Abstract
We report the discovery of novel small molecule inhibitors of platelet type 12-human lipoxygenase, which display nanomolar activity against the purified enzyme, using a quantitative high throughput screen (qHTS) on a library of 153,607 compounds. These compounds also exhibit excellent specificity, >50-fold selectivity vs. the paralogs, 5-human lipoxygenase, reticulocyte 15-human lipoxygenase type-1, and epithelial 15-human lipoxygenase type-2, and >100-fold selectivity vs. ovine cyclooxygenase-1 and human cyclooxygenase-2. Kinetic experiments indicate this chemotype is a non-competitive inhibitor that does not reduce the active site iron. Moreover, chiral HPLC separation of two of the racemic lead molecules revealed a strong preference for the (–)-enantiomers (IC50 of 0.43 +/- 0.04 and 0.38 +/- 0.05 μM) compared to the (+)-enantiomers (IC50 of >25 μM for both), indicating a fine degree of selectivity in the active site due to chiral geometry. In addition, these compounds demonstrate efficacy in cellular models, which underscores their relevance to disease modification.
Keywords: Human 12-lipoxygenase, high-throughput, selective, inhibitor
Introduction
Lipoxygenases (LOXs) are non-heme, iron-containing enzymes that are ubiquitous in the plant and animal kingdoms. LOXs regio- and stereo-specifically peroxidate polyunsaturated fatty acids (e.g. arachidonic acid (AA) and linoleic acid (LA)) containing cis, cis-1,4-pentadiene moieties to form the corresponding hydroperoxy fatty acids.1 LOXs are the first committed step in a cascade of metabolic pathways that are implicated in the onset of inflammatory diseases such as cancers, heart disease and asthmas,2-5 making LOXs ideal candidates for pharmaceutical inhibitory treatment. However, the discovery of selective, potent inhibitors is critical to providing relevant chemical tools and probes to investigate LOXs involvement in inflammation and disease states.
Human LOXs are distributed among a variety of tissues and cellular locations and have been implicated in numerous disease states. 5-LOX shuttles between the cytosol and nuclear membrane6, 7 and has been found to be implicated in cancer8-10 and asthma.5, 7 Despite 5-LOX having been targeted by pharmaceutical companies for many years,11 Zileutin, developed by the Abbott laboratories, remains the only FDA approved drug which targets a human lipoxygenase.12 Both Pfizer and Merck have developed potent and selective inhibitors of 5-LOX (PF-419183413 and MK-063314 respectively), however, both of these appear to have been discontinued from further clinical development.15 Reticulocyte 15-LOX-1 has been implicated in colorectal16-18 and prostate19-21 cancers, while epithelial 15-LOX-2 is expressed in hair, prostate, lung and cornea22,23 and has been demonstrated to have an inverse correlation of expression and prostate cancer.24,25 Mutations in epidermis-type lipoxygenase-3 and 12-(R)-LOX, which are expressed in the skin, have been shown to cause non-bullous congenital ichthyosiform erythroderma.26 Human platelet type 12-(S)-LOX (12-LOX), for which the current study focuses on, has been implicated in skin disease,27 pancreatic,28 breast,29, 30 prostate cancers,21 diabetes31 and blood coagulation32 and platelet activation.33, 34
Previous studies have demonstrated elevated 12-LOX mRNA expression levels in cancerous prostate tissue cell lines35 and that addition of 12-S-hydroxyeicosatetraenoic acid(12-HETE) to human prostate adenocarcinoma cells increased their motility and ability to invade neighboring tissues. This study has been supplemented with recent data that demonstrates 12-LOX enhances the excretion and expression of vascular endothelial growth factor (VEGF)35 under hypoxic conditions by up-regulating the protein level, mRNA and functionality of hypoxia inducible factor-1 alpha (HIF-1α), a transcription factor that up-regulates VEGF activity.36 Taken together, these studies provide a case for the involvement of 12-LOX in prostate cancer. Additional reports have implicated 12-LOX in breast cancer. 12-LOX mRNA was found to have an increased expression in breast cancer tissue cell lines37 versus noncancerous breast epithelial cell lines, where the addition of the LOX inhibitor cinnamyl-3,4-dihydroxy-alpha-cyanocinnamate dramatically inhibited the growth of the breast cancer cell lines. Increased expression of 12-LOX mRNA has also been witnessed in tissues extracted from patients with breast cancer,38-40 versus healthy adjacent tissue. 12-LOX is also expressed in human pancreatic islets, which increases with inflammatory cytokines and the 12-HETE generated leads to reduced insulin secretion and pancreatic beta cell damage.31
There have been many attempts to discover selective and potent 12-LOX inhibitors with limited success. Past drug discovery efforts for human lipoxygenases have utilized traditional medicinal chemistry41-49 and natural product isolation,50-58 yielding compounds that are reductive or promiscuous phenolic/terpene-based inhibitors. Other reports have focused on pharmacophore virtual screening59 to yield novel, selective inhibitors for rabbit 15-LOX. However, there are few 12-LOX inhibitors that are selective or potent and even fewer which are both.58, 60-62
Several natural products have been shown to exhibit micromolar inhibition of 12-LOX, particularly Hinokitiol58 and the catechins from green tea leaf extract,62 albeit with moderate selectivity over other human LOXs. Our laboratory has discovered and investigated many non-selective 12-LOX inhibitors. Investigations of marine derived natural products yielded brominated aryl phenols,57 and an SAR study, with the known reductive LOX inhibitor, NDGA, yielded a variety of potent derivatives as shown in Figure 1.63 It should be noted that the mistakenly identified 12-LOX selective inhibitor, baicalein, was shown, through steady-state kinetics, to not be selective in vitro, and in fact is equipotent against both 15-LOX-1 and 12-LOX.64
Figure 1.
Previously reported 12-LOX inhibitors.
In an attempt to move from screening to more structure-based design, our laboratory recently identified several non-reductive inhibitors for 12-LOX and 15-LOX-1 by performing docking to homology models, however these compounds displayed weak potency and poor selectivity.65 Our laboratory has also recently discovered four potent and selective 12-LOX inhibitors while testing a high-throughput screen (HTS) against the National Cancer Institute repository and the UC Santa Cruz Marine Extract library.60 Unfortunately, these compounds were not drug-like and not amenable to modification.
Due to the dearth of selective and potent 12-LOX inhibitors and the potential therapeutic benefit for such compounds, herein we have performed a quantitative HTS (qHTS) against a large library of 153,607 small molecules66 in conjunction with the NIH Molecular Libraries Probe Production Center Network (MLPCN) in search of novel, potent and selective 12-LOX inhibitors. We report the discovery and SAR of an 8-hydroxyquinoline-based scaffold with nanomolar potency that is selective over the related human isoforms, 5-LOX, reticulocyte 15-LOX-1, epithelial 15-LOX-2, as well as ovine cyclooxygenase-1 and human cyclooxygenase-2. In addition, the mode of inhibition and the mechanism of action are discussed.
Chemistry
The synthesis of analogues (1-30: see Table 1) could be rapidly accomplished utilizing the Betti Reaction (a type of Mannich reaction, Scheme 1).67,68,69 After optimization of the reaction conditions it was found that the reaction worked best in the absence of solvent (neat) and at temperatures between 120 °C-160 °C (160 °C when R2 = F, 150 °C when R2 = Cl or NO2 and 120 °C when R2 = Br (with overnight heating). One of the initial challenges that had to be overcome was purification of these substrates. While ultimately these could be purified via standard reversed-phase prep-HPLC, we found them to be not well-behaved and often resulted in having to re-prep the samples to achieve >95% purity. However, we found that recrystallization using ethanol or a mixture of ethanol-DMF provided the pure product without any chromatographic purification. This method allowed for the rapid preparation of a wide variety of analogues for SAR investigations. The synthesis of 31 and 33 (see Table 2) were conveniently achieved utilizing a common intermediate 9. Accordingly, treatment of analogue 9 with iodomethane and KOH gave the methoxy derivative 31 in good yield. Piperidine analogue 33 was synthesized via a Rh-catalyzed reduction of 9 under an atmosphere of hydrogen using methanol as a solvent. Naphthanol derivatives, such as compound 32, were synthesized in a similar manner as analogues 1-30, except longer reaction time and lower temperatures were required for optimal yields (55 °C, 15 h., See Supporting Information for a detailed synthetic procedure for 31-33).
Table 1.
12-LOX inhibition of analogues (1-30).a
 | |||||
|---|---|---|---|---|---|
| Compound | R1 | R2 | R3 | R4 | IC50 (μM) [+/- SD (μM)] |
| 1 | H | NO2 | thiophene | CH2CH3 | 0.8 [0.2] |
| 2 | H | Cl | thiophene | CH2CH3 | 1.0 [0.3] |
| 3 | H | Cl | thiophene | CH3 | 1.0 [0.1] |
| 4 | H | Br | thiophene | CH2CH3 | 14 [3.0] |
| 5 | H | Br | thiophene | CH3 | 1.0 [0.2] |
| 6 | H | H | thiophene | CH2CH3 | 3.4 [0.6] |
| 7 | H | F | thiophene | CH3 | 2.0 [0.2] |
| 8 | H | NO2 | furan | CH2CH3 | 1.2 [0.4] |
| 9 | H | Cl | furan | CH2CH3 | 1.0 [0.2] |
| 10 | H | Cl | furan | CH3 | 3.0 [0.5] |
| 11 | H | Br | furan | CH2CH3 | 2.0 [0.5] |
| 12 | H | Br | furan | CH3 | 2.0 [0.3] |
| 13 | H | F | furan | CH3 | 5.0 [1] |
| 14 | H | Cl | cyclopropane | CH2CH3 | 1.6 [0.3] |
| 15 | H | Cl | cyclopropane | CH3 | 3.0 [0.6] |
| 16 | H | Cl | isopropyl | CH2CH3 | 1.2 [0.4] |
| 17 | H | CI | isopropyl | CH3 | 2.6 [0.4] |
| 18 | H | Cl | methyl | CH2CH3 | >50 |
| 19 | H | Cl | methyl | CH3 | >150 |
| 20 | H | F | methyl | CH3 | >75 |
| 21 | H | Cl | H | CH3 | >150 |
| 22 | H | Cl | 5-Me-thiophene | CH2CH3 | 3.5 [1] |
| 23 | H | Cl | 5-bromofuran | CH2CH3 | >75 |
| 24 | Cl | Cl | furan | CH3 | >75 |
| 25 | N(Me)2 | Cl | furan | CH3 | >75 |
| 26 | piperidine | Cl | furan | CH3 | >75 |
| 27 | H | Cl | 4-Me-Ph | CH2CH3 | >150 |
| 28 | H | Cl | 4-F-Ph | CH2CH3 | >50 |
| 29 | H | Cl | furan | Ph | >25 |
| 30 | H | Cl | furan | 4-Me-Ph | >25 |
The UV-Vis-based manual inhibition data (3 replicates) were fit as described in the methods section.
Scheme 1.
Synthesis of 8-hydroxyquinoline analogues (1-30).
Table 2.
12-LOX inhibition of Analogues (31-33).a
 | |
|---|---|
| Compound | IC50 (μM) [+/- SD (μM)] |
| 31 | >75 |
| 32 | >75 |
| 33 | 3.0 [0.7] |
The UV-Vis-based manual inhibition data were fit as described in the methods section.
Results and Discussion
To discover novel inhibitors of 12-LOX, we utilized a chromogenic assay to detect the hydroperoxide lipoxygenase reaction products, as described elsewhere.70 Briefly, after allowing the lipoxygenase reaction to proceed to the desired completion point, ferrous Xylenol Orange was added and the purple product, generated by the oxidation of the ferrous ions to ferric ions by the hydroperoxide, was quantitated using visible light absorbance detection. Prior to the screen, the reaction parameters (concentrations of substrate and enzyme, percent substrate conversion, reagent stability under working conditions) were tested and optimized to ensure adequate performance (data not shown). The screen of the entire library involved testing each of the 153,607 compounds as a dilution series of 7 or more concentrations spanning the 57 μM to 0.7 nM range. This was accomplished by testing 936 assay plates in a fully-automated robotic system (Materials and Methods). The screen was associated with a high signal-to-background average of 4.29 and an average Z’ factor of 0.69 (Supporting Information, Figure S1A). Nordihydroguaiaretic acid (NDGA, a known pan-lipoxygenase inhibitor), present on every screening plate as a 16-point dose-response, exhibited consistent IC50 throughout the screen.
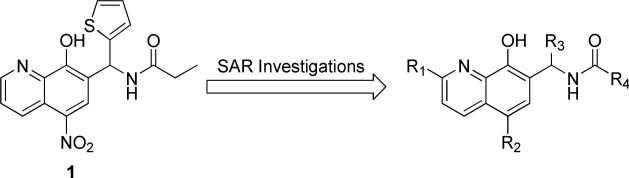
Screening data analysis included classification of the concentration responses based on curve shape, presence of asymptotes, efficacy, and IC50, as described elsewhere.71 In addition to 149,407 inactive samples and 2,295 inconclusive responses, samples belonging to 67 clusters of structurally similar primary screening hits and 124 singletons were associated with complete concentration-response curves (Supporting Information, Figure S1B). Further prioritization based on potency and synthetic tractability led to selection of 59 compounds for additional testing. Complete screening and follow-up data have been made available in PubChem (PubChem BioAssay identifier 2164). Orthogonal confirmation of the hits from the qHTS, using a UV-Vis-based manual assay, yielded an 8-hydroxyquinoline (8-HQ) based chemotype 1 (Figure 2) that displayed micromolar to submicromolar potency, with an IC50 of 0.8 +/- 0.2 μM. To investigate requirements for inhibition and potency, we prepared derivatives of the 8-HQ scaffold that were chemically modified at the R1, R2, R3 and R4 positions (see Table 1). In general, the core scaffold of 1 was tolerant to a variety of changes in size and electrostatics in the R2, R3 and R4 positions without dramatic changes in potency however, there were several combinations of modifications that were not well tolerated.
Figure 2.
Lead compound (1) and positions explored for SAR.
Investigations of various substituents at R3 revealed that compared to the thiophene or furan moiety, both smaller groups (R3 = Me, entries 18-20 and R3 = H, entry 21) and larger groups (R3 = 4-Me-Ph, entry 27 and R3 = 4-F-Ph, entry 28) resulted in essentially complete loss in activity. However, thiophene, furan, cyclopropyl, cyclopentyl, and isopropyl groups at this position all showed comparable activity.
While only limited investigation at R1 was conducted, all modifications at this position led to loss in activity (entries 24-26), suggesting that the binding pocket does not tolerate increased steric bulk in this position. Similarly, increasing the size of the substituents off the amide (R4) was not beneficial for improved activity, as shown with analogues 29 and 30 (R = Ph and 4-Me-Ph, respectively). Additionally, (R4 = CH(CH3)2 and (CH2)2CH3) were synthesized and these analogues also displayed weak potency (data not shown). In contrast, modifications to R2 was well tolerated, with H, Cl, Br, F, NO2 all showing comparable activity.
8-HQ scaffolds are known metal chelators,72 and chelation of the LOX active site iron has been documented in the literature previously.73, 74 Consequently, we sought to investigate the chemical requirements for metal chelation by modifying the oxygen and nitrogen ligands that could bind the active site iron (Table 2, see compounds 31-33). Both methylation of the phenol (entry 31) and removal of the ring nitrogen (analogue 32) resulted in a loss of potency. However, when the pyridine was reduced to the piperidine (analogue 33), a non-rigid heterocycle, the inhibition was unaffected (IC50 = 3.0 +/- 0.7 μM). This data is consistent with the inhibitors ligating the metal in a bidentate fashion, given that piperidines are comparable in ligand strength to pyridine75 and that neither of the monodentate modifications of the 8-HQ core inhibit 12-LOX. This hypothesis is supported by the fact that modifications at R1 (24, 25, 26) also lower potency, presumably due to sterics as the 8-HQ moiety binds the iron. Other small molecule 8-HQs, such as 8-HQ-5 sulfonic acid and 8-HQ-5 nitro, were found to be weak inhibitors (IC50 values > 80 μM against 12-LOX), however, 5,7-dichloro-8-HQ (chloroxine) was an exception. It effectively inhibited all LOX isozymes tested (IC50 values, 5-LOX = 3.6 +/- 0.8 μM, 12-LOX = 1.9 +/- 0.4 μM, 15-LOX-1 = 2.4 +/- 0.4 μM, 15-LOX-2 = 28 +/- 7 μM). Interestingly, chloroxine is a topical anti-microbial agent used in the treatment of seborrheic dermatitis and while no studies have linked lipoxygenase to this ailment, inflammation has been implicated in its treatment, suggesting a possible connection.76
We have attempted to characterize these inhibitors binding directly to the active site ferrous and ferric ion, using UV and fluorescence spectroscopy, however neither experiment was successful. 12-LOX loses activity dramatically as it concentrates, thus making it difficult to achieve enzyme concentrations that would yield a signal of sufficient intensity to observe.
Our final investigation into the SAR around the 8-HQ scaffold was to determine whether the two enantiomers had differential potency. As such, we utilized normal-phase chiral HPLC to effectively separate the enantiomers of both analogues 2 and 5 as shown in Table 3 (HPLC traces, Supporting Information, Figure S2 and S3). Both (–)-2 (aka 36) and (–)-5 (aka 34) displayed 12-LOX inhibition at approximately half the IC50 value of the racemic mixture (34 (–)-enantiomer, IC50 = 0.43 +/- 0.04 μM; (racemic-5) IC50 = 1.0 +/- 0.2 μM; 36 (–)-enantiomer, IC50 = 0.38 +/– 0.05 μM; (racemic-2), IC50 = 1.0 +/- 0.3 μM). In contrast, (+)-2 (aka 37) and (+)-5 (aka 35) displayed no inhibition (IC50 > 25 μM), indicating a fine degree of selectivity in the active site due to chiral geometry. We are currently attempting to crystallize the active enantiomers to determine their absolute stereochemistry.
Table 3.
12-LOX inhibition of enantiomerically pure analogues (34-37).a
 | |
|---|---|
| Compound | IC50 (μM) [+/- SD (μM)] |
| 34 | 0.43 [0.04] |
| 35 | >25 |
| 36 | 0.38 [0.05] |
| 37 | >25 |
The UV-Vis-based manual inhibition data were fit as described in the methods section.
The above data is suggestive that these inhibitors bind directly to the active site iron, which raises the possibility that these inhibitors could have the necessary redox potential to reduce the active ferric iron to the resting ferrous state, as has been reported previously.63, 64, 77, 78 To investigate the redox potential of this chemotype, DPPH, a free radical scavenger, was incubated with compound 1 and no reduction of DPPH was observed. Stoichiometric reduction of DPPH was achieved by the known reductive LOX inhibitor, NDGA, suggesting that these 8-HQ inhibitors are not reductive in nature. Unfortunately, measuring the oxidation of the inhibitor (reduction of the active site iron) with the pseudoperoxidase assay is not possible with 12-LOX, due to a lack of absorbance change at 234 nm. Another mechanism of inhibition seen for LOX is promiscuous inhibition due to small molecule aggregates.79 Compound 3 was therefore screened with increasing amounts of Triton X-100, from 0.005% to 0.02%, with negligible changes in the IC50, indicating that small molecule aggregates are not responsible for inhibition.
In addition to characterizing the inhibition constants, the mode of binding was investigated with steady state kinetics using compound 3 by monitoring the formation of 12-HPETE as a function of substrate and inhibitor concentration in the presence of 0.01% Triton-X-100. Replots of KM/kcat and 1/kcat versus inhibitor concentration (Supporting Information, Figure S4A and S4B) yielded linear plots, with Kic equaling 0.7 +/- 0.09 μM and Kiu equaling 0.8 +/- 0.1 μM, which are defined as the equilibrium constants of dissociation from the enzyme and enzyme substrate complex, respectively. The similar affinity of inhibitor binding to both the enzyme and the enzyme substrate complex (Kic is approximately Kiu) is a rare example of true non-competitive inhibition,80 whose inhibitor affinity is not affected by substrate binding. Regarding the chemical mechanism of inhibition, compound 2 did not display a time dependent inhibition when it was incubated with 12-LOX, unlike the time dependent inhibition seen for the bidentate catechol inhibitors against sLO-174 and the 8-HQ derivatives against macrophage migration inhibitory factor (MIF).81 This data may suggest that compound 2 is not an irreversible inhibitor, however, this could not be proven due to the inactivation of 12-LOX upon dialysis.
To determine the selectivity of this inhibitor chemotype, a subset of compounds was screened against a variety of LOX isozymes (Table 4). In general the selectivity was excellent, with the potency difference being 25- to 150-fold against 15-LOX-1, 45- to 1300-fold against 5-LOX, and 45- to 150-fold against 15-LOX-2. Chiral analogue, 36, was not only the most potent of the inhibitors but it also had some of the best selectivity relative to 5-LOX (1300-fold) and 15-LOX-1 (130-fold). Interestingly, the selectivity of this chemotype between 12-LOX and 15-LOX-1 is less than that between 12-LOX and 5-LOX, which may be accounted for by the similar active sites of 12-LOX and 15-LOX-1.82
Table 4.
Selectivity profile of representative analogues.a
| IC50 (μM) | ||||
|---|---|---|---|---|
| Compound | 12-LOX | 5-LOX | 15-LOX-1 | 15-LOX-2 |
| 1 | 0.8 | ND | >25 | ND |
| 3 | 1.0 | >100 | >25 | ND |
| 5 | 1.0 | >100 | >100 | >100 |
| 6 | 3.4 | >100 | >50 | >100 |
| 9 | 1.0 | >100 | >50 | >100 |
| 34 | 0.43 | >100 | >30 | ND |
| 36 | 0.38 | >100 | >50 | ND |
The UV-Vis-based manual inhibition data (3 replicates) were fit as described in the methods section.
Compound 5 was also screened against the 12/15-mouse lipoxygenase-(12/15-mLO), which generates predominately 12-HPETE, with a smaller percentage of 15-HPETE and had an IC50 value of 14 +/- 3.0 μM. While this value is over 10-fold larger than that for 12-LOX, it is still significantly lower than the IC50 of 5 against 15-LOX-1 (IC50 >150 μM), indicating the active site of 12/15-mLO is more similar to 12-LOX than 15-LOX-1. This was further supported when a highly specific inhibitor against 15-LOX-1 (IC50 < 10 nM against 15-LOX-1), showed an IC50 of greater than 75 μM against 12/15-mLO.70 In addition, compound 5, did not inhibit either cyclooxygenase-1 (COX-1), or COX-2 (IC50 > 100 μM for both), demonstrating a selectivity of greater than 100-fold for 12-LOX over both COX-1 and COX-2.
In order to demonstrate the potential for these compounds to be utilized in more advanced biological systems (e.g. cell-based assays), we investigated various in vitro ADME properties of a representative compound (analogue 34) as shown in Table 5. This chemotype was found to have acceptable kinetic solubility. It should be noted that these conditions are different from the conditions used for the IC50 determinations, which had detergent, lower salt concentrations and higher pH, all leading to greater inhibitor solubility. The inhibitor also showed good cell permeability and excellent stability in PBS buffer and mouse plasma. However, the compound was susceptible to metabolism by mouse liver microsomes with a T1/2 of under 10 minutes. Despite this result we were eager to determine the in vivo PK of this molecule to provide a basis for future investigations in disease relevant mouse models. As shown in Table 6, compound 34 had a reasonable plasma T1/2 of 3.5 h and a Cmax of 288 μM. Importantly, the exposure level exceeded the purified enzyme assay IC50 for the full 24 h period and IC50 in the platelet assay (vide infra) for 8 hours. Moreover, this compound does not efficiently cross the blood-brain-barrier (BBB), which for the treatment of diseases such as diabetes and thrombosis is considered a desirable result as CNS-active compounds could result in undesired side effects. These in vitro ADME and in vivo PK results suggest that the molecules described above should provide utility in both cell-based assays and possibility in vivo models probing the effects of 12-LOX inhibition.
Table 5.
In vitro ADME properties for representative analogue (compound 34).a
| Compound | aq. Kinetic sol. (PBS @ pH 7.4) | Caco-2 (Papp 10-6 m/s @ pH 7.4) | efflux ratio (B→A)/(A→B) | mouse liver microsome stability (T1/2) | PBS-pH 7.4 stability:% remaining after 48h | Mouse plasma stability:% remaining after 48h |
|---|---|---|---|---|---|---|
| 34 | 14.5 μM | 8.8 | 2.3 | <10 min. | 100 | 98.3 |
Kinetic solubility measurements were conducted at Analiza Inc. using nitrogen detection methodologies. Caco-2 permeability, microsomal stability and mouse plasma stability experiments were conducted at Pharmaron Inc.
Table 6.
in vivo PK data for representative analogue (compound 34)a
| compound | t1/2 (h) [plasma] | t1/2 (h) [brain] | [Brain/Plasma]b | Cmax (μM) [Plasma] | Cmax (μM) [Brain] | tmax (h) [Plasma] | tmax (h) [Brain] | cLogP |
|---|---|---|---|---|---|---|---|---|
| 34 | 3.5 | 1.7 | 0.01 | 288 | 5 | 0.25 | 0.5 | 2.8 |
Intraperitoneal (IP) administration (30 mg/kg body weight (mpk)), CD1 mice, n = 3, monitored at 8 time points (0.25 h, 0.5, 1, 2, 4, 8, 12, 24). Compound 34 formulated as a suspension in 50% PEG 200 and 10% Cremophor EL in saline solution
Calculated based on the average [b/p] ratio over 8 time points (24 h period). Experiments were performed by Pharmaron Inc.
Other groups have postulated that 7-substituted-8-HQ derivatives which contain substituted anilines in place of the amide moiety (at C-9) undergo a retro-Mannich reaction to afford a reactive species that can form a variety of covalent adducts (see Figure 3b).83 In agreement with these findings, we too found such analogues (aniline as opposed to amide group at C-9) were relatively unstable in both assay buffer and mouse plasma. However, as shown above, our lead compounds are completely stable in both PBS buffer (pH 7.4) and mouse plasma over a 48 h period. Moreover, this particular chemotype displayed favorable exposure levels in vivo (mouse) as described above. These findings suggest that the retro-Mannich pathway is much less facile for the amide-containing series potentially as a result of amide nitrogen being less basic than the corresponding aniline nitrogen. A comparable 8-HQ chemical series was reported by Wyeth researchers as ADAMTS inhibitors, which like our chemotype contains the amide moiety at C-9 (Figure 3a). They found that the compound displayed good ADME properties (CYP inhibition and microsomal stability), supporting the notion that this subtle structural difference may have a drastic effect on the overall stability of this class of compounds.84
Figure 3.
(a) Representative 8-HQ-based ADAMTS-5 inhibitor reported by Wyeth researchers with amide nitrogen at C-9. (b) Proposed mechanism of covalent modification for 8-HQs with aniline nitrogen at C-9.
A select group of inhibitors, 1, 34 and 35, were then tested for efficacy in a platelet cellular assay. Human platelets are known to express large amounts of 12-LOX upon stimulation of the protease-activated receptor (PAR) with its activating peptide, PAR1-AP.33 We therefore incubated our inhibitors with human platelets, followed by stimulation with PAR1-AP and measured the change in 12-HETE production. In three separate experiments, 100 μM of 1 and 34 showed significant inhibition of PAR1-AP-mediated 12-HETE production, while 35 showed no reduction in 12-HETE production. This data confirms the in vitro data, where both 1 and 34 are potent (IC50 values of 0.8 and 0.43 μM, respectively), while 35 is inactive (IC50 value > 25 μM) and supports the notion that the potent in vitro inhibitors inhibit 12-LOX intracellularly. A more thorough titration of inhibition in the platelet cells was then performed with compounds 1 and 34 (inhibitor concentration ranging from 1 to 100 μM). The IC50 values were determined to be 15 +/- 10 for 1 and 13 +/- 7 for 34 (Figure 4). These cell-based IC50 values are over 25-fold greater than the isolated enzyme IC50 values, which could be a result of limited solubility and/or intracellular metabolism of the inhibitors. We are currently testing various modifications of the inhibitor scaffold to improve the cellular potency with the hopes of applying these inhibitors as anti-coagulation therapeutics.
Figure 4.
Titration curves of 1 (Circles, solid line) and 34 (Triangles, dashed line), against platelets, measuring inhibition of 12-HETE concentration by LC-MS-MS. Fitting the data to a simple hyperbolic curve yielded IC50 values of 15 +/- 10 μM for 1 and 13 +/- 7 μM for 34.
The ability of compound 1 to inhibit the activity of 12-LOX in a diabetic disease relevant model was also assessed using primary human islets obtained from donated tissues. Upon stimulation with arachidonic acid and calcium ionophore, production of 12-HETE in donor islets is significantly increased (Figure 5). This increase was a consistent response across all donor islets tested, and the degree of response was compatible with donor variation. Inclusion of compound 1 at an assay concentration of 10 μM resulted in an uniform inhibition of 68.9 ± 14.5% across the three non-diabetic donors tested (Figure 5A). The basal and stimulated level of 12-HETE was elevated in islets from confirmed type 2 diabetic donors (Figure 5B). This is consistent with the relevance of 12-LOX to diabetes disease progression. Again, compound 1 resulted in an equivalent inhibition of stimulated 12-HETE production, 70 and 74 % respectively in the two diabetic donors assayed.
Figure 5.
Inhibition of 12-HETE production in human donor islets. (A) 12-HETE levels form three non diabetic donors (ND-1 to ND-3) either unstimulated (white bars) or stimulated for 60 minutes with 100μM arachidonic acid (AA) and 30 minutes with 5 μM A23187 (I) in the absence (gray bars) or presence of 10 μM compound 1 (black bars). (B) shows 12-HETE levels in human islets from two confirmed type 2 diabetic donors (T2D-1 and T2D-2). Conditions are as described in A.
Conclusion
Due to the absence of selective, drug-like inhibitors of 12-LOX in the literature, a HTS campaign of 153,607 compounds was performed that identified an 8-HQ chemotype as a novel, potent and selective inhibitor of 12-LOX. Modifications to various positions around the core scaffold led to the development of an SAR profile. In general, the dynamic interplay between the various substitutions is not well understood, and while the SAR was relatively flat, several changes are not at all tolerated. The most dramatic loss of inhibition was observed at the C-2 position of the 8-HQ core (R1), where all substitutions resulted in a complete loss of activity. Moreover, loss of activity was observed by removal of the nitrogen on the pyridine and/or methylation of the 8-hydroxy group, which strongly suggests the inhibitor is binding to the catalytic iron in a bidentate fashion. Further investigations demonstrated the (–)-enantiomers of this 8-HQ chemotype are seemingly responsible for the observed activity, with IC50 values approximately half of the racemic mixture (IC50 of 0.43 +/- 0.04 for 34 and 0.38 +/- 0.05 μM for 36). 5-LOX, 15-LOX-1, 15-LOX-2, COX-1 and COX-2 were not inhibited by this chemotype. Interestingly, this chemotype did show weak inhibition against the 12/15-mLO (IC50 = 14 μM), suggesting the active site of 12/15-mLO is more similar to 12-LOX than 15-LOX-1 (15-LOX-1 IC50 > 150 μM). This is consistent with the fact that 15-LOX-1 selective inhibitors were inactive against 12/15-mLO and indicates caution when performing lipoxygenase studies on particular diseases with mouse models. Investigating the mode of inhibition demonstrated that this class of compounds is thought to be non reductive and exhibits non-competitive inhibition. We are currently investigating these compounds in cell-based diabetes and blood coagulation models and it is our hope that this class of inhibitors will provide a molecular tool to determine the biological role of 12-LOX and further validate 12-LOX as a therapeutic target.
General Chemistry
Unless otherwise stated, all reactions were carried out under an atmosphere of dry argon or nitrogen in dried glassware. Indicated reaction temperatures refer to those of the reaction bath, while room temperature (rt) is noted as 25 °C. All solvents were of anhydrous quality purchased from Aldrich Chemical Co. and used as received. Commercially available starting materials and reagents were purchased from Aldrich and were used as received. Analytical thin layer chromatography (TLC) was performed with Sigma Aldrich TLC plates (5 × 20 cm, 60 Å, 250 μm). Visualization was accomplished by irradiation under a 254 nm UV lamp. Chromatography on silica gel was performed using forced flow (liquid) of the indicated solvent system on Biotage KP-Sil pre-packed cartridges and using the Biotage SP-1 automated chromatography system. 1H- and 13C NMR spectra were recorded on a Varian Inova 400 MHz spectrometer. Chemical shifts are reported in ppm with the solvent resonance as the internal standard (CDCl3 7.26 ppm, 77.00 ppm, DMSO-d6 2.49 ppm, 39.51 ppm for 1H, 13C respectively). Data are reported as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, brs = broad singlet, m = multiplet), coupling constants, and number of protons. Low resolution mass spectra (electrospray ionization) were acquired on an Agilent Technologies 6130 quadrupole spectrometer coupled to the HPLC system. High resolution mass spectral data was collected in house using an Agilent 6210 time-of-flight mass spectrometer, also coupled to an Agilent Technologies 1200 series HPLC system. If needed, products were purified via a Waters semi-preparative HPLC equipped with a Phenomenex Luna® C18 reverse phase (5 micron, 30 × 75 mm) column having a flow rate of 45 mL/min. The mobile phase was a mixture of acetonitrile (0.1% TFA) and H2O (0.1% TFA) at room temperature.
Samples were analyzed for purity on an Agilent 1200 series LC/MS equipped with a Luna® C18 reverse phase (3 micron, 3 × 75 mm) column having a flow rate of 0.8-1.0 mL/min over a 7-minute gradient and a 8.5 minute run time. The mobile phase was a mixture of acetonitrile (0.025% TFA) and H2O (0.05% TFA), and the temperature was maintained at 50 °C. Purity of final compounds was determined to be >95%, using a 3 μL injection with quantitation by AUC at 220 and 254 nm (Agilent Diode Array Detector).
Chiral HPLC separation of the enantiomers
Methanol was added to the sample until the sample was completely dissolved. Sample dissolution and analysis was performed at room temperature. The analytical analysis was performed on a Chiralcel OD column (4.6 × 150 mm, 5 micron). The mobile phase was 100% methanol at 1.0 mL/min. The sample was detected with a diode array detector (DAD) at 220 nm and 254 nm. Optical rotation (+/–) was determined with an in line polarimeter (PDR-Chiral). Preparative purification was performed on a Chiralcel OD column (2 × 25 cm, 5 micron). The mobile phase was 100% methanol at 4.5 mL/min. Fraction collection was triggered by UV absorbance (254 nm). The LC system was limited to 100 μSL injections. The fractions containing the requisite enantiomers were combined and concentrated under reduced pressure. The enantiomeric purity was determined by re-injection on the analytical column described above. The enantiomeric ratios (er) for all separated chiral compounds were found to be greater than 99:1. The optical rotations of the final compounds were obtained using a PerkinElmer model 341 polarimeter.
General procedure for the synthesis of 8-HQ analogues (Scheme 1)
Differentially substituted-quinolin-8-ol (1 equiv.), amide (1.05 equiv.) and the requisite aldehyde (1.1 equiv.) were stirred neat at 120-160 °C for 15 min. to 12 h (depending on the substituent at C-5). Upon heating, the reaction mixture melted and solid was formed after completion of the reaction. The solid product was washed with ethyl acetate and the crude product was purified by recrystallization from ethanol or an ethanol-DMF mixture.
Representative Characterization Data
N-((8-hydroxy-5-nitroquinolin-7-yl)(thiophen-2-yl)methyl)propionamide (1)
LC MS: rt (min) = 5.16; 1H NMR (DMSO-d6) δ 1.03 (t, J = 7.5 Hz, 3 H), 2.23 (qd, J = 7.6 Hz and 3.1 Hz, 2 H), 6.81 (dt, J = 3.5 Hz and 1.2 Hz, 1 H), 6.89 (dd, J = 8.6 Hz and 1.0 Hz, 1 H), 6.95 (dd, J = 5.1 Hz and 3.5 Hz, 1 H), 7.43 (dd, J = 5.1 Hz and 1.2 Hz, 1 H), 7.90 (dd, J = 8.8 Hz and 4.3 Hz, 1 H), 8.76 (s, 1 H), 9.02 (dd, J = 4.1 Hz and 1.6 Hz, 1 H), 9.09 (d, J = 8.8 Hz, 1 H), and 9.19 (dd, J = 8.8 Hz and 1.6 Hz, 1 H); 13C NMR (DMSOn-d6) δ 9.86, 28.38, 45.33, 121.73, 123.84, 125.21, 125.29, 125.35, 126.87, 127.23, 133.05, 134.37, 136.80, 145.17, 149.03, 157.34 and 172.29; HRMS (m/z): [M + H]+ calcd. for C17H16N3O4S, 358.0856; found, 358.0861.
N-((5-chloro-8-hydroxyquinolin-7-yl)(thiophen-2-yl)methyl)propionamide (2)
LC MS: rt (min) = 5.6; 1H NMR (DMSO-d6) δ 1.03 (t,-J = 7.6 Hz, 3 H), 2.16 - 2.28 (m, 2 H), 6.75 - 6.78 (m, 1 H), 6.89 - 6.96 (m, 2 H), 7.40 (dd, J = 5.1 Hz and 1.0 Hz, 1 H), 7.74 (dd, J = 8.6 Hz and 4.1 Hz, 1 H), 7.79 (s, 1 H), 8.50 (dd, J = 8.5 Hz and 1.5 Hz, 1 H), 8.91 (d, J = 8.8 Hz, 1 H), 8.98 (dd, J = 4.1 Hz and 1.4 Hz, 1 H) and 10.42 (brs, 1 H); HRMS (m/z): [M + H]+ calcd. for C17H16ClN2O2S, 347.0618; found, 347.0621. (–)-2 (e.g. 36) [α]D 23 = –24 (c = 0.6, CHCl3); (+)-2 (e.g. 37) [α]D 23 = +24 (c = 0.6, CHCl3).
N-((5-chloro-8-hydroxyquinolin-7-yl)(thiophen-2-yl)methyl)acetamide (3)
LC-MS: rt (min) = 5.28 ; 1H NMR (DMSO-d6) δ 1.94 (s, 3 H), 6.78 (d, J = 3.5 Hz, 1 H), 6.89 (d, J = 8.8 Hz, 1 H), 6.94 (dd, J = 5.1 Hz and 3.5 Hz, 1 H), 7.41 (dd, J = 5.1 Hz and 1.0 Hz, 1 H), 7.75 (dd, J = 8.5 Hz and 4.2 Hz, 1 H), 7.78 (s, 1 H), 8.51 (dd, J = 8.6 Hz and 1.4 Hz, 1 H), 8.96 - 9.02 (m, 2 H) and 10.43 (brs, 1 H); ); HRMS (m/z): [M + H]+ calcd. for C16H14ClN2O2S, 333.0459; found, 333.0460.
N-((5-bromo-8-hydroxyquinolin-7-yl)(thiophen-2-yl)methyl)propionamide (4)
LC-MS: rt (min) = 5.70; 1H NMR (DMSO-d6) δ 1.03 (t, J = 7.5 Hz, 3 H), 2.16 - 2.29 (m, 2 H), 6.76 (d, J = 3.3 Hz, 1 H), 6.87 - 6.96 (m, 2 H), 7.40 (dd, J = 5.1 Hz and 1.0 Hz, 1 H), 7.74 (dd, J = 8.5 Hz and 4.2 Hz, 1 H), 7.96 (s, 1 H), 8.43 (dd, J = 8.6 Hz and 1.4 Hz, 1 H), 8.86 - 8.99 (m, 2 H) and 10.45 (brs, 1 H); HRMS (m/z): [M + H]+ calcd. for C17H16BrN2O2S, 391.0105; found, 391.0108.
N-((5-bromo-8-hydroxyquinolin-7-yl)(thiophen-2-yl)methyl)acetamide (5) ML127
LC-MS: rt (min) = 5.36; 1H NMR (DMSO-d6) δ 1.94 (s, 3 H), 6.78 (dt, J = 3.5 Hz and 1.2 Hz, 1 H), 6.89 (dd, J = 8.9 Hz and 1.1 Hz, 1 H), 6.93 (dd, J = 5.1 Hz and 3.3 Hz, 1 H), 7.40 (dd, J = 5.1 Hz and 1.4 Hz, 1 H), 7.74 (dd, J = 8.6 Hz and 4.1 Hz, 1 H), 7.95 (s, 1 H), 8.43 (dd, J = 8.5 Hz and 1.5 Hz, 1 H), 8.95 (dd, J = 4.1 Hz and 1.6 Hz, 1 H), 9.00 (d, J = 8.8-Hz, 1 H) and 10.46 (brs, 1 H); 13C NMR (DMSO-d6) δ 22.55, 45.35, 108.50, 123.41, 124.84, 125.08, 125.68, 126.34, 126.78, 129.38, 134.97, 138.89, 145.91, 149.24, 149.71 and 168.39; HRMS (m/z): [M + H]+ calcd. for C16H14BrN2O2S, 376.9954; found, 376.9956. (–)-5 (e.g. 34) [α]D 23 = –18 (c = 0.3, CHCl3); (+)-5 (e.g. 35) [α]D 23 = +18 (c = 0.3, CHCl3)
N-((5-fluoro-8-hydroxyquinolin-7-yl)(thiophen-2-yl)methyl)acetamide (7)
LC-MS: rt (min) = 4.76; 1H NMR (DMSO-d6) δ 1.93 (s, 3 H), 6.77 (dt, J = 3.5 Hz and 1.2 Hz, 1 H), 6.88 - 6.95 (m, 2 H), 7.40 (dd, J = 5.0 Hz and 1.3 Hz, 1 H), 7.47 (d, J = 11.2 Hz, 1 H), 7.68 (dd, J = 8.5 Hz and 4.2 Hz, 1 H), 8.44 (dd, J = 8.5 Hz and 1.7 Hz, 1 H), 8.91 - 8.99 (m, 2 H) and 10.07 (brs, 1 H); 13C NMR (DMSO-d6) δ 22.55, 45.49, 104.54, 109.27, 109.48, 117.53, 117.72, 122.27, 123.67, 123.73, 124.86, 125.04, 126.73, 129.13, 129.16, 137.80, 137.83, 145.91, 146.04, 146.08, 148.17, 149.52, 150.60 and 168.37; HRMS (m/z): [M + H]+ calcd. for C16H14FN2O2S, 317.076; found, 317.0761.
N-(furan-2-yl(8-hydroxy-5-nitroquinolin-7-yl)methyl)propionamide (8)
LC-MS: rt (min) = 4.96; 1H NMR (DMSO-d6) δ 1.01 (t, J = 7.6 Hz, 3 H), 2.16 - 2.27 (m, 2 H), 6.13 (d, J = 3.3 Hz, 1 H), 6.38 (dd, J = 3.1 Hz and 1.8 Hz, 1 H), 6.69 (d, J = 8.41 Hz, 1 H), 7.61 (d, J = 1.0 Hz, 1 H), 7.90 (dd, J = 8.9 Hz and 4.2 Hz, 1 H), 8.68 (s, 1 H), 8.97 (d, J = 8.4 Hz, 1 H), 9.01 (dd, J = 4.1 Hz and 1.37 Hz, 1 H) and 9.17 – 9.21 (m, 1 H); HRMS (m/z): [M + H]+ calcd. for C17H16N3O5, 342.1084; found, 342.1082.
N-((5-chloro-8-hydroxyquinolin-7-yl)(furan-2-yl)methyl)propionamide (9)
LC-MS: rt (min) = 5.26; 1H NMR (DMSO-d6) δ 1.01 (t, J = 7.5 Hz, 3 H) 2.20 (qd, J = 7.5 Hz and 2.6 Hz, 2 H) 6.07 (d, J = 3.1 Hz, 1 H) 6.37 (dd, J = 3.0 Hz and 1.9 Hz, 1 H) 6.72 (d, J = 8.6 Hz, 1 H) 7.59 (s, 1 H) 7.66 - 7.89 (m, 2 H) 8.49 (dd, J = 8.5 Hz and 1.5 Hz, 1 H) 8.80 (d, J = 8.8-Hz, 1 H) 8.97 (dd, J = 4.2 Hz and 1.5 Hz, 1 H) and 10.38 (brs, 1 H); 13C NMR (DMSO-d6) δ 9.81, 28.28, 43.99, 106.95, 110.39, 118.47, 123.04, 123.07, 125.04, 126.15, 132.50, 138.62, 142.51, 149.16, 149.42, 153.95 and 172.21; HRMS (m/z): [M + H]+ calcd. for C17H16ClN2O3, 331.0844; found, 331.0849.
N-((5-chloro-8-hydroxyquinolin-7-yl)(furan-2-yl)methyl)acetamide (10)
LC-MS: rt (min) = 4.83; 1H NMR (DMSO-d6) δ 1.92 (s, 3 H), 6.06 - 6.09 (m, 1 H), 6.37 (dd, J = 3.2 Hz and 1.9 Hz, 1 H), 6.70 (d, J = 8.4 Hz, 1 H), 7.59 (dd, J = 1.9 Hz and 1.0 Hz, 1 H), 7.71 (s, 1 H), 7.74 (dd, J = 8.5 Hz and 4.2 Hz, 1 H), 8.50 (dd, J = 8.6 Hz and 1.6 Hz, 1 H), 8.88 (d, J = 8.6 Hz, 1 H), 8.97 (dd, J = 4.1 Hz and 1.6 Hz, 1 H) and 10.39 (brs, 1 H); 13C NMR (DMSO-d6) δ 22.51, 44.05, 106.99, 110.39, 118.48, 123.00, 123.05, 125.05, 126.14, 132.52, 138.62, 142.52, 149.19, 149.40, 153.86 and 168.47; HRMS (m/z): [M + H]+ calcd. for C16H14ClN2O3, 317.0687; found, 317.0689.
N-((5-bromo-8-hydroxyquinolin-7-yl)(furan-2-yl)methyl)propionamide (11)
LC-MS: rt (min) = 5.39; 1H NMR (DMSO-d6) δ 1.01 (t, J = 7.6 Hz, 3 H), 2.20 (qd, J = 7.5 Hz and 2.2 Hz, 2 H), 6.07 (d, J = 3.1 Hz, 1 H), 6.37 (dd, J = 3.2 Hz and 1.9 Hz, 1 H), 6.71 (d, J = 8.4 Hz, 1 H), 7.59 (dd, J = 1.8 Hz and 0.8 Hz, 1 H), 7.73 (dd, J = 8.6 Hz and 4.1 Hz, 1 H), 7.88 (s, 1 H), 8.42 (dd, J = 8.6 Hz and 1.6 Hz, 1 H), 8.81 (d, J = 8.8 Hz, 1 H), 8.94 (dd, J = 4.1 Hz and 1.6 Hz, 1 H) and 10.41 (brs, 1 H); 13C NMR (DMSO-d6) δ 9.81, 28.29, 43.96, 106.94, 108.33, 110.39, 123.38, 123.73, 126.35, 129.63, 134.95, 138.86, 142.51, 149.17, 150.03, 153.96 and 172.22; HRMS (m/z): [M + H]+ calcd. for C17H16BrN2O3, 375.0339; found, 375.0344.
N-((5-bromo-8-hydroxyquinolin-7-yl)(furan-2-yl)methyl)acetamide (12)
LC-MS: rt (min) = 5.03; 1H NMR (DMSO-d6) δ 1.92 (s, 3 H), 6.04 - 6.11 (m, 1 H), 6.37 (dd, J = 3.2 Hz and 1.9 Hz, 1 H), 6.70 (d, J = 8.6 Hz, 1 H), 7.56 - 7.62 (m, 1 H), 7.73 (dd, J = 8.5 Hz and 4.2 Hz, 1 H), 7.87 (s, 1 H), 8.43 (dd, J = 8.6 Hz and 1.6 Hz, 1 H), 8.89 (d, J = 8.6 Hz, 1 H), 8.94 (dd, J = 4.1 Hz and 1.6 Hz, 1 H) and 10.42 (brs, 1 H); 13C NMR (DMSO-d6) δ 22.51, 43.99, 106.96, 108.33, 110.39, 123.38, 123.65, 126.35, 129.61, 134.96, 138.86, 142.52, 149.18, 150.00, 153.87 and 168.47; HRMS (m/z): [M + H]+ calcd. for C16H14BrN2O3, 361.0182; found, 361.019.
Methods
Biological Reagents
All commercial fatty acids (Sigma-Aldrich Chemical Company) were re-purified using a Higgins HAIsil Semi-Preparative (5 μm, 250 × 10 mm) C-18 column. Solution A was 99.9% MeOH and 0.1% acetic acid; solution B was 99.9% H2O and 0.1% acetic acid. An isocratic elution of 85% A:15% B was used to purify all fatty acids, which were stored at – 80 °C for a maximum of 6 months.
Overexpression and Purification of 12-Human Lipoxygenase, 5-Human Lipoxygenase, 12/15-Mouse Lipoxygenase and the 15-Human Lipoxygenases
Human platelet 12-lipoxygenase (12-LOX), human reticulocyte 15-lipoxygenase-1 (15-LOX-1), human epithelial 15-lipoxygenase-2 (15-LOX-2), were expressed as N-terminally, His6-tagged proteins and purified to greater than 90% purity, as evaluated by SDS-PAGE analysis.50, 85, 86 Human 5-lipoxygenase was expressed as a non-tagged protein and used as a crude ammonium sulfate protein fraction, as published previously.87 Iron content of 12-LOX was determined with a Finnegan inductively coupled plasma mass spectrometer (ICP-MS), using cobalt-EDTA as an internal standard. Iron concentrations were compared to standardized iron solutions and used to normalize enzyme concentrations.
High-throughput Screen: Materials
Dimethyl sulfoxide (DMSO) ACS grade was from Fisher, while ferrous ammonium sulfate, Xylenol Orange (XO), sulfuric acid, and Triton X-100 were obtained from Sigma-Aldrich.
Compound library
A 153607 compound library was screened in 7 to 15 concentrations ranging from 0.7 nM to 57 μM. The library included 139798 diverse small drug-like molecules that are part of the NIH Small Molecule Repository. A collection of 2893 compounds from the Centers of Methodology and Library Development at Boston University (BUCMLD) and University of Pittsburgh (UPCMLD) were added to the library. Several combinatorial libraries from Pharmacopeia, Inc. totaled 1649 compounds. An additional 1981 compounds from the NCI Diversity Set were included. Lastly, 7286 compounds with known pharmacological activity were added to provide a large and diverse screening collection.
High-throughput screening protocol and HTS analysis
All screening operations were performed on a fully integrated robotic system (Kalypsys Inc, San Diego, CA) as described elsewhere.88 Three μL of enzyme (approximately 80 nM 12-LOX, final concentration) was dispensed into 1536-well Greiner black clear-bottom assay plate. Compounds and controls (23 nL) were transferred via Kalypsys PinTool equipped with 1536-pin array. The plate was incubated for 15 min at room temperature, and then a 1 μL aliquot of substrate solution (50 μM arachidonic acid final concentration) was added to start the reaction. The reaction was stopped after 6.5 min by the addition of 4 μL FeXO solution (final concentrations of 200 μM Xylenol Orange (XO) and 300 μM ferrous ammonium sulfate in 50 mM sulfuric acid). After a short spin (1000 rpm, 15 sec), the assay plate was incubated at room temperature for 30 min. The absorbances at 405 and 573 nm were recorded using ViewLux high-throughput CCD imager (Perkin-Elmer, Waltham, MA) using standard absorbance protocol settings. During dispense, enzyme and substrate bottles were kept submerged into +4 °C recirculating chiller bath to minimize degradation. Plates containing DMSO only (instead of compound solutions) were included approximately every 50 plates throughout the screen to monitor any systematic trend in the assay signal associated with reagent dispenser variation or decrease in enzyme specific activity.
Data were analyzed in a similar method as described elsewhere.71 Briefly, assay plate-based raw data were normalized to controls and plate-based data corrections were applied to filter out background noise. All concentration response curves (CRCs) were fitted using in-house developed software (http://ncgc.nih.gov/pub/openhts/). Curves were categorized into four classes: complete response curves (Class 1), partial curves (Class 2), single point actives (Class 3) and inactives (Class 4). Compounds with the highest quality, Class 1 and Class 2 curves, were prioritized for follow-up.
Lipoxygenase UV-Vis-based Manual Assay
The initial one-point inhibition percentages were determined by following the formation of the conjugated diene product at 234 nm (ε = 25,000 M-1cm-1) with a Perkin-Elmer Lambda 40 UV/Vis spectrophotometer at one inhibitor concentration. All reactions were 2 mL in volume and constantly stirred using a magnetic stir bar at room temperature (23 °C) with approximately 40 nM for 12-LOX (by iron content), 20 nM of 15-LOX-1 (by iron content), 1 μM for 15-LOX-2 (by Bradford). Reactions with 12-LOX were carried out in 25 mM HEPES (pH 8.0) 0.01% Triton X-100 and 10 μM AA. Reactions with the crude, ammonium sulfate precipitated 5-LOX were carried out in 25 mM HEPES (pH 7.3), 0.3 mM CaCl2, 0.1 mM EDTA, 0.2 mM ATP, 0.01% Triton X100 and 10 μM AA. Reactions with 15-LOX-1 and 15-LOX-2 were carried out in 25 mM HEPES buffer (pH 7.5), 0.01% Triton X-100 and 10 μM AA. The concentration of AA (for 5-LOX, 12-LOX and 15-LOX-2) was quantitatively determined by allowing the enzymatic reaction to go to completion. IC50 values were obtained by determining the enzymatic rate at various inhibitor concentrations and plotted against inhibitor concentration, followed by a hyperbolic saturation curve fit. The data used for the saturation curves were performed in duplicate or triplicate, depending on the quality of the data.
Fluorescence Iron Binding Assay
Fluorescence readings were taken with a Perkin Elmer Luminescence Spectrometer LS 50 B in the presence and absence of iron with the excitation maximum λex = 360 nm and the emission maximum λem = 410 nm. Excitation slit was set at 5 nm and the emission slit was set at 10 nm. Due to the low intensity of the fluorophore, the filter was kept open. As a control, 1 mM 8-HQ-5-sulfonic acid was dissolved in 25 mM HEPES buffer (pH 8.5) and aliquoted to a 2 mL cuvette. In a separate cuvette, 1 mM 8-HQ-5-sulfonic acid and either 10 μM Fe2+ (as [NH4]2[Fe][SO4]2·6H2O) or Fe3+ (as Fe2(SO4)3) in 25 mM HEPES buffer (pH 8.5) was added and allowed to equilibrate for 5 minutes. For the experimental, 2 mL of 1 mM 1 was dissolved in 25 mM HEPES buffer (pH 8.5) and the fluorescence intensity taken. In a separate cuvette, 1 mM 1 was dissolved in 25 mM HEPES buffer (pH 8.5) and was incubated with less than 1 μM (by iron content) of 12-LOX and allowed to equilibrate for 5 min.
DPPH Antioxidant Test
Compound 1 was dissolved in dimethyl sulfoxide (DMSO) at 1-20 mM concentration (1,000-fold concentrated). The antioxidant activity of this compound was assayed by monitoring the quenching of the standard free radical 1,1-diphenyl-2-picrylhydrazyl (DPPH) upon reaction with the testing compounds.89, 90 A known free radical scavanger, nordihydroguaiaretic acid (NDGA), was used as a positive control. Ten microliters of the testing reagent (1 mM), to achieve a final concentration of 5 μM, was added to 2 mL of 500 μM DPPH, stirring in a cuvette. Optical absorbance was monitored and recorded at 25-sec intervals as described elsewhere.89, 90 The decrease in optical absorbance at 517 nm was monitored using a Perkin-Elmer Lambda 40 spectrometer. The rate of reaction is proportional to the antioxidant potency of the test compounds.
Steady-State Inhibition Kinetics
Lipoxygenase rates were determined by monitoring the formation of the conjugated product, 12-HPETE, at 234 nm (ε = 25 000 M-1 cm-1) with a Perkin-Elmer Lambda 40 UV/Vis spectrophotometer. Reactions were initiated by adding approximately 80 nM 12-LOX to a constantly stirring 2 mL cuvette containing 3 – 40 μM AA in 25 mM HEPES buffer (pH 7.5), in the presence of 0.01% Triton X-100. The substrate concentration was quantitated by allowing the enzymatic reaction to proceed to completion. Kinetic data were obtained by recording initial enzymatic rates, at varied inhibitor concentrations, and subsequently fitted to the Henri-Michaelis-Menten equation, using KaleidaGraph (Synergy) to determine the microscopic rate constants, Vmax (μmol/min/mg) and Vmax/KM (μmol/min/mg/μM). These rate constants were subsequently replotted and normalized to the iron content (120 nM) as 1/kcat and KM/kcat versus inhibitor concentration, to yield Kiu and Kic, respectively.
Incubation Inhibition Kinetics
12-LOX rates were determined as above, with the following modifications. One μL of 1 mM compound 2, in DMSO, was added to 30 μL of 12-LOX (approximately 5 μM), and allowed to sit on ice for intervals of 2 minutes, upwards to 10 minutes. The mixture was added at designated time periods to a constantly stirring 2 mL cuvette, containing 10 μM AA in 25 mM HEPES buffer (pH 8.0), in the presence of 0.01% Triton X-100. The control to this reaction was the same as above, but only DMSO was added. Incubated samples with inhibitor, were also dialyzed to demonstrate reversible inhibition, but the enzyme died in the 2 hour timeframe of the experiment.
Cyclooxygenase Assay
Ovine COX-1 (Cat. No. 60100) and human COX-2 (Cat. No. 60122) were purchased from Cayman chemical. Approximately 2 μg of either COX-1 or COX-2 were added to buffer containing 100 μM AA, 0.1 M Tris-HCl buffer (pH 8.0), 5 mM EDTA, 2 mM phenol and 1 μM hematin at 37 °C. Data was collected using a Hansatech DW1 oxygen electrode chamber. Inhibitors were incubated with the respective COX for 20 minutes and added to the reaction mixture and the consumption of oxygen was recorded. Ibuprofen and the carrier solvent, DMSO, were used as positive and negative controls, respectively.
12/15 Mouse Lipoxygenase Expression, Purification and IC50 Assay
12/15-mouse lipoxygenase (12/15-mLO), with an N-terminus His-tag was expressed in SF9 insect cells. 12/15-mLO was PCR amplified from the pGFP-ALOX15 vector (Gift of Dr. Jerry Nadler) using the following primers, 5’-GGCGCGCGTCGACATGCACCACCATCACCATCATCACGGTGTCTACCGCATCCGCGTCTC-3’, 5’-GGCTCGAGTCTAGATCATTATATGGCCACGCTGTTTTCTACCAG-3’, yielding a fragment of approximately 2 kb. The PCR fragment was cut with SalI and XhoI and ligated into a pFastBac1 vector, cut with the same enzymes. 12/15mLO-pFastBac was then transposed into a recombinant pFastBac bacmid using DH10Bac cells and transfected into SF9 cells, as described in the product literature for pFastBac1. For purification, infected SF9 cells were harvested after 72 hrs by centrifugation and stored at -20 °C. Thawed cells were dounced in 25mM HEPES pH 7.5 and extract was loaded onto a Bio Rad UNO Q1 ion exchange column. 12/15-mLO was eluded with a 0-1M sodium chloride linear gradient using 25mM HEPES running buffer. Glycerol was added (15%) to active fractions, which were stored at -80C. It should be noted that the His-tag purification method could not be used due to rapid inactivation of the protein. It was unclear as to the cause of this inactivation, but it appears that certain salts lead to the inactivation. Inhibitor assays were performed on a Perkin Elmer Lambda 45 UV/Vis spectrophotometer. Assay buffer consisted of 10 μM arachidonic acid added to 25 mM HEPES (pH 7.5). These assays were performed in the absence of Triton X-100 because it was found to inactivate 12/15-mLO. 12/15-mLO was defrosted on ice and added to the cuvette containing substrate and inhibitor. Conjugated product formation was monitored at 234nm. The one-point inhibition percentage for weak inhibitors was calculated as 1- (Maximal experimental rate/maximal control rate) x 100%. The multi-point IC50 value was determined by plotting the percent inhibition vs. [inhibitor] and fitting with a hyperbolic equation, as defined before.70
Cellular Inhibition Assay
In order to determine if these compounds were effective in a cellular environment, a select few were tested against 12-LOX in platelets. Human platelets were obtained from healthy volunteers at Thomas Jefferson University and this study was approved by the Thomas Jefferson University Institutional Review Board. Informed consent was obtained from all donors prior to blood draw. 20 mLs of blood was drawn and centrifuged at 200 × g for 15 min. at room temperature. To the plasma, 10% ACD (1:10) and Apyrase (1 μL/2.5 mL) were added and the solution centrifuged at 2000 × g for 15 min. at room temperature. The platelets were resuspended with Tyrode's buffer and adjusted to a concentration of 3.0 × 108 platelets/mL. The platelets were subsequently treated with inhibitor for 10 min. at 37 °C prior to stimulation with 20 μM PAR1-AP (SFLLRN) under stirring conditions in an aggregometer. Following aggregation, samples were centrifuged at 2,000 × g for 10 min., the supernatant transferred to scintillation vials, extracted with methylene chloride, reduced with triphenylphosphine and evaporated to dryness. The dry samples were then re suspended in methanol and stored at -20 ºC until analyzed by Finnigan LTQ liquid chromatography−tandem mass spectrometry (LC−MS/MS) system. An internal control, 12-deuterated(d8)- HETE (12-d8-HETE) was added to each sample prior to injection on LC-MS to account for the variation in the detector response, since it was assumed that the change in detector response for 12-HETE and 12-d8-HETE would be similar. A Thermo Electron Corp. Aquasil (3 μm, 100 mm × 2.1 mm) C-18 column was used to detect the HETEs with an elution protocol consisting of 0.2 mL/min flow rate and a linear gradient from 54.9% ACN, 45% H2O, and 0.1% THF to 69.9% ACN, 30% H2O, and 0.1% THF. The corresponding 12-HETE and 12-d8-HETE compounds were detected using selective ion monitoring analysis (m/z =318.7 to 319.7 and 326.8 to 327.7) in negative ion mode and then identified by fragmentation pattern (12-HETE, parent ion at m/z 319 and fragments at m/z 179 and 163; 12-d8-HETE, parent ion at m/z 327 and fragments at m/z 184 and 214) from MS-MS.91 The electrospray voltage was set to 5.0 kV and a global acquisition MS mode was used. The MS-MS scan was performed for the five most abundant precursor ions. The Collision Induced Dissociation (CID) was used for MS-MS with a collision energy of 35 eV. The peak intensities of 12-HETEs were normalized to the 12-d8-HETE intensities. The amount of 12-HETE in samples was estimated using a standard curve generated from pure 12-HETE with concentrations ranging from 0 μM to 2.5 μM along with a constant amount of 12-d8-HETE added in each sample. The Assay is linear in the concentration range used for the 12-HETE standard curve with a R2 value of 0.997.
Inhibition of 12-HETE Production in Human Donor Islets
Studies with human donor islets had institutional approval. Islets were obtained from the Integrated Islet Distribution Program. Human donor islets were incubated overnight in CMRL media (Cat# 15-110-CV MediaTech, Inc. Manassas, VA) containing 10% Fetal Bovine Serum + Pen/Strep. The islets were serum-starved by incubating in serum free media, CMRL containing Pen/Strep and 1% Fatty acid Free Human Serum Albumin (Cat# A1887 Sigma, St. Louis, MO), for 1 h. Islets were incubated in serum free media containing 100μM Arachadonic Acid (Cat# BML-FA003-0100, Enzo Life Sciences Plymouth Meeting, PA), with and without 10 SM of compound #1 for 60 min. at 37 °C. Calcium ionophore, 5 μM of A23187 (Cat# C7522, Sigma, St. Louis, MO), was added for an additional 30 min. Samples were harvested, centrifuged @ 1000RPM for 5 min. and supernatant drawn off and islet pellet stored at -80 °C. Cell pellets were extracted using CHCl3/ MeOH and samples dried under Nitrogen gas before reconstitution in 250 μL of ELISA sample buffer. 12-HETE levels in samples were determined using a 12-HETE ELISA kit (Cat# 901-050, Assay Design, Plymouth Meeting, PA).
Supplementary Material
ACKNOWLEDGMENT
The authors thank Eric Hoobler for the 5-LOX assay, Norine Kuhn for islet incubations and the 12-HETE Elisa, Sam Michael for assistance with the primary HTP screen, and Paul Shinn, Danielle van Leer for assistance with compound management and purification. We also thank Christina Greco for critical reading of the manuscript. Financial support was from the National Institute of Health (R01 GM56062 (TRH), R00 HL089457 (MH), R01 DK 55240 (DTF, JLN)), the Juvenile Diabetes Research Foundation (DTF, TRH, JLN), and the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research (R03 MH081283 (TRH)). Additional financial support was from NIH (S10-RR20939 (TRH)) and the California Institute for Quantitative Biosciences for the UCSC MS Facility (TRH).
Footnotes
ABBREVIATIONS. LOX, lipoxygenase; soybean LOX 1, soybean lipoxygenase 1; 15 LOX-1, human reticulocyte 15-lipoxygenase-1; 15-LOX-2, human epithelial 15-lipoxygenase-2; 12-LOX, human platelet-type 12-lipoxygenase; rabbit 15-LOX, rabbit reticulocyte 15-lipoxygenase; COX, cyclooxygenase; NDGA, nordihydroguaiaretic acid; AA, arachidonic acid; 15-HPETE, 15-(S)-hydroperoxyeicosatetraenoic acid; 15-HETE, 15-(S)-hydroxyeicosatetraenoic acid; 12-HPETE, 12-(S)-hydroperoxyeicosatetraenoic acid; 12-HETE, 12-(S)-hydroxyeicosatetraenoic acid; LA, linoleic acid; 13-HPODE, 13-(S)-hydroperoxyoctadecadienoic acid; 13-HODE, 13-(S)-hydroxyoctadecadienoic acid; ALA, alpha-linolenic acid; 13-HPOTrE, 13-(S)-hydroperoxyoctadecatrienoic acid; 8-HQ, 8-hydroxyquinoline; mRNA, messenger ribonucleic acid; Vmax, maximal velocity (mmol/min); kcat, first order catalytic rate constant (Vmax/[E] (s-1)); KM, Henri-Michaelis-Menten Constant (mM); [E], total active enzyme concentration; IC50, inhibitor constant at 50% inhibition; DPPH, 1.1-diphenyl-2-picrylhydrazyl; XO, xylenol orange; HTS, high-throughput screening; MLSMR, Molecular Libraries Small Molecule Repository; qHTS, quantitative high-throughput screening; CRC, concentration response curve; SD, standard deviation; MLPCN, Molecular Libraries Probe Production Center Network.
Supporting Information Available: Additional experimental procedures and spectroscopic data (1H NMR, LC/MS and HRMS) for representative compounds. Analytical methods for the chiral HPLC separation of enantiomers is also included. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Solomon EI, Zhou J, Neese F, Pavel EG. New Insights from Spectroscopy into the Structure/Function Relationships of Lipoxygenases. Chem. Biol. 1997;4:795–808. doi: 10.1016/s1074-5521(97)90113-7. [DOI] [PubMed] [Google Scholar]
- 2.Ford-Hutchinson AW, Gresser M, Young RN. 5-Lipoxygenase. Annu. Rev. Biochem. 1994;63:383–417. doi: 10.1146/annurev.bi.63.070194.002123. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn H, Chaitidis P, Roffeis J, Walther M. Arachidonic Acid Metabolites in the Cardiovascular System: The Role of Lipoxygenase Isoforms in Atherogenesis with Particular Emphasis on Vascular Remodeling. J. Cardiovasc. Pharmacol. 2007;50:609–620. doi: 10.1097/FJC.0b013e318159f177. [DOI] [PubMed] [Google Scholar]
- 4.Pace-Asciak CR, Asotra S. Biosynthesis, Catabolism, and Biological Properties of Hpetes, Hydroperoxide Derivatives of Arachidonic Acid. Free Radical Biology and Medicine. 1989;7:409–433. doi: 10.1016/0891-5849(89)90125-1. [DOI] [PubMed] [Google Scholar]
- 5.Radmark O, Samuelsson B. 5-Lipoxygenase: Regulation and Possible Involvement in Atherosclerosis. Prostaglandins Other Lipid Mediat. 2007;83:162–174. doi: 10.1016/j.prostaglandins.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Brock TG. Regulating Leukotriene Synthesis: The Role of Nuclear 5-Lipoxygenase. J. Cell. Biochem. 2005;96:1203–1211. doi: 10.1002/jcb.20662. [DOI] [PubMed] [Google Scholar]
- 7.Newcomer ME, Gilbert NC. Location, Location, Location: Compartmentalization of Early Events in Leukotriene Biosynthesis. J. Biol. Chem. 2010;285:25109–25114. doi: 10.1074/jbc.R110.125880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh J. Inhibition of Arachidonate 5-Lipoxygenase Triggers Prostate Cancer Cell Death through Rapid Activation of C-Jun N-Terminal Kinase. Biochem. Biophys. Res. Commun. 2003;307:342–349. doi: 10.1016/s0006-291x(03)01201-4. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh J, Myers CE. Inhibition of Arachidonate 5-Lipoxygenase Triggers Massive Apoptosis in Human Prostate Cancer Cells. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13182–13187. doi: 10.1073/pnas.95.22.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakano H, Inoue T, Kawasaki N, Miyataka H, Matsumoto H, Taguchi T, Inagaki N, Nagai H, Satoh T. Synthesis and Biological Activities of Novel Antiallergic Agents with 5-Lipoxygenase Inhibiting Action. Bioorg. Med. Chem. 2000;8:373–380. doi: 10.1016/s0968-0896(99)00291-6. [DOI] [PubMed] [Google Scholar]
- 11.Radmark O, Werz O, Steinhilber D, Samuelsson B. 5-Lipoxygenase: Regulation of Expression and Enzyme Activity. Trends Biochem. Sci. 2007;32:332–341. doi: 10.1016/j.tibs.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Berger W, De Chandt MT, Cairns CB. Zileuton: Clinical Implications of 5-Lipoxygenase Inhibition in Severe Airway Disease. Int J Clin Pract. 2007;61:663–676. doi: 10.1111/j.1742-1241.2007.01320.x. [DOI] [PubMed] [Google Scholar]
- 13.Masferrer JL, Zweifel BS, Hardy M, Anderson GD, Dufield D, Cortes Burgos L, Pufahl RA, Graneto M. Pharmacology of Pf-4191834, a Novel, Selective Non-Redox 5-Lipoxygenase Inhibitor Effective in Inflammation and Pain. J. Pharmacol. Exp. Ther. 2010;334:294–301. doi: 10.1124/jpet.110.166967. [DOI] [PubMed] [Google Scholar]
- 14.Ducharme Y, Blouin M, Brideau C, Chateauneuf A, Gareau Y, Grimm EL, Juteau H, Laliberte S, MacKay B, Masse F, Ouellet M, Salem M, Styhler A, Friesen RW. The Discovery of Setileuton, a Potent and Selective 5-Lipoxygenase Inhibitor. ACS Med. Chem. Lett. 2010;1:170–174. doi: 10.1021/ml100029k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Company Information: www.pfizer.com, www.merck.com.
- 16.Jones R, Adel Alvarez LA, Alvarez OR, Broaddus R, Das S. Arachidonic Acid and Colorectal Carcinogenesis. Mol. Cell. Biochem. 2003;253:141–149. doi: 10.1023/a:1026060426569. [DOI] [PubMed] [Google Scholar]
- 17.Kelavkar UP, Cohen C, Kamitani H, Eling TE, Badr KF. Concordant Induction of 15-Lipoxygenase-1 and Mutant P53 Expression in Human Prostate Adenocarcinoma: Correlation with Gleason Staging. Carcinogenesis. 2000;21:1777–1787. doi: 10.1093/carcin/21.10.1777. [DOI] [PubMed] [Google Scholar]
- 18.Shureiqi I, Lippman SM. Lipoxygenase Modulation to Reverse Carcinogenesis. Cancer Res. 2001;61:6307–6312. [PubMed] [Google Scholar]
- 19.Hsi LC, Wilson LC, Eling TE. Opposing Effects of 15-Lipoxygenase-1 and -2 Metabolites on Mapk Signaling in Prostate Alteration in Peroxisome Proliferator-Activated Receptor Gamma. J. Biol. Chem. 2002;277:40549–40556. doi: 10.1074/jbc.M203522200. [DOI] [PubMed] [Google Scholar]
- 20.Shappell SB, Manning S, Boeglin WE, Guan YF, Roberts RL, Davis L, Olson SJ, Jack GS, Coffey CS, Wheeler TM, Breyer MD, Brash AR. Alterations in Lipoxygenase and Cyclooxygenase-2 Catalytic Activity and Mrna Expression in Prostate Carcinoma. Neoplasia. 2001;3:287–303. doi: 10.1038/sj.neo.7900166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shappell SB, Olson SJ, Hannah SE, Manning S, Roberts RL, Masumori N, Jisaka M, Boeglin WE, Vader V, Dave DS, Shook MF, Thomas TZ, Funk CD, Brash AR, Matusik RJ. Elevated Expression of 12/15-Lipoxygenase and Cyclooxygenase-2 in a Transgenic Mouse Model of Prostate Carcinoma. Cancer Res. 2003;63:2256–2267. [PubMed] [Google Scholar]
- 22.Brash AR, Boeglin WE, Chang MS. Discovery of a Second 15s-Lipoxygenase in Humans. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6148–6152. doi: 10.1073/pnas.94.12.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez AL, Roberts RL, Massion PP, Olson SJ, Shyr Y, Shappell SB. 15-Lipoxygenase-2 Expression in Benign and Neoplastic Lung: An Immunohistochemical Study and Correlation with Tumor Grade and Proliferation. Hum Pathol. 2004;35:840–849. doi: 10.1016/j.humpath.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Suraneni MV, Schneider Broussard R, Moore JR, Davis TC, Maldonado CJ, Li H, Newman RA, Kusewitt D, Hu J, Yang P, Tang DG. Transgenic Expression of 15-Lipoxygenase 2 (15-Lox2) in Mouse Prostate Leads to Hyperplasia and Cell Senescence. Oncogene. 2010;29:4261–4275. doi: 10.1038/onc.2010.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang DG, Bhatia B, Tang S, Schneider-Broussard R. 15-Lipoxygenase 2 (15-Lox2) Is a Functional Tumor Suppressor That Regulates Human Prostate Epithelial Cell Differentiation, Senescence, and Growth (Size). Prostaglandins Other Lipid Mediat. 2007;82:135–146. doi: 10.1016/j.prostaglandins.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Jobard F, Lefevre C, Karaduman A, Blanchet-Bardon C, Emre S, Weissenbach J, Ozguc M, Lathrop M, Prud'homme JF, Fischer J. Lipoxygenase-3 (Aloxe3) and 12(R)-Lipoxygenase (Alox12b) Are Mutated in Non Bullous Congenital Ichthyosiform Erythroderma (Ncie) Linked to Chromosome 17p13.1. Hum Mol Genet. 2002;11:107–113. doi: 10.1093/hmg/11.1.107. [DOI] [PubMed] [Google Scholar]
- 27.Hussain H, Shornick LP, Shannon VR, Wilson JD, Funk CD, Pentland AP, Holtzman MJ. Epidermis Contains Platelet-Type 12-Lipoxygenase That Is Overexpressed in Germinal Layer Keratinocytes in Psoriasis. Am J Physiol. 1994;266:C243–253. doi: 10.1152/ajpcell.1994.266.1.C243. [DOI] [PubMed] [Google Scholar]
- 28.Ding XZ, Iversen P, Cluck MW, Knezetic JA, Adrian TE. Lipoxygenase Inhibitors Abolish Proliferation of Human Pancreatic Cancer Cells. Biochem Biophys Res Commun. 1999;261:218–223. doi: 10.1006/bbrc.1999.1012. [DOI] [PubMed] [Google Scholar]
- 29.Connolly JM, Rose DP. Enhanced Angiogenesis and Growth of 12-Lipoxygenase Gene-Transfected Mcf-7 Human Breast Cancer Cells in Athymic Nude Mice. Cancer Lett. 1998;132:107–112. doi: 10.1016/s0304-3835(98)00171-2. [DOI] [PubMed] [Google Scholar]
- 30.Natarajan R, Nadler J. Role of Lipoxygenases in Breast Cancer. Front Biosci. 1998;3:E81–88. doi: 10.2741/a369. [DOI] [PubMed] [Google Scholar]
- 31.Ma K, Nunemaker CS, Wu R, Chakrabarti SK, Taylor-Fishwick DA, Nadler JL. 12-Lipoxygenase Products Reduce Insulin Secretion and {Beta}-Cell Viability in Human Islets. J. Clin. Endocrinol. Metab. 2010;95:887–893. doi: 10.1210/jc.2009-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas CP, Morgan LT, Maskrey BH, Murphy RC, Kuhn H, Hazen SL, Goodall AH, Hamali HA, Collins PW, O'Donnell VB. Phospholipid-Esterified Eicosanoids Are Generated in Agonist-Activated Human Platelets and Enhance Tissue Factor-Dependent Thrombin Generation. J. Biol. Chem. 2010;285:6891–6903. doi: 10.1074/jbc.M109.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holinstat M, Boutaud O, Apopa PL, Vesci J, Bala M, Oates JA, Hamm HE. Protease-Activated Receptor Signaling in Platelets Activates Cytosolic Phospholipase A2alpha Differently for Cyclooxygenase-1 and 12-Lipoxygenase Catalysis. Arterioscler Thromb Vasc Biol. 2011;31:435–442. doi: 10.1161/ATVBAHA.110.219527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaur G, Jalagadugula G, Mao G, Rao AK. Runx1/Core Binding Factor A2 Regulates Platelet 12-Lipoxygenase Gene (Alox12): Studies in Human Runx1 Haplodeficiency. Blood. 2010;115:3128–3135. doi: 10.1182/blood-2009-04-214601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Catalano A, Procopio A. New Aspects on the Role of Lipoxygenases in Cancer Progression. Histol Histopathol. 2005;20:969–975. doi: 10.14670/HH-20.969. [DOI] [PubMed] [Google Scholar]
- 36.Krishnamoorthy S, Jin R, Cai Y, Maddipati KR, Nie D, Pages G, Tucker SC, Honn KV. 12-Lipoxygenase and the Regulation of Hypoxia-Inducible Factor in Prostate Cancer Cells. Exp Cell Res. 316:1706–1715. doi: 10.1016/j.yexcr.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steele VE, Holmes CA, Hawk ET, Kopelovich L, Lubet RA, Crowell JA, Sigman CC, Kelloff GJ. Lipoxygenase Inhibitors as Potential Cancer Chemopreventives. Cancer Epidemiol Biomarkers Prev. 1999;8:467–483. [PubMed] [Google Scholar]
- 38.Jiang WG, Douglas Jones A, Mansel RE. Levels of Expression of Lipoxygenases and Cyclooxygenase-2 in Human Breast Cancer. Prostaglandins Leukot Essent Fatty Acids. 2003;69:275–281. doi: 10.1016/s0952-3278(03)00110-8. [DOI] [PubMed] [Google Scholar]
- 39.Mohammad AM, Abdel HA, Abdel W, Ahmed AM, Wael T, Eiman G. Expression of Cyclooxygenase-2 and 12-Lipoxygenase in Human Breast Cancer and Their Relationship with Her-2/Neu and Hormonal Receptors: Impact on Prognosis and Therapy. Indian J Cancer. 2006;43:163–168. doi: 10.4103/0019-509x.29421. [DOI] [PubMed] [Google Scholar]
- 40.Zeeneldin AA, Mohamed AM, Abdel HA, Taha FM, Goda IA, Abodeef WT. Survival Effects of Cyclooxygenase-2 and 12-Lipooxygenase in Egyptian Women with Operable Breast Cancer. Indian J Cancer. 2009;46:54–60. doi: 10.4103/0019-509x.48597. [DOI] [PubMed] [Google Scholar]
- 41.Agrawal VK, Bano S, Khadikar PV. Qsar Study on 5-Lipoxygenase Inhibitors Using Distance-Based Topological Indices. Bioorg. Med. Chem. 2003;11:5519–5527. doi: 10.1016/j.bmc.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 42.Arockia Babu M, Shakya N, Prathipati P, Kaskhedikar SG, Saxena AK. Development of 3d-Qsar Models for 5-Lipoxygenase Antagonists: Chalcones. Bioorg. Med. Chem. 2002;10:4035–4041. doi: 10.1016/s0968-0896(02)00313-9. [DOI] [PubMed] [Google Scholar]
- 43.Basha A, Ratajczyk JD, Dyer RD, Young P, Carter GW, Brooks CDW. Structure-Activity Relationships of Pyrimido-Pyrimidine Series of 5-Lipoxygenase Inhibitors. Med. Chem. Res. 1996;6:61–67. [Google Scholar]
- 44.Fleiseher R, Frohberg P, Buge A, Nuhn P, Wiese M. Qsar Analysis of Substituted 2-Phenylhydrazonoacetamides Acting as Inhibitors of 15-Lipoxygenase. Quantitative Structure-Activity Relationships. 2000;19:162–172. [Google Scholar]
- 45.Kim KH, Martin YC, Brooks CDW. Quantitative Structure-Activity Relationships of 5-Lipoxygenase Inhibitors Inhibitory Potency of Triazinone Analogues in a Broken Cell. Quantitative Structure-Activity Relationships. 1996;15:491–497. doi: 10.1002/jps.2600830334. [DOI] [PubMed] [Google Scholar]
- 46.Kim KH, Martin YC, Brooks DW, Dyer RD, Carter GW. Quantitative Structure-Activity Relationships of 5-Lipoxygenase Inhibitors Inhibitory Potency of Pyridazinone Analogues. J. Pharm. Sci. 1994;83:433–438. doi: 10.1002/jps.2600830334. [DOI] [PubMed] [Google Scholar]
- 47.Mano T, Stevens RW, Ando K, Nakao K, Okumura Y, Sakakibara M, Okumura T, Tamura T, Miyamoto K. Novel Imidazole Compounds as a New Series of Potent, Orally Active Inhibitors of 5-Lipoxygenase. Bioorg. Med. Chem. 2003;11:3879–3887. doi: 10.1016/s0968-0896(03)00436-x. [DOI] [PubMed] [Google Scholar]
- 48.Redrejo-Rodriguez M, Tejeda-Cano A, Pinto MD, Macias P. Lipoxygenase Inhibition by Flavonoids: Semiempirical Study of the Structure-Activity Relation. Journal of Molecular Structure-Theochem. 2004;674:121–124. [Google Scholar]
- 49.Stewart AO, Bhatia PA, Martin JG, Summers JB, Rodriques KE, Martin MB, Holms JH, Moore JL, Craig RA, Kolasa T, Ratajczyk JD, Mazdiyasni H, Kerdesky FA, DeNinno SL, Maki RG, Bouska JB, Young PR, Lanni C, Bell RL, Carter GW, Brooks CD. Structure-Activity Relationships of N-Hydroxyurea 5-Lipoxygenase Inhibitors. J. Med. Chem. 1997;40:1955–1968. doi: 10.1021/jm9700474. [DOI] [PubMed] [Google Scholar]
- 50.Amagata T, Whitman S, Johnson TA, Stessman CC, Loo CP, Lobkovsky E, Clardy J, Crews P, Holman TR. Exploring Sponge-Derived Terpenoids for Their Potency and Selectivity against 12-Human, 15-Human, and 15-Soybean Lipoxygenases. J. Nat. Prod. 2003;66:230–235. doi: 10.1021/np020462l. [DOI] [PubMed] [Google Scholar]
- 51.Benrezzouk R, Terencio MC, Ferrandiz ML, Hernandez-Perez M, Rabanal R, Alcaraz MJ. Inhibition of 5-Lipoxygenase Activity by the Natural Anti-Inflammatory Compound Aethiopinone. Inflamm Res. 2001;50:96–101. doi: 10.1007/s000110050730. [DOI] [PubMed] [Google Scholar]
- 52.Bracher F, Krauss J, Laufer S. Effects of Natural Products Containing Acylresorcinol Partial Structures on Cyclooxygenases and 5-Lipoxygenase. Pharmazie. 2001;56:430. [PubMed] [Google Scholar]
- 53.Carroll J, Jonsson EN, Ebel R, Hartman MS, Holman TR, Crews P. Probing Sponge-Derived Terpenoids for Human 15-Lipoxygenase Inhibitors. J. Org. Chem. 2001;66:6847–6851. doi: 10.1021/jo015784t. [DOI] [PubMed] [Google Scholar]
- 54.Fu X, Schmitz FJ, Govindan M, Abbas SA, Hanson KM, Horton PA, Crews P, Laney M, Schatzman RC. Enzyme Inhibitors: New and Known Polybrominated Phenols and Diphenyl Ethers from Four Indo-Pacific Dysidea Sponges. J. Nat. Prod. 1995;58:1384–91. doi: 10.1021/np50123a008. [DOI] [PubMed] [Google Scholar]
- 55.Jiang ZD, Ketchum SO, Gerwick WH. 5-Lipoxygenase-Derived Oxylipins from the Red Alga Rhodymenia Pertusa. Phytochemistry. 2000;53:129–133. doi: 10.1016/s0031-9422(99)00445-8. [DOI] [PubMed] [Google Scholar]
- 56.Rao KCS, Divakar S, Rao AGA, Karanth NG, Suneetha WJ, Krishnakantha TP, Sattur AP. Asperenone: An Inhibitor of 15-Lipoxygenase and of Human Platelet Aggregation from Aspergillus Niger. Biotechnol. Lett. 2002;24:1967–1970. [Google Scholar]
- 57.Segraves EN, Shah RR, Segraves NL, Johnson TA, Whitman S, Sui JK, Kenyon VA, Cichewicz RH, Crews P, Holman TR. Probing the Activity Differences of Simple and Complex Brominated Aryl Compounds against 15-Soybean, 15-Human, and 12-Human Lipoxygenase. J. Med. Chem. 2004;47:4060–4065. doi: 10.1021/jm049872s. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki H, Ueda T, Juranek I, Yamamoto S, Katoh T, Node M, Suzuki T. Hinokitiol, a Selective Inhibitor of the Platelet-Type Isozyme of Arachidonate 12-Lipoxygenase. Biochem Biophys Res Commun. 2000;275:885–889. doi: 10.1006/bbrc.2000.3390. [DOI] [PubMed] [Google Scholar]
- 59.Weinstein DS, Liu W, Gu Z, Langevine C, Ngu K, Fadnis L, Combs DW, Sitkoff D, Ahmad S, Zhuang S, Chen X, Wang FL, Loughney DA, Atwal KS, Zahler R, Macor JE, Madsen CS, Murugesan N. Tryptamine and Homotryptamine-Based Sulfonamides as Potent and Selective Inhibitors of 15-Lipoxygenase. Bioorg Med Chem Lett. 2005;15:1435–1440. doi: 10.1016/j.bmcl.2004.12.081. [DOI] [PubMed] [Google Scholar]
- 60.Deschamps JD, Gautschi JT, Whitman S, Johnson TA, Gassner NC, Crews P, Holman TR. Discovery of Platelet-Type 12-Human Lipoxygenase Selective Inhibitors by High-Throughput Screening of Structurally Diverse Libraries. Bioorg. Med. Chem. 2007;15:6900–6908. doi: 10.1016/j.bmc.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vasquez-Martinez Y, Ohri RV, Kenyon V, Holman TR, Sepulveda-Boza S. Structure-Activity Relationship Studies of Flavonoids as Potent Inhibitors of Human Platelet 12-Hlo, Reticulocyte 15-Hlo-1, and Prostate Epithelial 15-Hlo-2. Bioorg Med Chem. 2007;15:7408–7425. doi: 10.1016/j.bmc.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto S, Katsukawa M, Nakano A, Hiraki E, Nishimura K, Jisaka M, Yokota K, Ueda N. Arachidonate 12-Lipoxygenases with Reference to Their Selective Inhibitors. Biochem Biophys Res Commun. 2005;338:122–127. doi: 10.1016/j.bbrc.2005.08.214. [DOI] [PubMed] [Google Scholar]
- 63.Whitman S, Gezginci M, Timmermann BN, Holman TR. Structure-Activity Relationship Studies of Nordihydroguaiaretic Acid Inhibitors toward Soybean, 12-Human, and 15-Human Lipoxygenase. J. Med. Chem. 2002;45:2659–2661. doi: 10.1021/jm0201262. [DOI] [PubMed] [Google Scholar]
- 64.Deschamps JD, Kenyon VA, Holman TR. Baicalein Is a Potent in Vitro Inhibitor against Both Reticulocyte 15-Human and Platelet 12-Human Lipoxygenases. Bioorg. Med. Chem. 2006;14:4295–42301. doi: 10.1016/j.bmc.2006.01.057. [DOI] [PubMed] [Google Scholar]
- 65.Kenyon V, Chorny I, Carvajal WJ, Holman TR, Jacobson MP. Novel Human Lipoxygenase Inhibitors Discovered Using Virtual Screening with Homology Models. J. Med. Chem. 2006;49:1356–1363. doi: 10.1021/jm050639j. [DOI] [PubMed] [Google Scholar]
- 66.These compounds are a part of the NIH Small Molecule Repository: see http://mlsmr.glpg.com/MLSMR_HomePage/
- 67.Betti M. General Condensation Reaction between B-Naphthol, Aldehydes and Amines. Gazz. Chim. Ital. 1900;30:310–316. [Google Scholar]
- 68.Betti M. General Condensation Reaction between B-Naphthol, Aldehydes and Amines. Gazz. Chim. Ital. 1901;31:377–393. [Google Scholar]
- 69.Betti M. General Condensation Reaction between B-Naphthol, Aldehydes and Amines. Gazz. Chim. Ital. 1901;31:170–184. [Google Scholar]
- 70.Rai G, Kenyon V, Jadhav A, Schultz L, Armstrong M, Jameson JB, Hoobler E, Leister W, Simeonov A, Holman TR, Maloney DJ. Discovery of Potent and Selective Inhibitors of Human Reticulocyte 15-Lipoxygenase 1. J. Med. Chem. 2010;53:7392–7404. doi: 10.1021/jm1008852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative High Throughput Screening: A Titration-Based Approach That Efficiently Identifies Biological Activities in Large Chemical Libraries. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pierre JL, Baret P, Serratrice G. Hydroxyquinolines as Iron Chelators. Curr. Med. Chem. 2003;10:1077–1084. doi: 10.2174/0929867033457584. [DOI] [PubMed] [Google Scholar]
- 73.Liu ZD, Kayyali R, Hider RC, Porter JB, Theobald AE. Design, Synthesis, and Evaluation of Novel 2-Substituted 3-Hydroxypyridin-4-Ones: Structure-Activity Investigation of Metalloenzyme Inhibition by Iron Chelators. J Med Chem. 2002;45:631–639. doi: 10.1021/jm010817i. [DOI] [PubMed] [Google Scholar]
- 74.Nelson MJ, Brennan BA, Chase DB, Cowling RA, Grove GN, Scarrow RC. Structure and Kinetics of Formation of Catechol Complexes of Ferric Soybean Lipoxygenase-1. Biochemistry. 1995;34:15219–15229. doi: 10.1021/bi00046a030. [DOI] [PubMed] [Google Scholar]
- 75.Hahn C, Sieler J, Taube R. Synthesis of 2,6-Bis(Diphenylphosphinomethyl)Pyridine-Monoligand-Rhodium(I) Complexes [Rh(Pnp)L]X with L = Pyridine, Ch3cn, Dmso and X = Cf3so3, Bf4 from the Corresponding Ethylene Complex and Comparison of the Structures to the Piperidine Complex (L = Piperidine, X = Bf4). Polyhedron. 1998;17:1183–1193. [Google Scholar]
- 76.Gupta AK, Bluhm R. Seborrheic Dermatitis. J Eur Acad Dermatol Venereol. 2004;18:13–26. doi: 10.1111/j.1468-3083.2004.00693.x. quiz 19-20. [DOI] [PubMed] [Google Scholar]
- 77.Kemal C, Louis-Flamberg P, Krupinski-Olsen R, Shorter AL. Reductive Inactivation of Soybean Lipoxygenase 1 by Catechols: A Possible Mechanism for Regulation of Lipoxygenase Activity. Biochemistry. 1987;26:7064–7072. doi: 10.1021/bi00396a031. [DOI] [PubMed] [Google Scholar]
- 78.McMillan RM, Walker ER. Designing Therapeutically Effective 5-Lipoxygenase Inhibitors. Trends Pharmacol. Sci. 1992;13:323–330. doi: 10.1016/0165-6147(92)90100-k. [DOI] [PubMed] [Google Scholar]
- 79.McGovern SL, Helfand BT, Feng B, Shoichet BK. A Specific Mechanism of Nonspecific Inhibition. J Med Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 80.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. Portland Press, Ltd.; 1995. p. 343. [Google Scholar]
- 81.Crichlow GV, Lubetsky JB, Leng L, Bucala R, Lolis EJ. Structural and Kinetic Analyses of Macrophage Migration Inhibitory Factor Active Site Interactions. Biochemistry. 2009;48:132–139. doi: 10.1021/bi8014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sloane D, Leung R, Barnett J, Craik C, Sigal E. Conversion of Human 15-Lipoxygenase to an Efficient 12-Lipoxygenase the Side Chain Geometry of Amino Acids 417 and 418 Determine Positional Specificity. Prot Eng. 1995;8:275–282. doi: 10.1093/protein/8.3.275. [DOI] [PubMed] [Google Scholar]
- 83.McLean LR, Zhang Y, Li H, Li Z, Lukasczyk U, Choi YM, Han Z, Prisco J, Fordham J, Tsay JT, Reiling S, Vaz RJ, Li Y. Discovery of Covalent Inhibitors for Mif Tautomerase Via Cocrystal Structures with Phantom Hits from Virtual Screening. Bioorg. Med. Chem. Lett. 2009;19:6717–6720. doi: 10.1016/j.bmcl.2009.09.106. [DOI] [PubMed] [Google Scholar]
- 84.Gilbert AM, Bursavich MG, Lombardi S, Georgiadis KE, Reifenberg E, Flannery CR, Morris EA. N-((8-Hydroxy-5-Substituted-Quinolin-7-Yl)(Phenyl)Methyl)-2-Phenyloxy/Amin O-Acetamide Inhibitors of Adamts-5 (Aggrecanase-2). Bioorg. Med. Chem. Lett. 2008;18:6454–6457. doi: 10.1016/j.bmcl.2008.10.065. [DOI] [PubMed] [Google Scholar]
- 85.Ohri RV, Radosevich AT, Hrovat KJ, Musich C, Huang D, Holman TR, Toste FD. A Re(V)-Catalyzed C-N Bond Forming Route to Human Lipoxygenase Inhibitors. Org. Lett. 2005;7:2501–2504. doi: 10.1021/ol050897a. [DOI] [PubMed] [Google Scholar]
- 86.Chen XS, Brash A, Funk C. Purification and Characterization of Recombinant Histidine-Tagged Human Platelet 12-Lipoxygenase Expressed in a Baculovirus/Insect Cell System. Eur. J. Biochem. 1993;214:845–852. doi: 10.1111/j.1432-1033.1993.tb17988.x. [DOI] [PubMed] [Google Scholar]
- 87.Robinson SJ, Hoobler EK, Riener M, Loveridge ST, Tenney K, Valeriote FA, Holman TR, Crews P. Using Enzyme Assays to Evaluate the Structure and Bioactivity of Sponge-Derived Meroterpenes. J Nat Prod. 2009;72:1857–1863. doi: 10.1021/np900465e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Michael S, Auld D, Klumpp C, Jadhav A, Zheng W, Thorne N, Austin CP, Inglese J, Simeonov A. A Robotic Platform for Quantitative High-Throughput Screening. Assay Drug Dev Technol. 2008;6:637–657. doi: 10.1089/adt.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Leyen K, Arai K, Jin G, Kenyon V, Gerstner B, Rosenberg PA, Holman TR, Lo EH. Novel Lipoxygenase Inhibitors as Neuroprotective Reagents. J. Neurosci. Res. 2008;86:904–909. doi: 10.1002/jnr.21543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang H, Li J, Follett PL, Zhang Y, Cotanche DA, Jensen FE, Volpe JJ, Rosenberg PA. 12-Lipoxygenase Plays a Key Role in Cell Death Caused by Glutathione Depletion and Arachidonic Acid in Rat Oligodendrocytes. Eur J Neurosci. 2004;20:2049–2058. doi: 10.1111/j.1460-9568.2004.03650.x. [DOI] [PubMed] [Google Scholar]
- 91.Kim HY, Sawazaki S. Structural Analysis of Hydroxy Fatty Acids by Thermospray Liquid Chromatography/Tandem Mass Spectrometry. Biol. Mass Spectrom. 1993;22:302–310. doi: 10.1002/bms.1200220506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.