, which encodes a potent osteoinductive cytokine that appears to be up-regulated in FOP lesions (5), was significantly down-regulated in the traumatized tissue, as was CHRD (chordin), a BMP antagonist. Although not significant, there was substantial down-regulation of NOG (noggin) and GREM (gremlin), two additional BMP antagonists.
Figure 1.
Relative gene expression analysis of genes associated with heterotopic ossification measured using quantitative real-time RT-PCR (*p<0.05, Student’s t-tests with N=7). Abbreviations: BMP2 and BMP4 (bone morphogenetic proteins-2 and -4), ACTA (activin), ACVR1 (activin receptor type-1), NOG (noggin), GREM (gremlin) and CHRD (chordin).
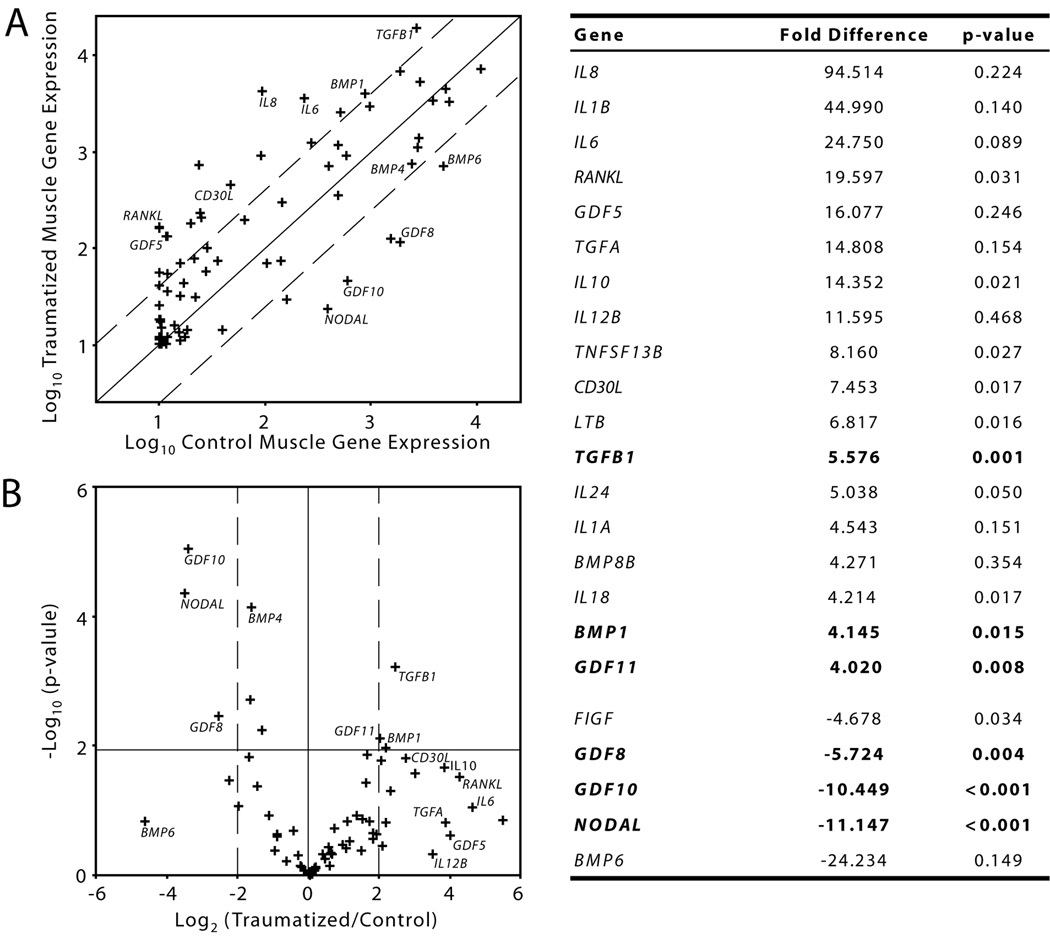
Differential regulation of cytokines between the traumatized and control muscle were represented using a scatter plot (Figure 2A) and a volcano plot (Figure 2B). All of the cytokines that were up- or down-regulated greater than four-fold were listed in Figure 2 with their associated p-value. Three of these genes were significantly up-regulated in the traumatized muscle (TGFB1, GDF11 and BMP1) and three genes were significantly down-regulated (NODAL, GDF10 and GDF8). Although other genes were substantially up-regulated in the traumatized muscle, the statistical significance associated with these genes was not as great as for the smaller set of genes that were down-regulated. The genes bounded in the lower right-hand region of the volcano plot were substantially up-regulated, but with a high degree of variability between patients.
Figure 2.
Cytokine gene expression profile of traumatized and control muscle. (A) The differential gene expression of 84 cytokines and cytokine receptors between traumatized and control muscle tissues with bounds (dashed lines) indicating 4-fold differential expression. (B) A volcano plot comparing the fold-difference in cytokine gene expression (x-axis; dashed lines indicating 4-fold differential expression) to the statistical significance (y-axis; hairline indicating p<0.015). Abbreviations: BMPs (bone morphogenetic proteins), TGFs (transforming growth factors), GDFs (growth and differentiation factors), TNFs (tumor necrosis factors), ILs (interleukins), RANKL (receptor activator for nuclear factor-κ ligand), CD30L (CD30 ligand), LTB (lymphotoxin-β) and FIGF (c-fos induced growth factor).
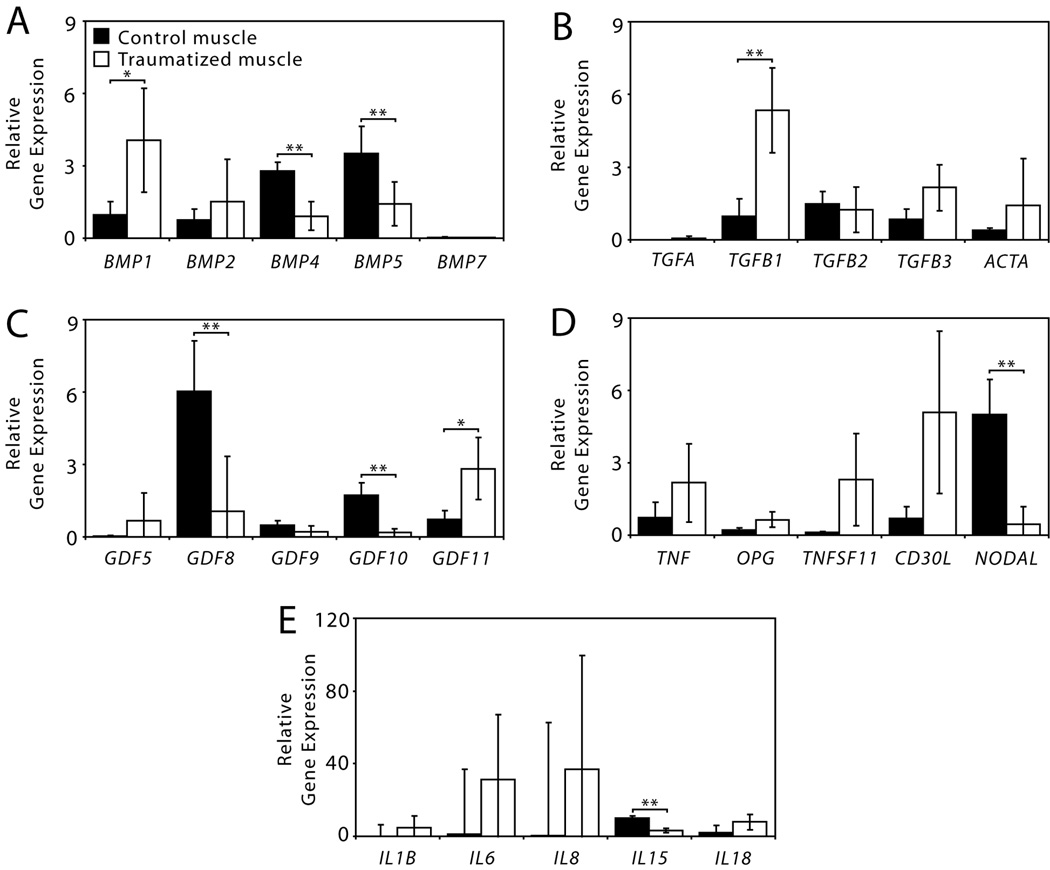
The PCR array data was then broken down into cytokine protein families (Figure 3). BMP1, a metalloproteinase, and GDF11 (growth and differentiation factor-11, also called BMP11) were the only BMP family members that were significantly up-regulated in the traumatized tissue, whereas BMP4, BMP5 and GDF10 (also called BMP3B) were significantly down-regulated (Figure 3A and C). TGFB1 and TGFB3 (transforming growth factors) were both substantially up-regulated in the traumatized muscle, although only TGFB1 was statistically significant, as there was a great deal of variability in the expression of TFGB3 between traumatized muscle samples (Figure 3B). GDF8, the gene that encodes myostatin, was significantly down-regulated in the traumatized muscle tissue (Figure 3C). A number of genes in the TNF (tumor necrosis factor) family and interleukins were substantially up-regulated (Figure 3D and E), although the only statistically significant difference in expression was down-regulation of NODAL and IL15 in the traumatized muscle tissue.
Figure 3.
Relative gene expression of cytokines measured with a semi-quantitative real-time RT-PCR array. (A) Bone morphogenetic proteins (BMPs), (B) Transforming growth factors (TGFs), (C) Growth and differentiation factors (GDFs), (D) Tumor necrosis factors, and (E) Interleukins (ILs). (*p<0.015; **p<0.0036, Student’s t-tests with N=12).
Immunohistochemistry of Traumatized Muscle
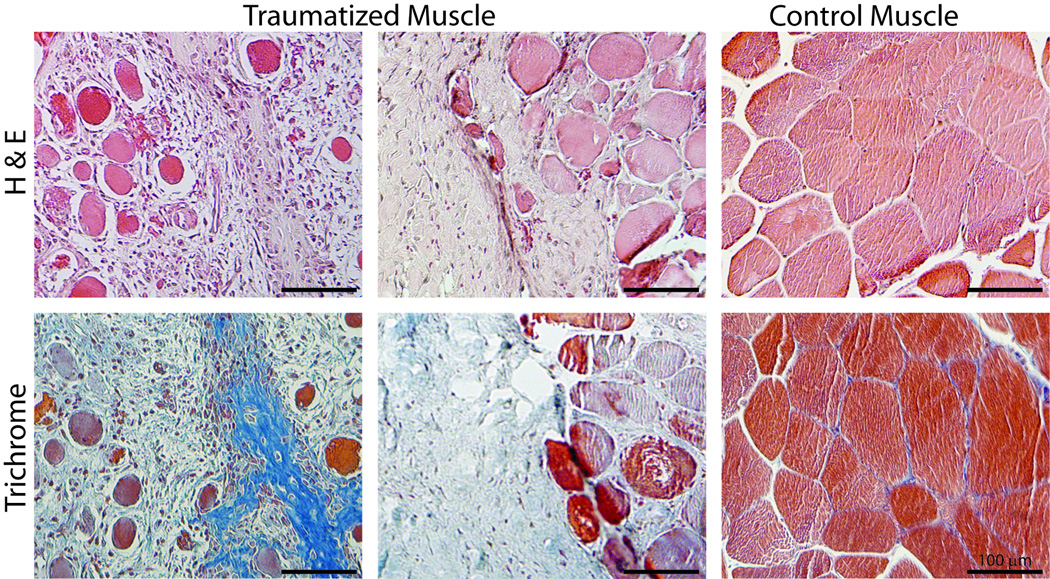
H&E staining showed significant loss of normal tissue architecture in the traumatized muscle samples compared to the controls (Figure 4). The regular orientation and structure of the muscle fibers was disrupted and there was an infiltration of mononuclear cells, including plump fibroblastic cells, indicative of a reactive or reparative response in the tissue. Mallory’s trichrome staining revealed regions interspersed between the remaining muscle fibrils that stained blue, which indicated fibrous tissue with increased collagen content, such as that found during the formation of fibroproliferative and osteochondral tissue (1).
Figure 4.
Histochemical analysis of traumatized muscle tissue compared to control muscle. Hematoxylin and Eosin (H & E) staining demonstrates the gross tissue morphology. Mallory’s trichrome staining differentiates between muscle fibers (deep red), regions rich with extracellular matrix (pale blue) and fibroproliferative lesions (deep blue). Scale bars = 100 µm.
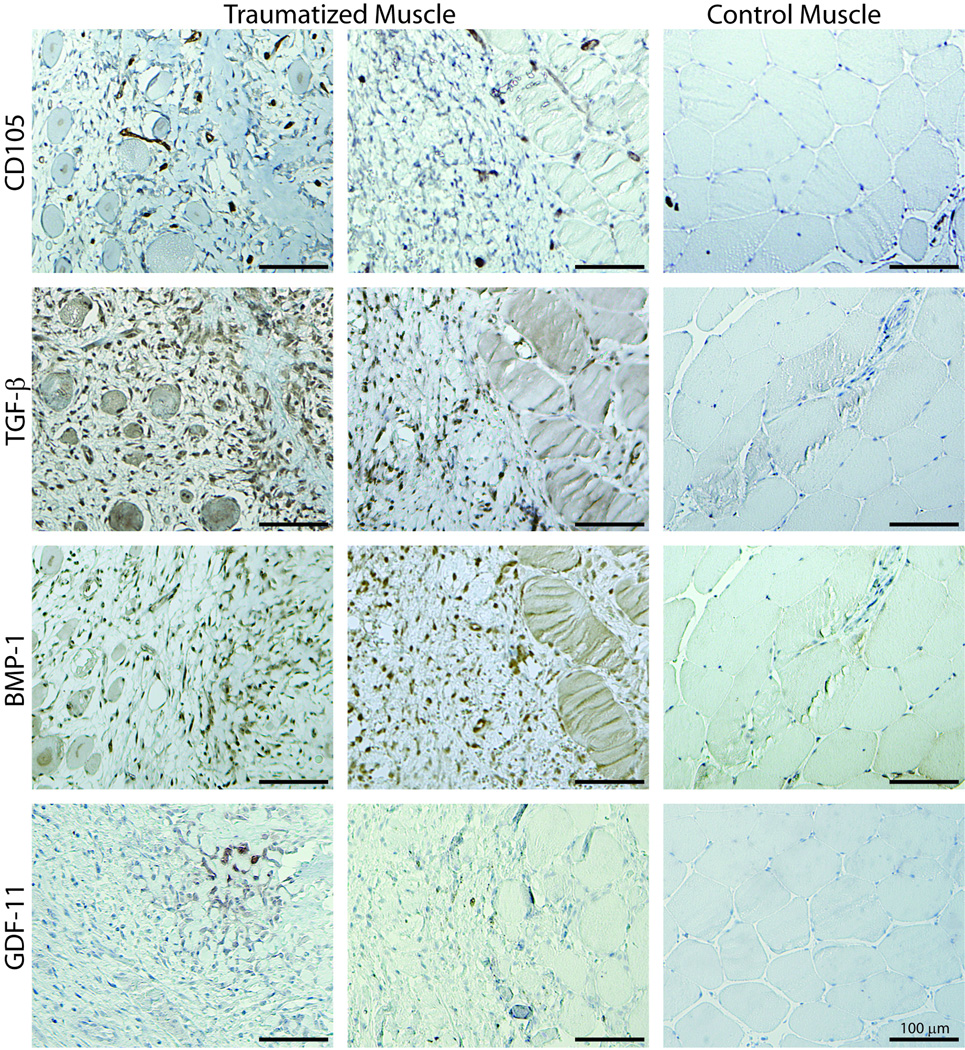
Immunohistochemistry was used to localize the statistically significant and substantially up-regulated cytokines in the muscle tissue (Figure 5). CD105, which has been recently shown to be positive on a population of mesenchymal progenitor cells within traumatized muscle tissue (9), was detected within the population of mononuclear cells in the damaged muscle tissue. In some tissue sections, these appeared to be present in clusters and co-localized with the fibrous tissue lesions. CD105 positive cells did not appear to populate the control muscle tissue. BMP-1 and TGF-β1 were also detected throughout the damaged muscle tissue, but appeared to be concentrated near the fibrous tissue lesions. GDF-11 was detected in the tissue, but it appeared to be concentrated in focal regions rather than being widely dispersed in the tissue. In some cases, these focal regions were coincident with the fibrous tissue lesions. BMP-1, GDF-11 and TGF-β1 were not present at detectable levels in the control muscle tissue.
Figure 5.
Immunohistochemical analysis to localize TGF-β, BMP-1 and GDF-11 relative to the CD105 positive cells in traumatized and control muscle tissue. Positive staining is indicated with DAB (brown) and counterstained with hematoxylin QS (pale blue). Scale bar = 100 µm.
Discussion
The goal of this study was to identify factors associated with wound healing pathologies in the differential gene expression profile of traumatized muscle compared to control muscle tissue. Our findings revealed a uniform up-regulation of cytokines associated with fibrosis (i.e., TGFB1) (14) and osteogenic induction (i.e., BMP1) (15), and thus define a novel cytokine gene expression profile to characterized severely injured muscle tissue. The up-regulated expression of these cytokines was verified and localized in the traumatized tissue relative to the resident CD105 positive MPCs. A second set of cytokines was variably up-regulated (i.e., BMP2, TGFB3, GDF5, IL6, etc.), exhibiting a wide range of expression between patients. The differential regulation of these cytokines between the traumatized muscle tissues could indicate differences in temporal expression in the tissue, which needs to be investigated to better define the wound healing pathologies. There was significant down-regulation of BMP-4, a cytokine that is up-regulated during the process of HO formation observed in FOP (5), which suggests that the etiology of post-traumatic HO may not recapitulate that of the genetic forms of the disease. Taken together, the results of this study describe the cytokine signaling profile of traumatized muscle tissue, which includes a set of cytokines that are variably up-regulated in response to the trauma and should be investigated further to determine whether they have a role in the formation of traumatic HO.
The findings reported here are strengthened by several aspects of our study. First, we investigated the differential expression of BMP4, NOG, and ACVR1 to evaluate the extent to which similarities may be inferred between the etiologies of FOP and traumatic HO. However, our study was not limited to these genes. By using a robust RT-PCR array system, we were able to investigate 84 cytokines commonly associated with trauma and chronic inflammation, and we established a comprehensive profile of muscle tissue following injury, including a novel set of candidate genes for further investigation to determine their potential role in traumatic HO. Secondly, after identifying the genes that appear to be significantly up-regulated in the traumatized tissue, we also used immunohistochemistry to verify that these cytokines are indeed produced in the muscle tissue. In addition to corroborating the gene expression results, the immunohistological analysis reveals the location of the gene products within the tissue constitution and relative to the CD105+ cells, which include the MPCs that are the putative population of HO osteoprogenitor cells (10). Taken together, our findings have provided intriguing indications of the potential interactions occurring at both the cell and tissue-levels, which may eventually be applied to predict the etiology of HO. Finally, all of the assays in this study have been performed on human tissues and do not involve animal models of traumatic injury. As a result, the conclusions of this study are directly applicable to the study of traumatized muscle tissue and the development of traumatic HO.
Despite these strengths, there are several caveats to this study that should be noted. First, the sample size of our gene expression assays is relatively small, which has limited the statistical power of our experiments. Second, the present study has not taken into consideration the amount of time between the injury and collection of the tissue for gene expression and histological analysis. Finally, the present study does not include any clinical data to differentiate between traumatized muscle from patients that develop HO and those who do not. The results presented here will be instructive to formulate larger studies to correlate the time-course of cytokine expression with the clinical outcomes of HO severity. However, the present study provides an initial look into the cell and molecular interactions in traumatized muscle tissue that is likely to form HO. As additional clinical data become available, the cytokine signaling profile identified in this study will enable us to prioritize further investigations to focus on specific candidate genes that are differentially regulated at specific time points after injury in patients that form HO.
While limited in scope, the gene-expression profile of traumatized muscle tissue presented here appears to deviate somewhat from the wealth of etiological data on the genetic disease FOP. Following minor trauma in patients with FOP, the expression of BMP-4 appears to be up-regulated prior to the formation of ectopic bone (5). We did not find data to demonstrate that BMP-4 was up-regulated in the traumatized muscle tissue; instead, we observed a significant decrease in the gene expression of BMP4. Similarly, a mutation in the BMP receptor AVCR1 is thought to play a substantial role in FOP, as it leads to constitutive activation of the associated downstream BMP signaling events (4). We did not find any evidence of a constitutive change in AVCR1 function via up-regulation of this receptor on the cell surface following musculoskeletal trauma, although it is possible that ACVR1 activation and/or signal transduction may be altered by other mechanisms that were not investigated in our study. It is also possible that aberrant signaling related to BMP-4 and ACVR-1 occurs transiently in post-traumatic HO, and outside the post-injury interval observed in this investigation, while it appears to occur constitutively in FOP. However, taken together, these data suggest that caution should be exercised in attributing the pathological mechanisms associated with FOP to the etiology of HO following traumatic injury, as post-traumatic HO may not completely recapitulate the etiology of FOP.
Our data show that BMP1 is significantly up-regulated in traumatized muscle at risk for developing HO, whereas the majority of the BMPs were down-regulated. BMP-1 is unique among the BMPs in that it is a metalloproteinase rather than a TGF-β-like protein. It acts by processing collagen precursors, proteolytically deactivating BMP antagonists, such as chordin (15), and activating latent TGF-β1 (16). BMP-1 may also be inducible by TGF-β1, making this a positive feedback loop of interest in pathologic fibrosis (16).
TGFB1 was the most significantly up-regulated gene in the traumatized muscle, and its expression in muscle injury has been previously documented. TGF-β1 has been identified within injured muscle fibers after strain injury (17), and treatment of the muscle with TGF-β1 antagonists is sufficient to prevent muscle fibrosis after injury (14). TGF-β1 has been reported to induce endothelial cells to undergo transdifferentiation into mesenchymal cells (18), which may occur prior to ectopic bone formation in FOP (11). TGF-β1 may also enhance the multipotency of adjacent mesenchymal progenitor cells (19), as well as initiate chemotaxis and proliferation of osteoblasts (20). Given its pro-fibrotic and osteoinductive effects, TGF-β1 is likely to have a multifaceted role in the etiology of HO. TGF-β3 may be able to attenuate the pro-fibrotic effects of TGF-β1 (21), but the up-regulation of TGFB3 was highly variable in the traumatized muscle tissue. Therefore, the ratio of TGFB1 to TFGB3 may provide a better predictor for identifying patients that are susceptible to fibrosis and osteoinductive fibroproliferative lesions, the putative initiation sites of HO (7; 12).
We have identified fewer genes that were down-regulated in traumatized muscle tissues, but the results are highly statistically significant. Although the functions of GDF10 and NODAL in skeletal muscle are not well understood, GDF8 (also known as myostatin) is a negative regulator of muscle formation (22). The uniform response of these genes to down-regulate in the traumatized muscle is in contrast to the high degree of variability in the genes that are up-regulated (See Figure 2), and indicates that multiple cytokine signaling mechanisms are activated over time in the traumatized muscle tissue. Similarly, GDF11 was significantly up-regulated in the traumatized tissue, but it was localized to focal regions that were unevenly distributed through the tissue. Given the varying levels of their temporal expression and localization, there is reason to suspect that one or more of these cytokines may participate in the pathological mechanisms that lead to the formation of HO. Although GDF-5 has been shown to be osteoinductive (23), the ability of the other genes to promote fibrogenesis or induce osteogenesis is unknown. Further work is required to determine whether the differential regulation of these cytokines may be correlated to the incidence or severity of HO.
The results of this study provide a valuable description of the biochemical environment in muscle tissue following severe injury. We have verified that trauma may generate fibrosis and fibrotic lesions in the muscle, which are assumed to be a necessary step in HO formation, although the tissue-level gene expression profile suggests that there will likely be significant distinctions between post-traumatic HO and genetic forms of the disease, such as FOP. We have identified a set of differentially regulated cytokines in the traumatized muscle that may have a role in the mechanisms of tissue fibrosis and ectopic bone formation. We are currently assessing the fibrogenicity and osteoinductivity of these candidate cytokines using novel, high-throughput in vitro assays for fibrosis and osteogenic differentiation of fibroblasts and traumatized-muscle derived MPCs. We are also continuing to correlate these cytokines to the incidence of HO as additional clinical outcomes data become available. Therefore, the results of this study provide useful guidance that should lead us to more focused future investigations to define key events leading to fibrosis and post-traumatic HO that may serve as novel targets for prophylactic or therapeutic interventions.
Acknowledgement
Support was provided by the Military Amputee Research Program at WRAMC (PO5-A011), the NIH Intramural Research Program (Z01 AR41131) and the Commonwealth of Pennsylvania, Department of Health.
Footnotes
Views expressed in this manuscript are that of the authors alone, and do not reflect that of the United States Government, the United States Army, or the Department of Defense
References
- 1.Kaplan FS, Glaser DL, Hebela N, Shore EM. Heterotopic ossification. J Am Acad Orthop Surg. 2004;12:116–125. doi: 10.5435/00124635-200403000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Pape HC, Marsh S, Morley JR, et al. Current concepts in the development of heterotopic ossification. J Bone Joint Surg Br. 2004;86:783–787. doi: 10.1302/0301-620x.86b6.15356. [DOI] [PubMed] [Google Scholar]
- 3.Potter BK, Burns TC, Lacap AP, et al. Heterotopic ossification following traumatic and combat-related amputations. Prevalence, risk factors, and preliminary results of excision. J Bone Joint Surg Am. 2007;89:476–486. doi: 10.2106/JBJS.F.00412. [DOI] [PubMed] [Google Scholar]
- 4.Shore EM, Xu M, Feldman GJ, et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet. 2006;38:525–527. doi: 10.1038/ng1783. [DOI] [PubMed] [Google Scholar]
- 5.Shafritz AB, Shore EM, Gannon FH, et al. Overexpression of an osteogenic morphogen in fibrodysplasia ossificans progressiva. N Engl J Med. 1996;335:555–561. doi: 10.1056/NEJM199608223350804. [DOI] [PubMed] [Google Scholar]
- 6.Ahn J, Serrano de la Pena L, Shore EM, Kaplan FS. Paresis of a bone morphogenetic protein-antagonist response in a genetic disorder of heterotopic skeletogenesis. J Bone Joint Surg Am. 2003;85-A:667–674. doi: 10.2106/00004623-200304000-00013. [DOI] [PubMed] [Google Scholar]
- 7.Shore EM, Kaplan FS. Insights from a rare genetic disorder of extra-skeletal bone formation, fibrodysplasia ossificans progressiva (FOP) Bone. 2008;43:427–433. doi: 10.1016/j.bone.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nesti LJ, Jackson WM, Shanti RM, et al. Differentiation potential of multipotent progenitor cells derived from war-traumatized muscle tissue. J Bone Joint Surg Am. 2008;90:2390–2398. doi: 10.2106/JBJS.H.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson WM, Aragon AB, Djouad F, et al. Mesenchymal progenitor cells derived from traumatized human muscle. J Tissue Eng Regen Med. 2009;3:129–138. doi: 10.1002/term.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson WM, Aragon AB, Nesti LJ, Tuan RS. Putative heterotopic ossification progenitor cells derived from traumatized muscle. J Orthop Res. 2009 doi: 10.1002/jor.20924. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lounev VY, Ramachandran R, Wosczyna MN, et al. Identification of progenitor cells that contribute to heterotopic skeletogenesis. J Bone Joint Surg Am. 2009;91:652–663. doi: 10.2106/JBJS.H.01177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beiner JM, Jokl P. Muscle contusion injury and myositis ossificans traumatica. Clin Orthop Relat Res. 2002:S110–S119. doi: 10.1097/00003086-200210001-00013. [DOI] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 14.Shah M, Foreman DM, Ferguson MW. Neutralising antibody to TGF-beta 1,2 reduces cutaneous scarring in adult rodents. J Cell Sci. 1994;107:1137–1157. doi: 10.1242/jcs.107.5.1137. [DOI] [PubMed] [Google Scholar]
- 15.Ge G, Greenspan DS. Developmental roles of the BMP1/TLD metalloproteinases. Birth Defects Res C Embryo Today. 2006;78:47–68. doi: 10.1002/bdrc.20060. [DOI] [PubMed] [Google Scholar]
- 16.Bock O, Hoftmann J, Theophile K, et al. Bone morphogenetic proteins are overexpressed in the bone marrow of primary myelofibrosis and are apparently induced by fibrogenic cytokines. Am J Pathol. 2008;172:951–960. doi: 10.2353/ajpath.2008.071030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CA, Stauber F, Waters C, et al. Transforming growth factor-beta following skeletal muscle strain injury in rats. J Appl Physiol. 2007;102:755–761. doi: 10.1152/japplphysiol.01503.2005. [DOI] [PubMed] [Google Scholar]
- 18.Bhowmick NA, Ghiassi M, Bakin A, et al. Transforming Growth Factor-{beta}1 Mediates Epithelial to Mesenchymal Transdifferentiation through a RhoA-dependent Mechanism. Mol. Biol. Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simionescu A, Simionescu DT, Vyavahare NR. Osteogenic Responses in fibroblasts activated by elastin degradation products and transforming growth factor-beta1: role of myofibroblasts in vascular calcification. Am J Pathol. 2007;171:116–123. doi: 10.2353/ajpath.2007.060930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macdonald KK, Cheung CY, Anseth KS. Cellular delivery of TGF-beta1 promotes osteoinductive signalling for bone regeneration. J Tissue Eng Regen Med. 2007;1:314–317. doi: 10.1002/term.31. [DOI] [PubMed] [Google Scholar]
- 21.Shah M, Foreman DM, Ferguson MW. Neutralisation of TGF-beta 1 and TGF-beta 2 or exogenous addition of TGF-beta 3 to cutaneous rat wounds reduces scarring. J Cell Sci. 1995;108:985–1002. doi: 10.1242/jcs.108.3.985. [DOI] [PubMed] [Google Scholar]
- 22.Kirk S, Oldham J, Kambadur R, et al. Myostatin regulation during skeletal muscle regeneration. J Cell Physiol. 2000;184:356–363. doi: 10.1002/1097-4652(200009)184:3<356::AID-JCP10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 23.Spiro RC, Thompson AY, Poser JW. Spinal fusion with recombinant human growth and differentiation factor-5 combined with a mineralized collagen matrix. The Anat Rec. 2001;263:388–395. doi: 10.1002/ar.1119. [DOI] [PubMed] [Google Scholar]