Abstract
Malaria research often requires isolation of individually infected red blood cells (RBCs) or a homogenous parasite population derived from a single parasite (clone). Traditionally, isolation of individual, parasitized RBCs or parasite cloning is achieved by limiting dilution or micromanipulation. This protocol describes a method for more efficient cloning of the malaria parasite, which uses a cell sorter to rapidly isolate singly Plasmodium falciparum-infected RBCs. By gating the parameters of forward angle light scatter (FSC) and side angle light scatter (SSC) in a cell sorter, singly-infected RBCs can be isolated and automatically deposited into a 96-well culture plate within one minute. To include a Percoll purification step, the entire procedure to seed a 96-well plate with singly-infected RBCs takes less than 40 min. This highly efficient single-cell sorting protocol should be useful for cloning of both laboratory parasite populations from genetic manipulation experiments and clinical samples.
Keywords: malaria parasite, Plasmodium falciparum, parasite cloning, single-cell sorting
INTRODUCTION
Single, live malaria parasites or homogenous parasite populations are frequently needed for molecular, biochemical, immunological and drug sensitivity studies. For example, studies designed to elucidate the expression of multigene families of variant proteins (such as the var gene family in Plasmodium falciparum and vir gene family in P. vivax) in individual parasites have used single, infected red blood cells (RBCs)1,2. In addition, parasite cloning is also needed for in vitro monitoring of parasite’s sensitivity to antimalarial drugs, since in vitro drug assays using samples with mixed parasite strains are not accurate3, and clinical samples obtained from hyperendemic regions often contain mixed parasite strains4. Furthermore, genetic manipulation of the parasites by homologous recombination may result in a genetically heterogeneous parasite population as a result of integration of the plasmid at different genomic loci, and thus requires a cloning step to select parasite clones from the wanted recombination event.
For molecular and biochemical analysis, individual parasites in RBCs are mostly obtained by micromanipulation using micropipettes and an inverted light microscope connected to a micromanipulator system1,2. Although accurate, this procedure is rather time-consuming and may not be suitable for parasite cloning purpose. To obtain homogeneous parasite populations (clones) of the malaria parasite P. falciparum, limiting dilution is the most commonly-used method5. To do this, parasitemia is quantified by microscopy, and the culture is diluted to a desired parasitemia so that infected RBCs can be divided into single culture units. Although this method is simple without requiring sophisticated equipment, it is tedious, taking at least 1.5 h to complete. In addition, this method is not accurate and unable to distinguish multiply infected RBCs, which are common under stationary culture conditions. Therefore, subsequent molecular analysis is necessary to confirm the clonality of parasite populations arisen from individual culture wells.
Flow cytometry and cell sorting are essential tools in contemporary biological research, allowing rapid and highly accurate quantitation of individual cells based on differential fluorescence or light scattering properties induced by laser beams. The intensity of the light scattered in the forward direction (FSC) denotes cell size, while the 90° side scatter (SSC) is refracted in proportion to the granular content of the cell, thus denoting the intracellular complexity. The sorting function allows high-speed, accurate separation and collection of individual cells based on user-defined flow cytometry parameters. Beyond the exceptionally accurate and analytical nature, sorting function is non-destructive with little to no effect on cell viability or function. In malaria research, cell sorting has been used to isolate infected liver cells 6,7 and sexually differentiated gametocytes 8, which served as starting materials for subsequent molecular and biochemical analysis of the collected cell populations. In addition, a flow-sorting protocol has been used to isolate genetically transformed, green fluorescent protein (GFP)-expressing blood stages of the rodent malaria parasite P. berghei 9, which were used to generate stably transformed parasites without the use of drug selectable markers.
In this protocol, we have first determined the parameters for sorting P. falciparum-infected RBCs without using fluorescent dyes so that the sorted parasites can be cultured subsequently10. This capability is essential for cloning field clinical isolates and genetically transformed P. falciparum parasites that do not express a fluorescent marker. We have opted to select the late trophozoite stage of the parasite for sorting to facilitate the distinction of singly- from multiply-infected RBCs and for culturing the sorted parasites. In addition, a Percoll gradient purification step is added to highly enrich late trophozoite stage parasites before sorting, which significantly reduces the content of uninfected RBCs and cell debris in the pre-sorting materials, and shortens the sorting time. We have used this protocol for cloning genetically transformed GFP-expressing P. falciparum parasite10. An additional step of sorting GFP-expressing parasites allows more effective cloning of the GFP-expressing parasites. With the inclusion of a Percoll purification step, the entire sorting procedure to fill a 96-well plate is ~40 min, which is much more efficient than the traditional limiting dilution method. However, the current protocol has a few limitations. First, this single-cell sorting procedure requires a high speed cell sorter, which is not always within easy reach of the malaria parasite culture laboratories. Second, the current protocol is optimized for sorting trophozoite-infected RBCs, although the flow cytometric parameters for sorting other malaria parasite stages such as rings and schizonts could be determined. The protocol described in this paper is designed for the Cytopeia Influx™ high speed sorter (BD Biosciences). One can easily adjust the sorting conditions on other cell sorters, and the protocol should be equally adaptable for sorting singly infected RBCs by other human malaria parasites.
MATERIALS
REAGENTS
· Parasites The P. falciparum reference line 3D7A is used in this protocol. Other laboratory strains such as K1, D10, and Dd2, and clinical isolates can also be used. Parasites are routinely cultured at mixed stages with a <5% parasitemia. For sorting genetically transformed, GFP-expressing parasites, we used a 3D7 line expressing GFP under the PfGCN5 promoter10.
! CAUTION All experiments using P. falciparum parasites must be performed in biological safety level 2 laboratories.
-
·
AlbuMax II (Invitrogen; Cat. no.: 11021-045) (Store at 4°C. So far only Invitrogen produces the this reagent as serum replacement)
-
·
Gentamicin sulfate (EMD Chemicals, Cat. no.: EM-4730) (Prepare a stock solution of 100 mg/ml gentamicin sulfate in distilled water and store at −20°C)
-
·
Giemsa solution (Sigma-Aldrich, Cat. no.: 48900)
-
·
HEPES (J. T. Baker; Cat. no.: JT4018-04)
-
·
Human RBCs (type O) (ordered from a blood bank) (Store at 4°C and use within a month)
-
·
Human sera (type AB) (ordered from a blood bank) (For consistency, we normally order >10 units of sera, mix and aliquot before storage at −20°C)
-
·
Hypoxanthine (EMD Biosciences, Cat. no.: 80058-270)
-
·
Sodium bicarbonate (EMD Chemicals, Cat. no.: SX0320-1)
-
·
Percoll (Amersham, Cat. no.: 17-0891-01) (Store at 4°C)
-
·
Phosphate-buffered saline (PBS) (Prepare a 10 × stock solution and dilute to working solution. 10 X PBS solution can also be purchased, e.g., Fisher Scientific; Cat. no.: BP399-500).
-
·
Potassium chloride (KCl) (Fisher Scientific, Cat. no.: P217-500)
-
·
Potassium phosphate, monobasic (KH2PO4) (Fisher Scientific, Cat. no.: BP362-500)
-
·
RPMI 1640 (Invitrogen; Cat. no.: 31800-15)
-
·
Sodium chloride (NaCl) (EMD Chemicals, Cat. no.: SX0425-1)
-
·
Sodium phosphate, dibasic (Na2HPO4) (J. T. Baker; Cat. no.: JT4062-01)
-
·
Sorbitol (J. T. Baker Analytical, Cat. no.: JTV045-7)
EQUIPMENT
-
·
Cell culture flask (25 cm2 growth area and canted neck) (BD Falcon, Cat. no.: 353082)
-
·
Cell sorter (Cytopeia Influx high speed sorter, BD Biosciences)
-
·
Filter (Nalge Nunc International, Cat. no.: 595-3320)
-
·
Fluorescence microscope (Nikon, Eclipse E600)
-
·
15 ml Falcon tube (BD Falcon, Cat. no.: 352096)
-
·
Gas mixture (5% CO2, 3% O2 and 92% N2 in a gas cylinder)
-
·
Glass slides (VWR, Cat. no.: 16004-430)
-
·
Laminar flow hood (biological safety level 2) for cell culture
-
·
Light microscope (Nikon, Eclipse 50I)
-
·
Pipette 3 ml (BD Biosciences, Cat. no.: 357543)
-
·
Pyrex large-size desiccator with knob top (candle jar) (Corning, Cat. no.: 3080-250)
-
·
Shaking incubator (Barnstead, Lab-line incubator shaker)
-
·
37°C stationary incubator (Barnstead, Lab-line water-jacketed incubator)
-
·
Table top centrifuge (Beckman Coulter Allegra™ 6R)
-
·
96 well culture plate (BD Biosciences, Cat. no.: 353072)
REAGENT SETUP
10 X PBS
Add 80 g NaCl, 2 g KCl, 14.4 g Na2HPO4, and 2.4 g KH2PO4 to 1 L of distilled water, adjust pH to 7.2, and sterilize by filtration through a 0.22 µm filter or autoclaving at 120°C for 20 min. Store under room temperature.
Giemsa working solution
Add 19 volumes of phosphate buffer to 1 volume of Giemsa solution. For 1 L of phosphate buffer, add 11.54 ml of 1M Na2HPO4 and 8.46 ml of 1M NaH2PO4 to distilled water and adjust the volume with distilled water to 1 L. The Giemsa working solution is normally prepared fresh and used within one week.
Medium preparation
Incomplete culture medium: RPMI 1640 (10.4 g powder with L-glutamine) supplemented with 25 mM HEPES, pH 7.5, 25 mM sodium bicarbonate, 50 mg/L hypoxanthine, and 40 µg/ml gentamicin sulfate. Incomplete culture medium should be kept at 4°C and used within one month. Complete culture medium: For preparing complete medium supplemented with 10% (v/v) human sera, filtrate incomplete culture medium through a 0.22 µm filter, and add human sera directly. For complete medium supplemented with 0.5% (w/v) AlbuMax II, add AlbuMax II to the incomplete culture medium and sterilize the medium by filtration through a 0.22 µm filter. Complete culture medium should be kept at 4°C and used within 1–2 weeks.
Sorbitol
Prepare 5% (w/v) solution by dissolving 5 g of sorbitol in 100 ml of distilled water and sterilize by filtration through a 0.22 µm filter. Store at room temperature.
Percoll solution
Prepare 90% Percoll (200 ml) by mixing 20 ml of 10 X PBS with 180 ml of Percoll. Prepare 65% Percoll by combining 6.5 ml of 90% Percoll with 2.5 ml of incomplete culture medium, and prepare 35% Percoll by combining 3.5 ml of 90% Percoll with 5.5 ml of incomplete culture medium. Mix well and sterilize by filtration through a 0.22 µm filter. The sterile 90%, 65%, and 35% Percoll solutions are stored at 4°C. To cast a step Percoll gradient, use a 3 ml plastic pipette to load 35% Percoll on top of 65% Percoll. Hold the pipette so that the end is against the inner wall of the 15 ml Falcon tube. Slowly release the 35% Percoll on to the 65% Percoll layer so that a sharp interface forms between the two Percoll layers.
EUIPMENT SETUP
Light microscope
All light microscopes with a 100 × oil immersion objective are suitable for routine examination of Giemsa-stained blood smears.
Fluorescence microscope
Any fluorescence microscope equipped with a UV light source and a series of filters for detecting excitations for GFP, Hoechst or DAPI (4',6-diamidino-2-phenylindole) for staining the nuclei is suitable.
Cell sorter
In our laboratory, we use a Cytopeia Influx™ high speed sorter for single cell sorting. For other sorters, the setup is similar (see Box 2)
BOX 2. │ SETUP OF A CELL SORTER.
High-speed cell sorter is normally operated by a dedicated operator who is experienced in preparing the setting of the sorter and operating the machine.
If the purpose of single-cell sorting is for growing the sorted parasites, the plate-loading platform of the cell sorter needs to be decontaminated by UV light. Before performing parasite sorting, tubes or lines that deliver cells within the sorter should be rinsed with sterile PBS.
All performance of cell sorting is under room temperature.
Complete culture medium is used for cell sorting and collection.
The following parameters on the sorter are programmed: argon laser at 488 nm; flow chip diameter, 100 ml; sheath solution, PBS; sheath pressure, 52 PSI; crystal frequency, 85 KH; crystal drive, 12.2%; delay, 44 drops; sort mode, purity-yield; coincident abort, on. FSC and SSC are set at 24% and 44.25% photomultiplier tubes (PMT) power, respectively, to get maximum sensitivity.
For sorting GFP-expressing cells, another step is added in the sorting program. GFP excitation is performed at the wavelength of 488 nm and fluorescence is detected with a GFP filter (531/40 nm). Since GFP fluorescence intensity depends on factors such as promoters used to drive GFP expression, developmental stages, copy number of the expression plasmid, gating parameters for GFP fluorescence intensity (FL1) should be defined by the user. GFP-positive cells will be further selected based on the FSC and SSC scattergram to sort out singly infected RBCs.
PROCEDURE
All steps below must be performed under sterile conditions if the sorted parasites are used for culture purpose.
Percoll purification  TIMING 30 min
TIMING 30 min
-
1 │
Culture P. falciparum as described in Box 1. Transfer parasite culture from 25 cm2 flask to a 15 ml disposable Falcon tube. Centrifuge at 2000 rpm (~500 ×g) for 5 min at room temperature. Discard supernatant. Resuspend the cells with incomplete culture medium to 10% hematocrit (total ~2.5 ml).
-
2 │
Slowly layer cell suspension on top of the freshly prepared Percoll gradient (see REAGENT SETUP).
▲ CRITICAL STEP Layer the cell suspension in a similar manner as preparing the Percoll gradient. Make sure that the cell suspension is dispensed on top of the layer slowly so that the interface is not disturbed. Careful preparation of the Percoll gradient and parasite layer will help to reduce the amount of contaminants by dead cells or cell debris at the 35%/65% Percoll interface.
-
3 │
Centrifuge in a swing-out rotor at 1,500 × g at room temperature for 15 min (2,500 rpm in a Sorvall or Beckman tabletop centrifuge) without a brake.
-
4 │
Recover parasites from the 35%/65% Percoll interface and transfer it to a 15 ml disposable Falcon tube.
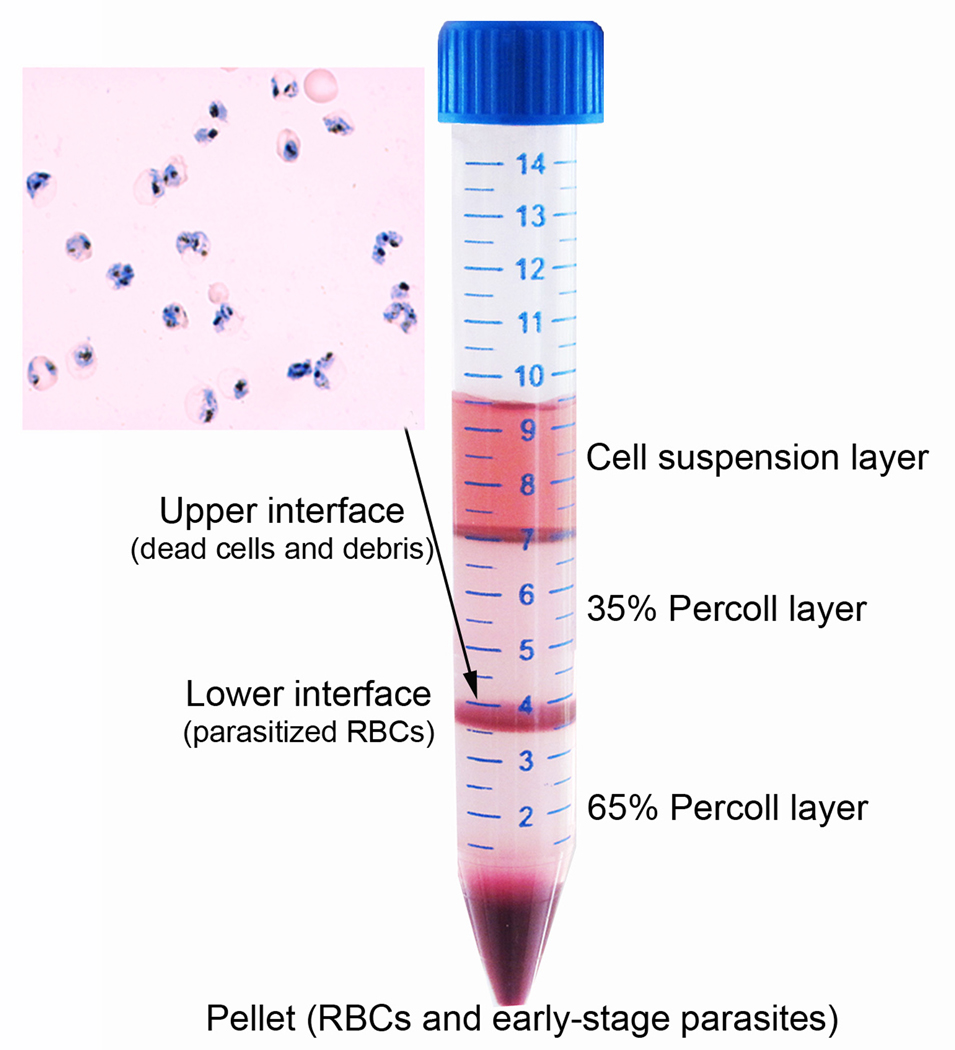
▲ CRITICAL STEP If above steps are performed properly, two thin, sharp interfaces between different layers and a large red pellet at the bottom of the tube will appear (Fig. 1). The upper interface mainly contains dead parasites and cell debris. The lower interface contains mostly late stage parasitized RBCs. The pellet consists of uninfected RBCs and early stage parasitized RBCs. To harvest the lower interface containing late stage parasitized RBCs, first use a pipette to remove and discard the top culture layer and most of 35% Percoll layer without touching the 35%/65% Percoll interface. Then, use a glass pipette to transfer content of the 35%/65% Percoll interface into a fresh 15 ml Falcon tube.
? TROUBLESHOOTING
-
5 │
Add 10 volumes of incomplete culture medium drop by drop to the recovered parasites, gently shaking after each addition.
-
6 │
Spin down the cells at 2,000 rpm (~500 ×g) for 5 min at room temperature and discard supernatant.
-
7 │
Wash once more with 10 volumes of incomplete culture medium by centrifugation at 2000 rpm (~500 ×g) for 5 min at room temperature and remove supernatant.
-
8 │
Resuspend parasitized RBCs with 1 ml of complete culture medium.
? TROUBLESHOOTING
BOX 1 │ PARASITE CULTURE.
We routinely culture P. falciparum parasites in 25 cm2 culture flasks in human type O RBCs at 5% hematocrit and <5% parasitemia11. For culture-adapted parasites, we use complete culture medium containing either 10% human sera or 0.5% AlbuMax II. For culturing malaria parasites from clinical samples, the complete medium containing 10% human sera is used. It is advised that parasites from clinical samples are pre-adapted to laboratory culture conditions before being subjected to cell sorting in order to increase cloning efficiency. Parasite culture is gassed with a gas mixture (5% CO2, 3% O2 and 92% N2) and medium is changed daily. Alternatively, parasites can be cultured in a Vaseline-sealed glass desiccator (candle jar). For setting up a candle jar for culturing malaria parasites (in 96-well plates or unsealed culture flasks), a candle is lit after placing the culture dishes in the candle jar, and the lid is closed and sealed with Vaseline. The candle jar is placed in a 37°C incubator. This creates an air-tight chamber with a low-oxygen atmosphere suitable for malaria parasite growth.
For sorting singly-infected RBCs, the parasites are cultured under slow shaking conditions (50 rpm/min) at <1% parasitemia. These conditions are necessary to reduce multiply-infected RBCs, which will result in a more concentrated FSC and SSC scattergram. Although culture under stationary conditions and at high parasitemias is still acceptable for sorting, we have found that cultures under shaking conditions and at low parasitemias consistently yielded satisfactory sorting results without the requirement of stringent selection of parasite fractions as described in this protocol.
Culture synchronization by sorbitol is necessary before sorting in order to obtain parasites of more homogeneous stages12. Briefly, culture with majority of the parasites at ring stage is centrifuged at 2,000 rpm (~500 × g) for 5 min to pellet the cells. Mix the cells with 5 volumes of 5% sorbitol for 5 min at 37°C. Centrifuge the suspension to pellet the cells and wash the pellet once with incomplete culture medium. Then the cells are resuspended in complete culture medium and cultured under standard culture conditions.
Parasite developmental stages are examined by Giemsa staining at different time intervals to select the mid-late trophozoite stage. Briefly, a drop of parasite culture is smeared on a glass slide, dried under room temperature, fixed with methanol for 10 sec, and stained in Giemsa working solution for 10 min. The slide is then rinsed briefly in tap water, dried and observed under a microscope with oil immersion and an objective at 100 ×. A 0.25 ml of trophozoite stage parasites cultured in a 25 cm2 flask at 0.5–1% parasitemia and 5% hematocrit is enough for sorting to fill more than ten 96-well plates.
Figure 1.
Purification of parasitized RBCs on a step Percoll gradient consisting of an upper 35% and a lower 65% Percoll layer. After centrifugation, the upper interface contains mostly cell debris, whereas the lower interface contains enriched parasitized RBCs. The bottom pellet contains mostly unparasitized RBCs and ring-stage parasites. Inset shows an image from Giemsa-stained thin smear from the lower interface, which contains mostly late-stage trophozoites.
Pre-load cell culture plates  TIMING 5 min
TIMING 5 min
-
9 │
Pre-load a 96-well plate with RBC suspension at 1% hematocrit in complete culture medium at 200 µl per well.
Cell Sorting  TIMING ~5 min
TIMING ~5 min
-
10 │
Set up the cell sorter as described in Box 2. Place the tube containing suspended parasites in Cytopeia Influx high speed sorter under sterile conditions at room temperature, and start sorting process. Adjust cell flow speed to 5,000 events/sec.
? TROUBLESHOOTING
-
11 │
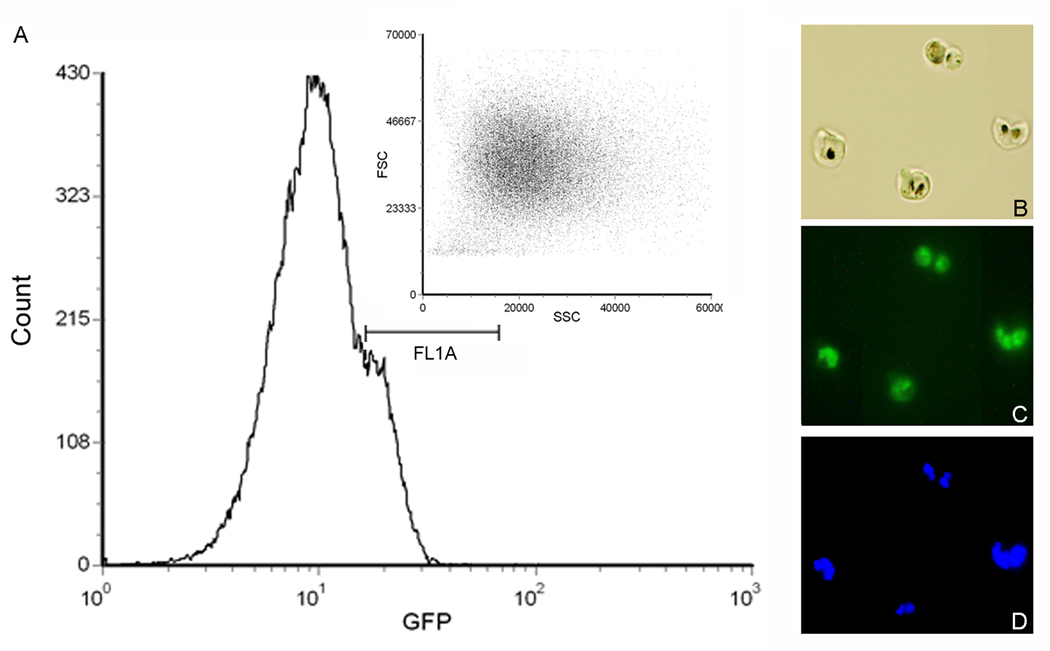
(optional) For sorting GFP-expressing parasites, first detect cells with GFP fluorescence and select cell populations with the desired intensity (FL1), and set up a gate to select GFP-positive cells (Fig. 2). Set up a scattergram of FSC and SSC in linear display to show distribution of gated GFP-positive cell.
-
12 │
Set up a two dimensional scattergram of FSC and SSC in linear display to show all cell distribution.
-
13 │
Choose gating parameters to select the area in the scattergram that contains high percentages of singly-infected RBCs (Fig. 3).
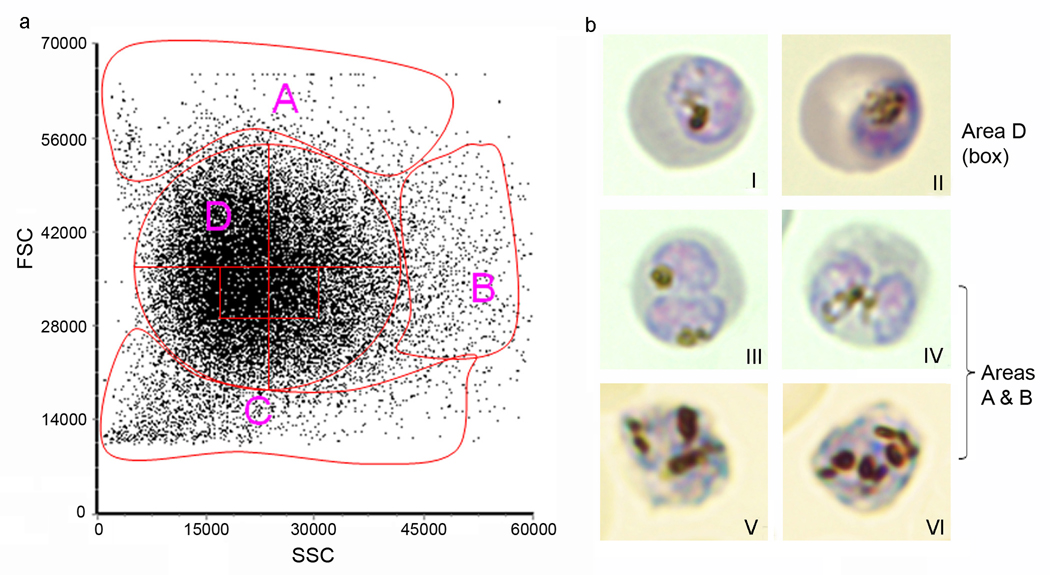
▲ CRITICAL STEP The actual sorting procedure is carried out by an experienced operator who programs and runs the sorter. It is critical to select the proper fractions with maximal percentage of singly-infected RBCs. With different sorters, a user will need to define the parameters and select the proper areas in the scattergram (see Step 14). Step 14 may take 20–40 min (<5 min for sorting a small portion of the culture; and 15–35 min for microscopic examination of Giemsa-stained slides to determine the area that contains singly-infected RBCs). Once the gating parameters are determined, they can be used in subsequent sorting experiments and the time to set up the sorter (Box 2) is normally less than 5 min. A typical scattergram generated from a FSC and SSC scans is shown in Fig. 3, which can be divided into four areas (Fig. 3a, A–D). Whereas Areas A and B contain many multiply-infected RBCs, Area C contains mostly uninfected RBCs and cell debris (Fig. 3b). Area D, the most condensed area of the scattergram, contains at least 70% singly parasitized RBCs (Fig. 3b). Within Area D, two small boxed areas just below the center contain the highest percentage of singly-infected cells (>98%). More cell debris occurs below the boxed areas, whereas more multiply-infected RBCs are found above the boxed areas. For parasite cloning purpose, one would rather select a slightly lower area to ensure that selected parasites are clonal.
-
14 │
(Optional) Put a glass slide on the stand of a receiving tray in the sorter. Sort 1000–2000 cells from the selected fractions to the slide, and stain the slide with Giemsa working solution. Examine the slides under a light microscope (oil immersion, 100 ×) to see whether the sorted cells are singly-parasitized RBCs.
-
15 │
Put the 96-well plate on the stand of a receiving tray in the sorter. Set the programmable micro-titer well tray configuration to deposit a single cell to each well, and start sorting each single particle (event) into a single well automatically.
? TROUBLESHOOTING
Figure 2.
Flow sorting of GFP-expressing cells. (A) Pre-sorting of GFP-positive cells by using a GFP filter and setting the gate on the GFP fluorescence intensity. In this case, the gate FL1A is used and cells with lower fluorescence intensity (unparasitized RBCs, parasites without GFP expression, etc.) are excluded from sorting. The inset shows the FSC and SSC scattergram of the selected parasitized RBCs, which is used for selection of singly-infected RBCs. (B–D) Gating for fluorescence intensity results in a parasite population made exclusively of GFP-expressing cells. (B) Light microscopic image of sorted parasitized RBCs by gating GFP fluorescence. (C) Fluorescent microscopic image of the same cells showing GFP fluorescence. (D) Fluorescent microscopic image of the same cells showing parasite nuclei stained with a fluorescent DNA dye, Hoechst-33258.
Figure 3.
Determination of singly infected RBCs from a typical scattergram. (a) A typical FSC and SSC scatter plot of P. falciparum trophozoite-stage culture after enrichment with a step Percoll gradient. Setting the gates to collect cells from the four major areas shows that Area A contains mostly singly-parasitized RBCs, whereas cells with high FSC or SSC (Areas A and B) are made of mostly multiply-parasitized RBCs. Area C contains mostly uninfected RBCs and cell debris. Further gating in the boxed areas of Area D results in >98% singly parasitized RBCs. (b) Giemsa-stained images of parasitized RBCs sorted from different areas of the scatter plot show singly-infected RBCs (images I and II) from Area D, doubly-infected RBCs (images III and IV) and triply-infected RBCs (images V and VI) from Areas A and B of the scatter plot.
Culturing sorted parasites  TIMING 2–3 weeks
TIMING 2–3 weeks
-
16 │
At this point, individual cells can be used for molecular and biochemical analysis. For parasite cloning, incubate the plate in a candle jar at 37°C (Box 1), and change culture medium on day 5 and 10. On day 12, make slides from the culture wells and examine parasite growth by Giemsa staining. Positive cultures are expanded first to 24-well plates and subsequently to 25 cm2 flasks. Parasites at this step are enough for long-term storage and subsequent analysis.
? TROUBLESHOOTING
● TIMING
Steps 1–8, Percoll purification: 30 min
Step 9, Pre-load cell culture plates with RBCs and medium: 5 min
Steps 10–15, Sorter setup (Box 2), 5 min; cell sorting: 1–5 min
Step 16, Culturing the sorted parasites: 2–3 weeks
? TROUBLESHOOTING
Table 1 │ Troubleshooting guide is listed in Table 1.
Table 1.
Troubleshooting table
| STEPS | PROBLEM | POSSIBLE CAUSE | SOLUTION |
|---|---|---|---|
| 1–4 | Low yield of parasites after Percoll purification | Parasitemia of culture is too low | Use culture with high parasitemias (normally 0.5–1% is enough) |
| 35% Percoll or culture suspension is added too fast, which strongly disturbs the 35/65 interface | Lean the pipette tip against the wall of tube and reach to the liquid surface and slowly add the 35% Percoll solution | ||
| 6–9 | Too much cell debris in the Percoll purification | Parasites culture suspension is added too fast, which strongly disturbs the 35/65 interface | Slowly add culture suspension to the 35% Percoll surface |
| 10 | The flow in the sorter is blocked | Cell aggregation occurs when cell concentration is too high | Lower cell density, complete separate cells by gently vortexing before sorting |
| 11–13 | Percentage of singly infected cells is low | Gating is not appropriate. You will get more doubly or triply infected RBCs or many uninfected cells and debris if the fractions chosen for sorting are higher or lower than the correct gate, respectively. | Pre-sorting is an option to seed cells on the glass slide, which are checked. Normally lower fractions are preferred though one might get more cell debris or uninfected RBCs |
| 15–16 | Single cell is not loaded into each well. | Plate is not placed in the right position | Make sure the plate is put on the receiving tray stand and reaches the end of frame |
| RBCs undergo lysis during culture within two weeks | RBCs are not fresh when put into plate | Use fresh RBCs (not more than one week) Add new RBCs at 0.5% hematocrit to each well when changing medium on day 5 and 10 |
ANTICIPATED RESULTS
After Percoll gradient purification, parasitized RBCs usually make up >95% (Fig. 1). For sorting of GFP-positive parasites, gating at a high fluorescence intensity (FL1) will result in >90% GFP-positive parasite population (Fig. 2). In a typical FSC and SSC scattergram, we normally observe RBCs parasitized by single, double and triple parasites in different areas (Fig. 3). Selection of the boxed fractions in Area D (Fig. 3) will result in ~90% of single cell units. If parasite cloning is the purpose, strict selection of an area in Area D results in almost 100% single clones 10. Under the specified culture conditions, parasite growth in the culture plate is detected in 2 weeks.
ACKNOWLEDGEMENTS
We would like to thank financial support from NIAID, NIH (R01AI064553 and U19AI089672).
Footnotes
Author contributions: J.M. designed and performed experiments, analysed data and wrote the paper; L.C. supervised the project and wrote the paper.
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
References
- 1.Chen Q, et al. Identification of Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) as the rosetting ligand of the malaria parasite P. falciparum. J. Exp. Med. 1998;187:15–23. doi: 10.1084/jem.187.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez-Becerra C, et al. Variant proteins of Plasmodium vivax are not clonally expressed in natural infections. Mol. Microbiol. 2005;58:648–658. doi: 10.1111/j.1365-2958.2005.04850.x. [DOI] [PubMed] [Google Scholar]
- 3.Liu S, Mu J, Jiang H, Su XZ. Effects of Plasmodium falciparum mixed infections on in vitro antimalarial drug tests and genotyping. Am. J. Trop. Med. Hyg. 2008;79:178–184. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong D, et al. Plasmodium falciparum genetic diversity in western Kenya highlands. Am. J. Trop. Med. Hyg. 2007;77:1043–1050. [PubMed] [Google Scholar]
- 5.Rosario V. Cloning of naturally occurring mixed infections of malaria parasites. Science. 1981;212:1037–1038. doi: 10.1126/science.7015505. [DOI] [PubMed] [Google Scholar]
- 6.Tarun AS, et al. Quantitative isolation and in vivo imaging of malaria parasite liver stages. Int. J. Parasitol. 2006;36:1283–1293. doi: 10.1016/j.ijpara.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Tarun AS, et al. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc. Natl. Acad. Sci. USA. 2008;105:305–310. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan SM, et al. Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell. 2005;121:675–687. doi: 10.1016/j.cell.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Janse CJ, Franke-Fayard B, Waters AP. Selection by flow-sorting of genetically transformed, GFP-expressing blood stages of the rodent malaria parasite, Plasmodium berghei. Nature Protocols. 2006;1:614–623. doi: 10.1038/nprot.2006.88. [DOI] [PubMed] [Google Scholar]
- 10.Miao J, Li X, Cui L. Cloning of Plasmodium falciparum by single-cell sorting. Exp. Parasitol. 2010;126:198–202. doi: 10.1016/j.exppara.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trager W, Jenson JB. Cultivation of malarial parasites. Nature. 1978;273:621–622. doi: 10.1038/273621a0. [DOI] [PubMed] [Google Scholar]
- 12.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]