Abstract
Objective: The gene for mast cell chymase (CMA1) is an ideal candidate for investigating the genetic predisposition to coronary heart disease (CHD), as activated mast cells have been found to be present in a greater proportion in the shoulder region of atheroma than in normal coronary intimae. Previous studies have indicated that CMA1 promoter polymorphism rs1800875 may be involved in regulating immunoglobulin E (IgE) levels in patients with eczema, and it is associated with the progression of immunoglobulin A nephropathy. Methods: The association between single nucleotide polymorphism (SNP) rs1800875, serum chymase, and serum IgE levels was examined in 175 CHD subjects and 95 non-CHD subjects. Results: Statistical analysis indicated no significant difference in allele frequency between CHD and non-CHD. However, a significant association was found between CMA1 genotypes and total IgE levels in CHD subjects. Meanwhile, crossover analysis revealed that, in GG homozygotes, CHD risk was nearly six times higher in those with IgE (U/ml) level <2.58 (natural logarithm conversion), while no association was found with chymase level. Conclusions: Polymorphism rs1800875 of CMA1 may be associated with serum IgE level in CHD subjects, but not with chymase level in both groups. In GG homozygotes, high IgE level is a protective factor against coronary disease.
Keywords: Coronary heart disease, CMA1, Serum immunoglobulin E (IgE), Serum chymase
1. Introduction
Mast cells contribute importantly to allergic and innate immune responses by releasing various preformed and newly synthesized mediators (Gurish and Austen, 2001; Galli et al., 2005). Previous studies have shown mast cell accumulation in human atherosclerotic lesions (Jeziorska et al., 1997). Recently, mast cells were reported to participate in atherosclerosis by releasing pro-inflammatory cytokines, chemokines, and proteases to induce inflammatory cell recruitment, cell apoptosis, angiogenesis, and matrix protein remodeling (Sun et al., 2007). Mast cell chymase and tryptase are unique to mast cells and have been directly and indirectly proven to participate in atherosclerosis and abdominal aortic aneurysms (Qin and Shi, 2011).
Mast cell chymase, a glycoprotein with a molecular weight of 29 kDa, is stored in high amounts within the secretary granules of mast cells. It is secreted into the intercellular substance by activated mast cells under strong stimulation (Schwartz and Austen, 1980). The enzyme is one of the important enzymes in generating angiotensin-II (Ang-II) from Ang-I. High levels of Ang-II forming activity and chymase expression have been demonstrated in human atherosclerotic lesions (Ihara et al., 1999). Mast cell chymase may contribute to plaque erosion and complications of atherosclerosis by inducing endothelial cell and smooth muscle cell apoptosis (Heikkila et al., 2008). Meanwhile, serum chymase levels have been demonstrated to be higher in patients with acute myocardial infarction or unstable angina pectoris than in patients with stable angina pectoris or those without significant coronary heart disease (CHD) (Xiang et al., 2011). The gene encoding mast cell chymase, CMA1, has been mapped within a cluster of genes for cellular proteases on chromosome 14q11.2 (Urata et al., 1991). This gene, embracing 5 exons and 4 introns, accounts for a total length of approximately 3 kb (Urata et al., 1991). Previous studies have focused on rs1800875 in the promoter of CMA1. Ortlepp et al. (2001) showed that the allele G of rs1800875 is a genetic risk factor for atherosclerosis in venous coronary artery bypass grafts. Meanwhile, in subjects with self-reported eczema or atopic asthma, a significant association was found between rs1800875 and total immunoglobulin E (IgE) levels (Iwanaga et al., 2004; Sharma et al., 2005). Therefore, the relationship between rs1800875 genotypes and IgE level remains unclear in the coronary patient, although recently increased serum IgE levels have been found in myocardial infarction (Szczeklik et al., 1993).
In the present study, we detected the genotype of single nucleotide polymorphism (SNP) rs1800875 in Chinese CHD population, and evaluated the association between this polymorphism and the quantitative traits associated with CHD, such as total serum chymase and IgE levels.
2. Materials and methods
2.1. Subjects
A total of 270 patients admitted to the Department of Cardiology at the Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China, were recruited consecutively from July to December 2008. All patients had no history of inflammation-associated diseases including asthma, history of allergic diseases, rheumatic heart disease, arthritis, cancer, chronic hepatic disease, renal failure, valvular heart disease, and other cardiac diseases. This study was conducted in conformity with the Helsinki Declaration and approved by the Hospital Review Committee. All subjects signed the informed consent and underwent coronary angiography. A total of 175 subjects with at least 50% stenosis of one or more main coronary arteries were selected as substantial CHD. The remaining 95 subjects, with the main coronary artery having less than 50% stenosis or without luminal narrowing, were diagnosed as unsubstantial CHD (non-CHD) controls. We recorded subject age, height, weight, body mass index (BMI), sex, diabetes, history of hypertension, smoking (consuming tobacco for at least three years), and family history of CHD. Arterial blood samples were extracted from the sheath in the radial or femoral artery during the interventional procedure. All serum sample aliquots were stored at −80 °C for routine chymase and IgE measurement.
2.2. PCR amplification and genotyping
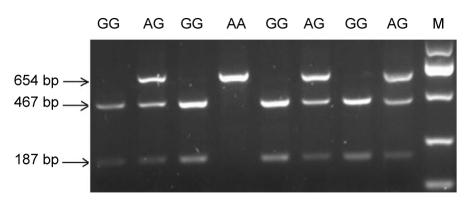
Genomic DNA was extracted from the peripheral blood lymphocytes following the previous procedure (Iwanaga et al., 2004). The SNP rs1800875 was investigated using primer pairs reported previously: forward primer 5′-TGCCCCACATCAACATTCATTC-3′ and reverse primer 5′ TCCGGAGCTGGAGAACTCTTGT-3′ (He et al., 2004). Polymerase chain reaction (PCR) amplifications were performed in a total volume of 20 μl containing 0.1 μmol/L of each primer, 1.5 mmol/L MgCl2, 200 μmol/L deoxyribonucleotide triphosphates (dNTPs), 50 ng DNA template, 0.03 U/μl Taq DNA polymerase, and 2× PCR buffer (TaKaRa, China). Reaction conditions used with the thermal cycler (Biometra, Germany) were as follows: an initial incubation at 94 °C for 3 min, 30 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 5 min (Liang et al., 2009). The PCR products were genotyped by restriction digestion with endonuclease BstXI (TaKaRa, China). The digestion products were then separated on a 2% agarose gel stained with gel red for visualization under ultraviolet (UV) light. PCR products from subjects with GG genotypes were refractory to digestion with BstXI while DNA from homozygote for the A allele (AA genotype) was completely digested into two fragments (Fig. 1). The accuracy of the restriction fragment length polymorphism (RFLP) genotyping was confirmed by direct sequencing of the random DNA samples (n=30) for all three respective genotypes.
Fig. 1.
Agarose gel electrophoresis analysis of restriction endonuclease digestion
DNA bands with sizes at 467 and 187 bp indicated homozygous G alleles, and homozygous A alleles were represented by an uncut fragment of 654 bp, whereas the heterozygous genotype displayed a combination of 654, 467 and 187 bp
2.3. Statistical analysis
Independent sample t-test was used for the comparison between two groups for normal distribution and homogeneity variance. For the ranked data, Fisher’s exact test was applied for the comparison between two groups. Allele and genotype distributions between non-CHD and CHD were compared by χ 2 test. Odds ratios (ORs) were calculated as a measure of the association of the genotype with the phenotype of CHD using two-by-two table/χ 2 analysis. Crossover analysis was used to discover the interactions of IgE, chymase, and CMA1 genotypes with CHD. For each OR, two-tailed P values and 95% confidence intervals (95% CI) were determined. SAS 9.1 version was used for analysis and P<0.05 was considered to be statistically significant.
3. Results
3.1. Baseline for CHD and non-CHD
Independent sample t-test showed that CHD subjects were older than non-CHD subjects ((65.09±9.79) vs. (57.93±10.60) years, P<0.0001, Table 1). There was no significant difference in BMI between the two groups. Fisher’s exact test indicated that both genders (P=0.0166) and history of diabetes mellitus (P=0.0344), rather than smoking, family history of CHD, or hypertension, affected CHD status (Table 1).
Table 1.
Clinical data comparisons between subjects with and without CHD
| Variables | Value |
P | |
| CHD | Non-CHD | ||
| Age (year) | 65.09±9.79 | 57.93±10.60 | <0.0001* |
| BMI (kg/m2) | 23.92±3.14 | 23.53±3.45 | 0.3498* |
| Sex | |||
| Male | 121 (67.22%) | 59 (32.78%) | 0.0166** |
| Female | 47 (52.22%) | 43 (47.78%) | |
| Smoking | |||
| No | 109 (61.24%) | 69 (38.76%) | 0.6420** |
| Yes | 59 (64.13%) | 33 (35.87%) | |
| Family history of CHD | |||
| No | 152 (64.14%) | 85 (35.86%) | 0.0823** |
| Yes | 16 (48.48%) | 17 (51.52%) | |
| Diabetes mellitus | |||
| No | 129 (59.17%) | 89 (40.83%) | 0.0344** |
| Yes | 39 (75.00%) | 13 (25.00%) | |
| Hypertension | |||
| No | 48 (55.17%) | 39 (44.83%) | 0.0995** |
| Yes | 120 (65.57%) | 63 (34.43%) | |
Total n=270. Values are expressed as mean±SD or n (%)
Independent sample t-test
Fisher’s exact test
P<0.05 was considered to be statistically significant
3.2. Genotype and allele frequencies
The genotype and allele frequencies in CHD and non-CHD subjects are listed in Table 2. No significant differences among these genotypes were observed between CHD and non-CHD (P=0.5326). The frequencies of alleles were consistent with Hardy-Weinberg expectations in the non-CHD and CHD groups (χ 2=1.23, P=0.2676; χ 2=0.08, P=0.7828).
Table 2.
Distribution of CMA1 genotypes between non-CHD and CHD subjects and exact test for Hardy-Weinberg equilibrium
| Group | n | Genotype frequency* |
P | Allele frequency* |
Hardy-Weinberg equilibrium |
||||
| AA | AG | GG | A | G | χ2 | P | |||
| CHD | 144 | 11 (7.64%) | 60 (41.67%) | 73 (50.69%) | 0.5326 | 82 (28.47%) | 206 (71.53%) | 0.08 | 0.7828 |
| Non-CHD | 76 | 9 (11.84%) | 28 (36.84%) | 39 (51.32%) | 46 (30.26%) | 106 (69.74%) | 1.23 | 0.2676 | |
Total n=220
Values are expressed as n (%)
3.3. Association between serum IgE and chymase levels and genotypes
In CHD subjects, the IgE (U/ml) level is substantially higher in AA/AG genotype than in GG (4.03±1.25 vs. 3.33±1.43 (natural logarithm conversion), P=0.0032). No parallel result was found in the non-CHD subjects (P=0.0745). Meanwhile, no difference of serum chymase levels was found between CMA1 genotypes (CHD: P=0.3641; non-CHD: P=0.2532; Table 3).
Table 3.
Serum chymase and IgE comparisons between CMA1 genotypes
| Group | IgE |
Chymase |
||||
| n | lnc*# | P | n | c (μg/ml)* | P | |
| CHD | 133 | 0.0032 | 130 | 0.3641 | ||
| GG | 3.33±1.43 | 16.58±4.29 | ||||
| AA/AG | 4.03±1.25 | 15.90±4.31 | ||||
| Non-CHD | 71 | 0.0745 | 70 | 0.2532 | ||
| GG | 4.10±1.24 | 18.01±7.37 | ||||
| AA/AG | 3.54±1.37 | 16.43±3.64 | ||||
Values are expressed as mean±SD
The natural logarithm conversion of IgE concentration (U/ml)
3.4. Association between CHD risk and genotypes, serum chymase and IgE levels
Stratified analysis was used to explore the interaction of IgE, chymase, and genotypes with CHD. Serum IgE and chymase data were classified into two groups by comparing to the median (Table 4). Crossover analysis was applied to investigate the interaction of chymase, IgE, and genotypes in CHD subjects. No significant difference was found between non-CHD and CHD with respect to the interaction of serum chymase with IgE (Interaction OR=0.684, P=0.51; Table 5). Surprisingly, our data indicated that, in subjects with GG genotype, CHD risk was nearly six times higher in IgE (U/ml) level <2.58 (natural logarithm conversion) group than IgE level ≥2.58 (OR=5.97, P=0.0006; Table 6), while the IgE level had no effect on CHD risk in subjects with AA/AG genotype (OR=3.22 (IgE level ≥2.58) vs. OR=3.33 (IgE level <2.58); Table 6). Therefore, we draw the conclusion that IgE level may have an interaction with GG for CHD risk, but not in AA/AG genotype. The whole interaction OR caused by IgE and genotypes is more notable (OR=5.77, P=0.0116; Table 6) than that calculated for IgE and chymase levels (OR=0.68, P=0.5143; Table 5).
Table 4.
Stratified analysis for IgE, chymase and CMA1 genotypes between non-CHD and CHD subjects
| Variables |
n |
OR (95% CI)c | P | |
| CHD | Non-CHD | |||
| IgE (n=252)a | ||||
| <2.58 | 81 (64.29%) | 45 (35.71%) | 1.00 | |
| ≥2.58 | 75 (59.52%) | 51 (40.48%) | 0.72 (0.40, 1.27) | 0.2546 |
| Chymase (n=247)b | ||||
| <15.81 μg/ml | 79 (63.71%) | 45 (36.29%) | 1.00 | |
| ≥15.81 μg/ml | 74 (60.16%) | 49 (39.84%) | 1.10 (0.62, 1.94) | 0.7514 |
| Genotypes (n=219) | ||||
| GG | 73 (65.18%) | 39 (34.82%) | 1.00 | |
| AA/AG | 71 (66.36%) | 36 (33.64%) | 1.27 (0.67, 2.39) | 0.4606 |
Expressed by median after natural logarithm conversion of IgE concentration (U/ml)
Expressed by median
Adjusted by age, BMI, sex, diabetic disease, smoking, family history, and hypertension
Table 5.
Crossover analysis for the interaction of IgE and chymase between non-CHD and CHD subjects
| IgEa | Chymaseb |
n |
OR (95% CI) | P | |
| CHD | Non-CHD | ||||
| ≥2.58 | <15.81 μg/ml | 39 (61.90%) | 24 (38.10%) | 1.00 | |
| ≥2.58 | ≥15.81 μg/ml | 33 (55.93%) | 26 (44.07%) | 0.91 (0.41, 2.02) | 0.8133 |
| <2.58 | <15.81 μg/ml | 40 (65.57%) | 21 (34.43%) | 1.22 (0.53, 2.79) | 0.6447 |
| <2.58 | ≥15.81 μg/ml | 41(64.06%) | 23 (35.94%) | 1.62 (0.71, 3.67) | 0.2523 |
| Interaction | 0.68 (0.22, 2.15) | 0.5143 | |||
Total n=247
Expressed by median after natural logarithm conversion of IgE concentration (U/ml)
Expressed by median
Table 6.
Crossover analysis for IgE and genotypes between non-CHD and CHD subjects
| IgEa | Genotypes |
n |
OR (95% CI) | P | |
| CHD | Non-CHD | ||||
| ≥2.58 | GG | 23 (50.00%) | 23 (50.00%) | 1.00 | |
| ≥2.58 | AA/AG | 37 (68.52%) | 17 (31.48%) | 3.22 (1.25, 8.31) | 0.0157 |
| <2.58 | GG | 47 (77.05%) | 14 (22.95%) | 5.97 (2.16, 16.49) | 0.0006 |
| <2.58 | AA/AG | 26 (61.90%) | 16 (38.10%) | 3.33 (1.21, 9.19) | 0.0201 |
| Interaction | 5.77 (1.48, 22.53) | 0.0116 | |||
Total n=203
Expressed by median after natural logarithm conversion of IgE concentration (U/ml)
4. Discussion
We examined the association of the SNP rs1800875, serum chymase, and IgE levels in non-CHD and CHD subjects in a Chinese population. The IgE level is substantially higher in AA/AG genotype than in GG homozygote in CHD patients. Meanwhile, in subjects with GG genotype, CHD risk was nearly six times higher in those with IgE level <2.58. Therefore, SNP rs1800875 may be associated with CHD risk and IgE level in CHD patients.
In this study, the allele and genotype distributions of SNP rs1800875 exhibited no significant association between the locus and CHD. Previous studies have shown that rs1800875 was not associated with hypertrophic or dilated cardiomyopathy (Pfeufer et al., 1998; Wu et al., 2002). Gardemann et al. (2000) showed that rs1800875 variation had no significant impact on the risk and extent of CHD after analyzing a population of more than 2 000 patients. Our study was consistent with these observations, which may substantiate the hypothesis that this SNP locus is not the crucial locus affecting chymase expression. Considering the important roles of chymase in the formation and progression of atherosclerosis plaques, it is highly possible that other gene variables in CMA1, such as SNP, insert and deletion, or repeat polymorphism, affect the serum chymase level. For instance, a novel (TG)n(GA)m repeat polymorphism 254-bp downstream of CMA1 is associated with atopic asthma and total serum IgE levels (Sharma et al., 2005).
Interestingly, we observed a significant association between the genotypes of SNP rs1800875 and serum IgE level in CHD patients. Our data suggested that IgE level is substantially higher in AA/AG genotypes than GG only in CHD patients, which is not consistent with two studies involving asthma patients, both of which had reported a higher total IgE level in the GG genotype (Iwanaga et al., 2004; Sharma et al., 2005). IgE levels are associated with many factors, and the difference in these studies may be attributed to the different subjects used, the ethnicity, and environmental exposure. In addition, it is also possible that some other genes or loci contribute to the high IgE levels. The actions of human chymase may partly contribute to the relationship of IgE responsiveness and rs1800875. On the one hand, in human atherosclerotic lesions, increased IgE levels and enhanced FcεRI expression may have adjuvant activity sufficient to activate mast cells. Interleukin-4 (IL-4) and IL-13, secreted by T helper-2 (Th-2) cells, provide the first signal to B cells to switch the IgE isotopes (Busse and Lemanske, 2001). On the other hand, a previous study demonstrated that IgE synthesis could be promoted by the addition of a rat chymase to a culture of murine spleen cells motivated by IL-4 and lipopolysaccharide (Yoshikawa et al., 2001). As well, administration of a synthetic chymase inhibitor (Y-40613) suppressed total IgE levels in a rat model of atopic dermatitis (Imada et al., 2002). However, this may not explain the relationship between CMA1 genotypes and high serum IgE level if one only considers the actions of human chymase. First, genetic susceptibility of IgE responsiveness is likely to be caused by a myriad of polymorphisms in multiple genes regulating immunologic responses (Xu et al., 2000). However, it is still unclear whether the association is due to the polymorphism altering gene expression, or another causal allele in linkage disequilibrium with the rs1800875 or other functional and established loci. Only a few loci could be established consistently and robustly, such as FCER1B, IL-13, and STAT6 (Vercelli, 2008). Meanwhile, it may be effective to investigate the gene-gene interactions in mast cell degranulation and combined effects on atherosclerosis, and gene-gene interactions involved in the biosynthesis of mediators, such as leukotrienes and prostaglandins. Secondly, we know that few B cells were found in human atherosclerotic intima, which appears paradoxical in the light of the mass staining IgE in the same region (Roselaar et al., 1996). Moreover, chymase can control the bioavailability of cytokines and growth factors, such as activating IL-1β (Mizutani et al., 1991), releasing membrane-bound stem cell factor (Longley et al., 1997), and degrading IL-4 (Tunon de Lara et al., 1994). However, as a key factor in the generation of IgE, IL-4 is deactivated by human chymase, which appears paradoxical with the view discussed above. Lastly, it is of importance that rs1800875 seems to have little or even no effect on the expression of serum chymase in our study. The relationship between IgE responsiveness and rs1800875 still remains unclear.
Subsequently, it is important to note that in subjects with GG homozygote, CHD risks was nearly six times in IgE level <2.58 group than IgE level ≥2.58, which indicates that a higher IgE level is protective for GG homozygotes. This observation is concordant with the two other studies (Criqui et al., 1987; Szczeklik et al., 1988). In white populations, IgE levels were found to be significantly higher in the patients with unstable angina and acute myocardial infarction, compared to the patients with stable angina pectoris and controls (Korkmaz et al., 1991). Szczeklik et al. (1988) suggest that CHD patients with high IgE levels might be protected against complications of infarction. Another study showed that patients with high serum IgE levels might be protected against sudden cardiac death (Szczeklik et al., 1993). In our study, the mean IgE level in CHD subjects amounts to the normal serum IgE level range in Chinese population. In subjects with GG homozygote, a corresponding higher IgE level (≥2.58) plays a protective role in terms of the CHD risk. The exact mechanism for this risk reduction remains poorly elucidated thus far. Perhaps, the net effect of the mediators released from activated mast cells is advantageous to the local blood flow environment at the region of necrotic myocardium (Szczeklik et al., 1988). Actually, using antiplatelet therapy is effective to reduce the risks associated with percutaneous coronary intervention, such as restenosis and ischemia-reperfusion injury (Ying et al., 2010). Besides, impaired platelet aggregability and an extended bleeding time were regularly found in subjects with high IgE (Szczeklik et al., 1986). This mild homeostatic imbalance might explain the phenomenon that death from myocardial infarction in patients with atopic bronchial asthma appears to be rare (Szczeklik et al., 1977).
Large population-based prospective studies with ethnically diverse populations are needed to further elucidate the relationships of SNP rs1800875 with IgE and chymase levels in CHD patients.
Acknowledgments
We thank all the patients and their families who agreed to participate in this study.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 30670867) to Mei-xiang XIANG, and the Major Program of Science and Technology Department of Zhejiang Province, China (No. 2007C13058) to Mei-xiang XIANG
References
- 1.Busse WW, Lemanske RF. Advances in immunology: asthma. N Engl J Med. 2001;2001(344):350–362. doi: 10.1056/NEJM200102013440507. [DOI] [PubMed] [Google Scholar]
- 2.Criqui MH, Lee ER, Hamburger RN, Klauber MR, Coughlin SS. IgE and cardiovascular disease. Results from a population-based study. Am J Med. 1987;82(5):964–968. doi: 10.1016/0002-9343(87)90159-8. [DOI] [PubMed] [Google Scholar]
- 3.Galli SJ, Kalesnikoff J, Grimbaldeston MA, Piliponsky AM, Williams CMM, Tsai M. Mast cells as “tunable” effector and immunoregulatory cells: recent advances. Annu Rev Immunol. 2005;23(1):749–786. doi: 10.1146/annurev.immunol.21.120601.141025. [DOI] [PubMed] [Google Scholar]
- 4.Gardemann A, Harnami M, Katz N, Tillmann H, Haberbosch W. The chyrnase A(−1903)G gene polymorphism is not associated with the risk and extent of coronary heart disease. Atherosclerosis. 2000;150(2):445–446. doi: 10.1016/S0021-9150(00)00387-7. [DOI] [PubMed] [Google Scholar]
- 5.Gurish MF, Austen KF. The diverse roles of mast cells. J Exp Med. 2001;194(1):F1–F6. doi: 10.1084/jem.194.1.F1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He H, Li LM, Cao WH, Sun NL, liu MZ, Hu YH. Association between the I/D polymorphism of the ACE gene, A/B polymorphism of the chymase gene and the left ventricular hypertrophy in essential hypertension. Chin J Hypertens. 2004;12(1):39–43. (in Chinese) [Google Scholar]
- 7.Heikkila HM, Latti S, Leskinen MJ, Hakala JK, Kovanen PT, Lindstedt KA. Activated mast cells induce endothelial cell apoptosis by a combined action of chymase and tumor necrosis factor-α. Arterioscler Thromb Vasc Biol. 2008;28(2):309–314. doi: 10.1161/ATVBAHA.107.151340. [DOI] [PubMed] [Google Scholar]
- 8.Ihara M, Urata H, Kinoshita A, Suzumiya J, Sasaguri M, Kikuchi M, Ideishi M, Arakawa K. Increased chymase-dependent angiotensin II formation in human atherosclerotic aorta. Hypertension. 1999;33(6):1399–1405. doi: 10.1161/01.hyp.33.6.1399. [DOI] [PubMed] [Google Scholar]
- 9.Imada T, Komorita N, Kobayashi F, Naito K, Yoshikawa T, Miyazaki M, Nakamura N, Kondo T. Therapeutic potential of a specific chymase inhibitor in atopic dermatitis. Jpn J Pharmacol. 2002;90(3):214–217. doi: 10.1254/jjp.90.214. [DOI] [PubMed] [Google Scholar]
- 10.Iwanaga T, McEuen A, Walls AF, Clough JB, Keith TP, Rorke S, Barton SJ, Holgate ST, Holloway JW. Polymorphism of the mast cell chymase gene (CMA1) promoter region: lack of association with asthma but association with serum total immunoglobulin E levels in adult atopic dermatitis. Clin Exp Allergy. 2004;34(7):1037–1042. doi: 10.1111/j.1365-2222.2004.02000.x. [DOI] [PubMed] [Google Scholar]
- 11.Jeziorska M, McCollum C, Woolley DE. Mast cell distribution, activation, and phenotype in atherosclerotic lesions of human carotid arteries. J Pathol. 1997;182(1):115–122. doi: 10.1002/(SICI)1096-9896(199705)182:1<115::AID-PATH806>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Korkmaz ME, Oto A, Saraclar Y, Oram E, Oram A, Ugurlu S, Karamehmetoglu A, Karaagaoglu E. Levels of IgE in the serum of patients with coronary arterial disease. Int J Cardiol. 1991;31(2):199–204. doi: 10.1016/0167-5273(91)90216-C. [DOI] [PubMed] [Google Scholar]
- 13.Liang YH, Chen XL, Yu ZS, Chen CY, Bi S, Mao LG, Zhou BL, Zhang XN. Deletion analysis of SMN1 and NAIP genes in southern Chinese children with spinal muscular atrophy. J Zhejiang Univ-Sci B. 2009;10(1):29–34. doi: 10.1631/jzus.B0820125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longley BJ, Tyrrell L, Ma Y, Williams DA, Halaban R, Langley K, Lu HS, Schechter NM. Chymase cleavage of stem cell factor yields a bioactive, soluble product. PNAS. 1997;94(17):9017–9021. doi: 10.1073/pnas.94.17.9017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizutani H, Schechter N, Lazarus G, Black RA, Kupper TS. Rapid and specific conversion of precursor interleukin 1 β (IL-1β) to an active IL-1 species by human mast cell chymase. J Exp Med. 1991;174(4):821–825. doi: 10.1084/jem.174.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortlepp JR, Janssens U, Bleckmann F, Lauscher J, Merkelbach-Bruse S, Hanrath P, Hoffmann R. A chymase gene variant is associated with atherosclerosis in venous coronary artery bypass grafts. Coron Artery Dis. 2001;12(6):493–497. doi: 10.1097/00019501-200109000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Pfeufer A, Busjahn A, Vergopoulos A, Knoblauch H, Urata H, Osterziel KJ, Menz M, Wienker TF, Faulhaber HD, Steinmetz A, et al. Chymase gene locus is not associated with myocardial infarction and is not linked to heart size or blood pressure. Am J Cardiol. 1998;82(8):979–981. doi: 10.1016/S0002-9149(98)00518-9. [DOI] [PubMed] [Google Scholar]
- 18.Qin Y, Shi GP. Cysteinyl cathepsins and mast cell proteases in the pathogenesis and therapeutics of cardiovascular diseases. Pharmacol Ther. 2011;131(3):338–350. doi: 10.1016/j.pharmthera.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roselaar SE, Kakkanathu PX, Daugherty A. Lymphocyte populations in atherosclerotic lesions of apoE −/− and LDL receptor −/− mice. Decreasing density with disease progression. Arterioscler Thromb Vasc Biol. 1996;16(8):1013–1018. doi: 10.1161/01.ATV.16.8.1013. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz LB, Austen KF. Enzymes of the mast cell granule. J Invest Dermatol. 1980;74(5):349–353. doi: 10.1111/1523-1747.ep12543620. [DOI] [PubMed] [Google Scholar]
- 21.Sharma S, Rajan UM, Kumar A, Soni A, Ghosh B. A novel (TG)n(GA)m repeat polymorphism 254 bp downstream of the mast cell chymase (CMA1) gene is associated with atopic asthma and total serum IgE levels. J Hum Genet. 2005;50(6):276–282. doi: 10.1007/s10038-005-0252-x. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, Sukhova GK, Wolters PJ, Yang M, Kitamoto S, Libby P, MacFarlane LA, Mallen-St Clair J, Shi GP. Mast cells promote atherosclerosis by releasing proinflammatory cytokines. Nat Med. 2007;13(6):719–724. doi: 10.1038/nm1601. [DOI] [PubMed] [Google Scholar]
- 23.Szczeklik A, Nizankowski R, Mruk J. Myocardial infarction in status asthmaticus. Lancet. 1977;1(8012):658–659. doi: 10.1016/S0140-6736(77)92102-X. [DOI] [PubMed] [Google Scholar]
- 24.Szczeklik A, Milner PC, Birch J, Watkins J, Martin JF. Prolonged bleeding time, reduced platelet aggregation, altered PAF-acether sensitivity and increased platelet mass are a trait of asthma and hay fever. Thromb Haemost. 1986;56(3):283–287. [PubMed] [Google Scholar]
- 25.Szczeklik A, Sladek K, Szczerba A, Dropinski J. Serum immunoglobulin E response to myocardial infarction. Circulation. 1988;77(6):1245–1249. doi: 10.1161/01.CIR.77.6.1245. [DOI] [PubMed] [Google Scholar]
- 26.Szczeklik A, Dropinski J, Gora PF. Serum immunoglobulin E and sudden cardiac arrest during myocardial infarction. Coron Artery Dis. 1993;4(11):1029–1032. doi: 10.1097/00019501-199311000-00012. [DOI] [PubMed] [Google Scholar]
- 27.Tunon de Lara JM, Okayama Y, McEuen AR, Heusser CH, Church MK, Walls AF. Release and inactivation of interleukin-4 by mast cells. Ann N Y Acad Sci. 1994;725:50–58. doi: 10.1111/j.1749-6632.1994.tb39789.x. [DOI] [PubMed] [Google Scholar]
- 28.Urata H, Kinoshita A, Perez DM, Misono KS, Bumpus FM, Graham RM, Husain A. Cloning of the gene and cDNA for human heart chymase. J Biol Chem. 1991;266(26):17173–17179. [PubMed] [Google Scholar]
- 29.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8(3):169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 30.Wu GR, Ma AQ, Li ZH, Geng T. Study of the association of the I/D polymorphism of the ACE gene, A/G polymorphism of the heart chymase gene and idiopathetic dilated cardiomyopathy. J Clin Cardiol. 2002;18(3):100–103. (in Chinese) [Google Scholar]
- 31.Xiang M, Sun J, Lin Y, Zhang J, Chen H, Yang D, Wang J, Shi GP. Usefulness of serum tryptase level as an independent biomarker for coronary plaque instability in a Chinese population. Atherosclerosis. 2011;215(2):494–499. doi: 10.1016/j.atherosclerosis.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Postma DS, Howard TD, Koppelman GH, Zheng SL, Stine OC, Bleecker ER, Meyers DA. Major genes regulating total serum immunoglobulin E levels in families with asthma. Am J Hum Genet. 2000;67(5):1163–1173. doi: 10.1086/321190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying SQ, Xiang MX, Fang L, Wang JA. Temporal changes in circulating P-selectin, plasminogen activator inhibitor-1, magnesium, and creatine kinase after percutaneous coronary intervention. J Zhejiang Univ-Sci B. 2010;11(8):575–582. doi: 10.1631/jzus.B1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshikawa T, Imada T, Nakakubo H, Nakamura N, Naito K. Rat mast cell protease-I enhances immunoglobulin E production by mouse B cells stimulated with interleukin-4. Immunology. 2001;104(3):333–340. doi: 10.1046/j.1365-2567.2001.01320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]