Abstract
Rationale
Increasing evidence suggests that misfolded proteins and intracellular aggregates contribute to cardiac disease and heart failure. Several cardiomyopathies, including the αB-crystallin R120G mutation (CryABR120G) model of desmin-related cardiomyopathy, accumulate cytotoxic misfolded proteins in the form of pre-amyloid oligomers (PAOs) and aggresomes. Impaired autophagic function is a potential cause of misfolded protein accumulations, cytoplasmic aggregate loads and cardiac disease. Atg7, a mediator of autophagosomal biogenesis, is a putative regulator of autophagic function.
Objective
To determine whether autophagic induction by Atg7 is sufficient to reduce misfolded protein and aggregate content in protein misfolding-stressed cardiomyocytes.
Methods and Results
To define the gain and loss of function effects of Atg7 expression on CryABR120G protein misfolding and aggregates, neonatal rat cardiomyocytes (RNC) were infected with adenoviruses expressing either wild-type CryAB or CryABR120G, and co-infected with Atg7 adenovirus or with Atg7 silencing siRNAs to produce gain- or loss-of Atg7 function. Atg7 overexpression effectively induced basal autophagy with no detrimental effects on cell survival, suggesting that Atg7 can activate autophagy with no apparent cytotoxic effects. Autophagic flux assays on CryABR120G expressing cardiomyocytes revealed reduced autophagic function, likely contributing to the failure of misfolded proteins and aggregates to be cleared. Co-expression of Atg7 and CryABR120G significantly reduced PAO staining, aggregate content and cardiomyocyte cytotoxicity. Conversely, Atg7 silencing in the CryABR120G background significantly inhibited the already reduced rate of autophagy and increased CryABR120G aggregate content and cytotoxicity.
Conclusions
Atg7 induces basal autophagy, rescues the CryABR120G autophagic deficiency, and attenuates the accumulation of misfolded proteins and aggregates in cardiomyocytes.
Keywords: autophagy, Atg7, aggregate, amyloid
Introduction
Cellular homeostasis is necessary for cell survival. To maintain homeostasis, a cell must balance synthetic with catabolic processes. Eukaryotic cells have two major protein degradation pathways: the ubiquitin-proteasome system and the autophagy-lysosomal pathway. The proteasome specializes in the selective degradation of short-lived proteins.1,2 Macroautophagy, hereafter referred to as autophagy, is a process of bulk protein degradation, which functions to engulf and degrade damaged or long-lived proteins and organelles.3,4
Autophagy is initiated by elongation of an initial isolation membrane, the phagopore, which surrounds cytoplasmic material for sequestration.5 As the membrane expands, it forms a double-membrane vesicle referred to as an autophagosome. Autophagosomes completely surround the cytoplasmic products and act as a delivery vehicle for cargo degradation. Autophagosomes often fuse with other vacuolar structures such as endocytic vesicles to form amphisomes. Finally, the outer membrane of an amphisome fuses with the lysosome. The hydrolytic enzymes within the autolysosome degrade the inner membrane and subsequently, the cytoplasmic contents. The resulting macromolecules are released from the lysosome and recycled for use by the cell. This process is associated with both normal homeostasis and for countering cell stress.
Autophagy occurs at a basal level in most tissues, including the heart, contributing to the routine turnover of cellular trash. Under conditions of stress, such as nutrient starvation, autophagy is upregulated as an energy salvage process. In addition to energetic homeostasis, autophagy is involved in tissue development, differentiation and remodeling.6 Autophagy is also associated with several human diseases and models of cell death,7 and data indicate that autophagy can be either beneficial or detrimental, depending upon the cellular context.8,9
Gene ablation of critical components in the autophagic pathway has shown that autophagy is clearly necessary for mammalian survival. Beclin 1−/− mice die in early embryogenesis,10 and Atg5−/− and Atg7−/− mice do not survive the neonatal starvation period.11,12 Conditional models of autophagic impairment cause accumulation of ubiquitinated protein aggregates in cells.12 Although the effects of autophagic deficiency differ from tissue to tissue, a consensus phenotype is the formation of intracellular protein aggregates. Presumably, these protein aggregates represent undigested protein byproducts that accumulate due to the loss of autophagic function. Numerous human diseases accumulate protein aggregates, leading to the general hypothesis that at least some of the pathology associated with aggregation-based diseases is due to insufficient autophagic function.
To date, few models of autophagic induction have been developed and most are limited to starvation-induced autophagy. Transgenic overexpression of Beclin1-GFP had no effect on basal autophagy.13 Cardiomyocyte autonomous overexpression of Beclin 1 also failed to induce basal autophagy, but did significantly increase starvation-induced autophagy.14 FoxO1, FoxO3 and Sirt1 studies in cardiomyocytes have also been limited to starvation-induced autophagy.15,16 We hypothesized that Atg7 might be a viable target through which to activate basal autophagic function. Atg7 has dual functions in autophagosomal biogenesis. First, Atg7 acts as an E1-like ligase, which conjugates Atg5 to Atg12, a necessary step for formation of a functional autophagosome.17,18 Second, Atg7 converts Atg8 (LC3) from an immature, cytosolic form to a mature autophagosomal membrane protein by adding a phosphatidylethanolamine group.19 Genetic deletion of Atg7 causes an overt loss of autophagy, proving that Atg7 is necessary for autophagic function.12
In this manuscript, we test the hypothesis that Atg7 overexpression is sufficient to induce increased basal levels of autophagic function. We hypothesized that, if Atg7 did induce autophagy, we could then use this to evaluate whether autophagic upregulation is beneficial or detrimental to a cardiomyocyte under different, stressed conditions or in the unstressed cell. We tested this hypothesis by expressing Atg7 in RNCs, and by determining the ability of increased levels of Atg7 to clear protein aggregates formed by CryABR120G expression.
Methods
An expanded Methods sections is available in the Online Data Supplement at http://circres.ahajournals.org
Primary Cultures and Adenoviral Infection
Primary RNCs were isolated from the ventricles of 1–2-day old Sprague-Dawley pups and plated on 10 cm2 plates at a density of 1.5×106 cells in 10% FBS in DMEM. Twenty-four hours after plating, cells were infected with adenoviral constructs (10MOI, unless otherwise noted) for 2 hours in DMEM media. Adenoviral constructs expressing LacZ, Atg7, CryAB, CryABR120G and GFP-LC3 were employed. Post-infection cells were maintained in 2% FBS, 1% Penicillin/Streptomycin in high glucose DMEM until fixed or harvested.
Atg7 Adenovirus Construction
The mouse coding sequence for Atg7 (BC058597) was subcloned into a Blunt TOPO vector (Invitrogen) and a COOH-terminal FLAG tag added. The Flag-tagged Atg7 was then subcloned into the pCMV-Shuttle vector (Stratagene) for adenovirus synthesis. The adenovirus was expanded in HEK cells and CsCl purified prior to use.
siRNAKnockdown of Atg7
To specifically silence Atg7 expression, a pool of siRNAs (Invitrogen) was tested for their capacity to reduce Atg7 mRNA and protein levels in RNC transfections. The most potent silencing siRNA was used for all subsequent experiments. A non-specific siRNA was used for a negative control siRNA in all silencing experiments. Twenty-four hours after plating, cells were transfected with 100μmol/L siRNA, with Lipofectamine 2000 (Invitrogen), in OptiMem (Invitrogen) media overnight. In experiments using both siRNAs and adenoviruses, cells were transfected first, returned to growth media for 6 hours and then infected for 2 hours.
Autophagic Flux Assays
To measure autophagic synthesis versus degradation, LC3-II protein levels were quantified with and without lysosomal inhibition for each genetic condition. To inhibit lysosomal function, media containing 50 nmol/L Bafilomycin A1 (Sigma) was added to the cells for 4 hours. An equal volume of DMSO was used as a vehicle control. Ad GFP-LC3 (provided by J. Sadoshima, University of New Jersey Medical School) was used to quantify GFP-LC3 positive puncta under different experimental conditions as a reporter of autophagy levels. Fluorescent images were captured at 60X magnification on a BX60 microscope (Olympus), using NIS elements software (Nikon). The number of GFP-LC3 positive puncta per cell and treatment were quantified with Image J software (NIH). Images from 30–50 different cells were captured per well of a chamber slide and 4 different wells per genetic condition were quantified. GFP-LC3 analyses were not done on cells expressing CryABR120G because GFP-LC3 became trapped in the aggregate structures, preventing puncta-specific quantification.
Aggregate-Filter Trap Assays
To quantify changes in aggregate content RIPA-insoluble proteins were treated with DNAse (1mg/mL in 10mM Tris, 15mM MgCl2) (Roche) for 1 hour and protein quantitated with a modified Bradford assay. The insoluble protein was then diluted to 0.015μg/μL with 2% SDS, 20mM EDTA, 50mM DTT dissolved in TBS. Five micrograms of resuspended insoluble protein was dotted onto a nitrocellulose membrane (BioRad), which was blocked and immunoblotted with appropriate antibodies, as outlined above.
Statistical Analyses
Data are expressed as mean +/− standard error of the mean. All statistical tests were done with SigmaPlot 9.0 software. Comparisons between two groups were analyzed with Student’s t-test (P<0.05). Comparisons between multiple groups were analyzed with one-, two-, or three-way ANOVAs followed by Tukey’s post-hoc correction (P<0.05).
The authors had full access to and take full responsibility for the integrity of the data. The authors have read and agree to the manuscript as written.
RESULTS
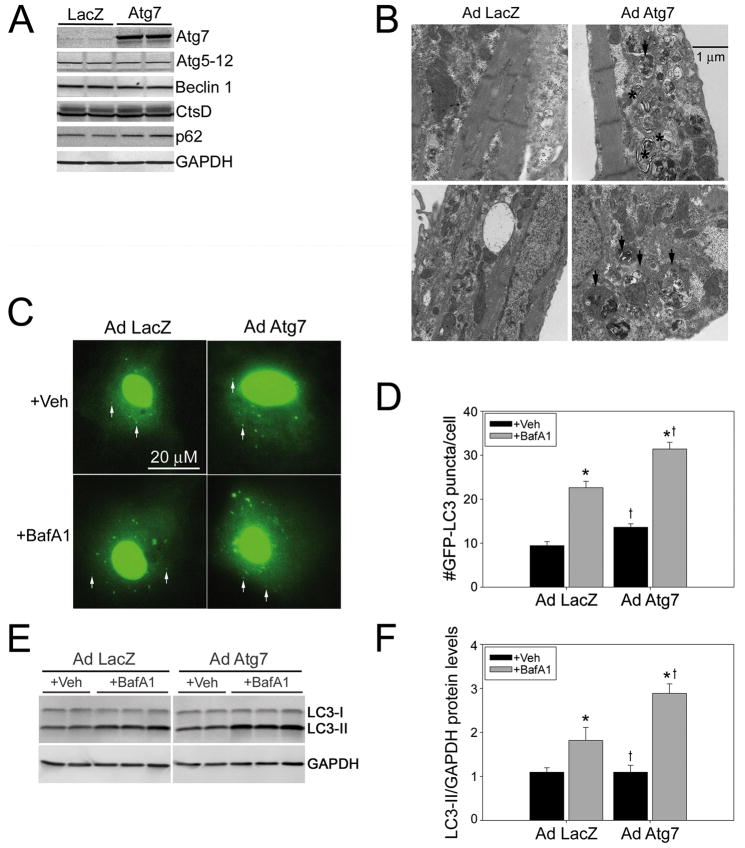
To test the hypothesis that increased levels of Atg7 are sufficient to induce autophagy, we generated an adenovirus in which the CMV promoter was used to drive Atg7 expression in RNCs (Figure 1). AdAtg7 induced Atg7 protein levels by 7.8-fold, while leaving other autophagic markers such as Beclin1, Atg5–12, p62 and cathepsin D unchanged (Figure 1A). These data suggest that any effects of Atg7 overexpression subsequently detected would not be due to changes in lysosomal content or the upregulation of Beclin1. Localization of Atg7 was examined by immunocytochemistry, which showed a diffuse cytoplasmic staining pattern with stronger perinuclear staining upon Atg7 overexpression (Supplementary Figure 1A). To determine whether Atg7 overexpression induces the intracellular structures characteristic of autophagy we evaluated the ultrastructure of the infected RNCs (Figure 1B) and observed a marked upregulation in the number of autophagic structures. Most notably there was a large number of amphisomes, an intermediate structure that develops after an autophagosome engulfs its cargo and fuses with other vesicular structures such as multi-vesicular endosomes, which have yet to fuse with and be degraded by lysosomes. In order to evaluate the extent of autophagy, autophagic flux assays were performed using a GFP-LC3 reporter. The active form of LC3 (LC3-II) localizes in autophagosomes, which appear as punctate structures, while inactive LC3 (LC3-I) stains diffusely in the cytoplasm. Increased levels of LC3-II could be due to either increased LC3 synthesis or impaired LC3 degradation and the current gold standard for evaluating autophagic function is the autophagic flux assay,20,21 which uses the lysosomal inhibitor Bafilomycin A1 (BafA1) to block lysosomal function and thus LC3 degradation. If LC3-II levels increase as a result of Atg7 expression, and in the presence of BafA1 relative to the LacZ controls, then the increase in LC3-II levels must be due to an increase in LC3 synthesis (increased autophagy). Conversely if gene expression with BafA1 blockade results in less LC3-II accumulation, the decrease would be due to impaired LC3 synthesis (decreased autophagy). Atg7 overexpression significantly increased the number of GFP-LC3 puncta/cell with Bafilomycin treatment over LacZ-expressing controls (Figure 1C and 1D).
Figure 1. Atg7 induces basal autophagy.
A, Representative immunoblots show a 7.8-fold increase in Atg7 protein levels in Ad Atg7 versus Ad LacZ, without induction of other autophagic proteins (n=4/treatment). B, Ultrastructural analyses confirmed an increase in autophagic structures with Atg7 expression. Amphisomes are denoted by black arrows. Multilamellar bodies are denoted by black asterisks. C, Autophagosomes are apparent as GFP-LC3-positive puncta (white arrows; n=4 wells/group). D, Autophagic flux assay shows a significant increase in GFP-LC3 puncta in Ad Atg7-infected cardiomyocytes in the presence of the lysosomal inhibitor BafA1. E, Autophagic flux assay shows increased LC3-II levels in representative immunoblots in Ad Atg7 versus Ad LacZ infected RNCs treated with BafA1 (n=6/treatment). F, LC3-II synthesis is significantly increased by Ad Atg7 expression with lysosomal inhibition. *P<0.05, significant difference Veh versus BafA1 treatment. †P<0.05, significant difference Ad Atg7 versus Ad LacZ infection.
Flux assays were performed on mock-infected versus AdLacZ-infected cells to show that the infection process does not alter autophagy (Supplementary Figure 1B). Alterations in autophagic flux were further confirmed by immunoblotting. AdAtg7 and AdLacZ-infected cells exhibit similar LC3-II levels under control (+Veh) conditions, while lysosomal inhibition with BafA1 differentially increased LC3-II levels 1.7-fold as compared to AdLacZ control-infected RNCs (Figure 1E, 1F). Thus Atg7 significantly induces autophagic flux (LC3-II synthesis) above control levels. Because these data were performed on cardiomyocytes grown in media with high glucose and 2% serum and not in starved cells, the data confirm that Atg7 overexpression is sufficient to induce basal autophagy and Atg7 induction is not limited to starvation-induced autophagy.
As autophagy has been associated with both beneficial and detrimental outcomes, we directly tested the hypothesis that Atg7-induced autophagy is not detrimental by carrying out cytotoxicity assays. At five days post-infection, Atg7 expression did not lead to any increased cytotoxicity as measured by both the adenylate kinase and lactate dehydrogenase release assays relative to LacZ expressing or mock infected controls (Supplementary Figure 1C).
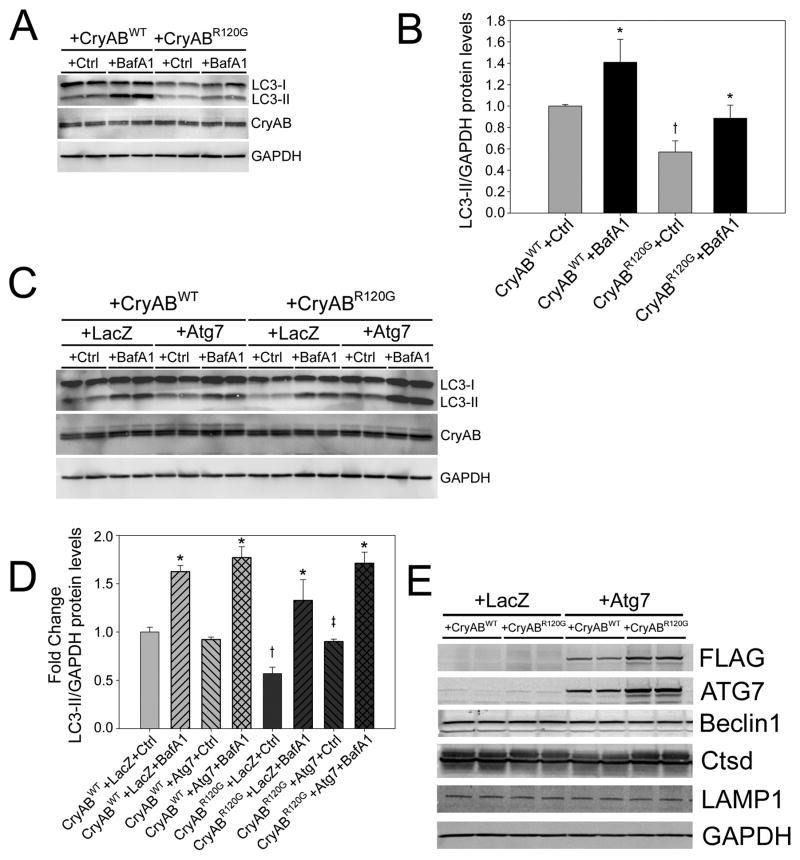
It has been hypothesized that models accumulating misfolded proteins and protein aggregates may have impaired autophagy.22–24 Previously, we studied a model of desmin-related cardiomyopathy, which occurred as result of cardiomyocyte-specific expression of a CryAB carrying a mutation causative for the disease.25–28 CryABR120G expression results in the accumulation of cytotoxic pre-amyloid oligomers (PAOs) and aggresomes, which cause cardiomyocyte toxicity in vitro and heart failure in vivo.27,28 To test the hypothesis that CryABR120G expression also results in impaired autophagy, we measured autophagic flux in RNCs transfected with adenoviruses overexpressing either wild-type CryAB (AdCryAB) or CryABR120G (AdCryABR120G). CryABR120G expression resulted in reduced LC3-II levels under basal and lysosome-inhibited conditions (Figure 2A). CryABR120G expression causes a significant reduction in LC3 synthesis (−37%) relative to CryABWT expressing cells (Figure 2B). Thus CryABR120G expression leads to decreased autophagic function. CryABWT flux did not differ from flux levels in mock-infected cells (Supplementary Figure 2A). Next, we tested the hypothesis that AdAtg7 overexpression could rescue the autophagic deficiency present in the CryABR10G expressing cardiomyocytes. Autophagic flux assays showed that enhanced levels of Atg7 rescued the autophagic deficiency observed in CryABR120G cardiomyocytes (Figure 2C and 2D). As with Atg7 expression alone, when Atg7 is co-expressed with CryABR120G no alterations in the protein levels of lysosomal markers or Beclin 1 were observed (Figure 2E). Induction of autophagy by Atg7 was further confirmed by EM analyses, which showed an increase in autophagic structures in AdAtg7 and AdCryABR120G co-expressing cardiomyocytes (Supplementary Figure 2B).
Figure 2. Atg7 rescues autophagic inhibition of CryABR120G.
A, Representative immunoblots of the autophagic flux assay shows reduced LC3-II levels with CryABR120G expression relative to CryABWT with and without lysosomal inhibitor BafA1 (n=4/treatment). B, LC3-II synthesis is significantly decreased with CryABR120G expression. *P<0.05, significant difference Veh versus BafA1. †P<0.05, significant difference CryABR120G versus CryABWT. C, A Representative immunoblots of the autophagic flux assay illustrating that Atg7 rescues the LC3-II deficiency of CryABR120G in the presence of BafA1 (n=6/treatment). D, LC3-II levels are elevated with CryABR120G and Atg7 co-expression. *P<0.05, significant difference between BafA1 versus Veh. †P<0.05, significant difference between LacZ+CryABR120G versus LacZ+CryABWT. ‡P<0.05, significant difference between Atg7+CryABR120G versus LacZ+CryABR120G. E, A representative immunoblot shows autophagic marker levels with CryABR120G and Atg7 co-expression (n=4/treatment).
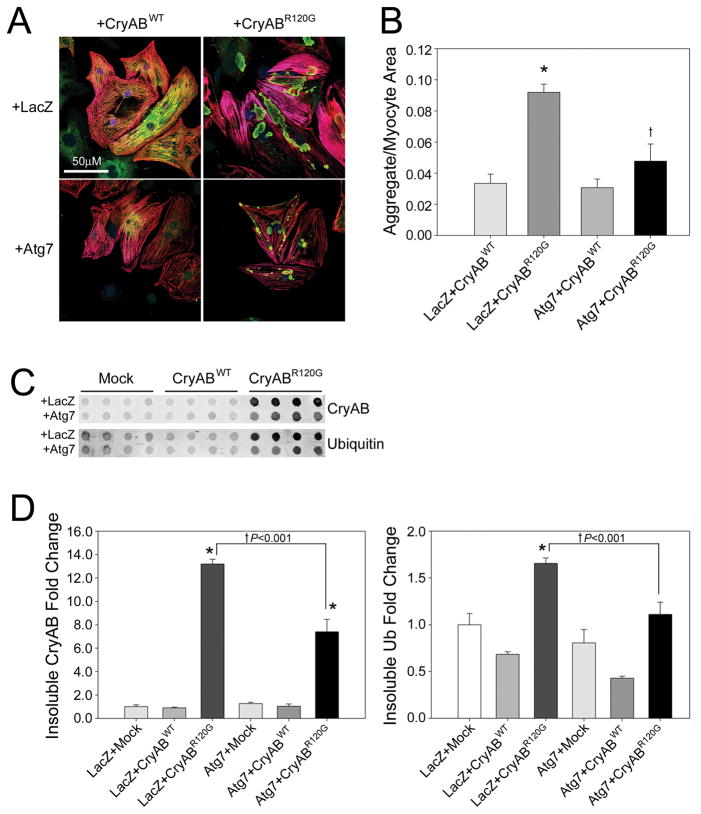
To test the hypothesis that aggregate accumulation was a result of decreased autophagy, we co-expressed CryABR120G with Atg7 to rescue autophagic function and determine if aggregate accumulation correspondingly decreased. Aggresomal accumulation was quantitated by CryAB staining (Figure 3A).25 Atg7 expression resulted in a significant reduction in aggregate content (Figure 3B). To quantitate aggregate content, we measured the area occupied by CryAB-positive aggregate staining (green) relative to TnI cardiomyocyte staining (red). We found a significant, 48% reduction in aggregate area when Atg7 was co-expressed with CryABR120G. A second method of quantifying the reduction in aggregate content was through use of filter trap assays (Figure 3C), with significant reductions of −44% and −36% in insoluble CryAB and ubiquitin, respectively when Atg7 was co-expressed with CryABR120G (Figure 3D).
Figure 3. Atg7 expression reduces CryABR120G aggregate content.
A, Immunofluorescent staining shows decreased CryAB (green) aggregate content when Atg7 is co-expressed with CryABR120G. Cardiomyocytes counterstained by a TnI antibody (red). B, Aggregate area relative to the cardiomyocyte area (n=4/treatment). *P<0.05, significant difference between CryABR120G versus CryABWT. †P<0.05, significant difference CryABR120G+Atg7 versus CryABR120G+LacZ. C, The filter trap assay shows reduced insoluble CryAB and Ubiquitin with CryABR120G and Atg7 co-expression. D, Densitometry for insoluble CryAB and Ubiquitin (n=4/treatment). *P<0.05, significant difference between CryABR120G versus CryABWT. †P<0.001, significant difference between CryABR120G+Atg7 versus CryABR120G+LacZ.
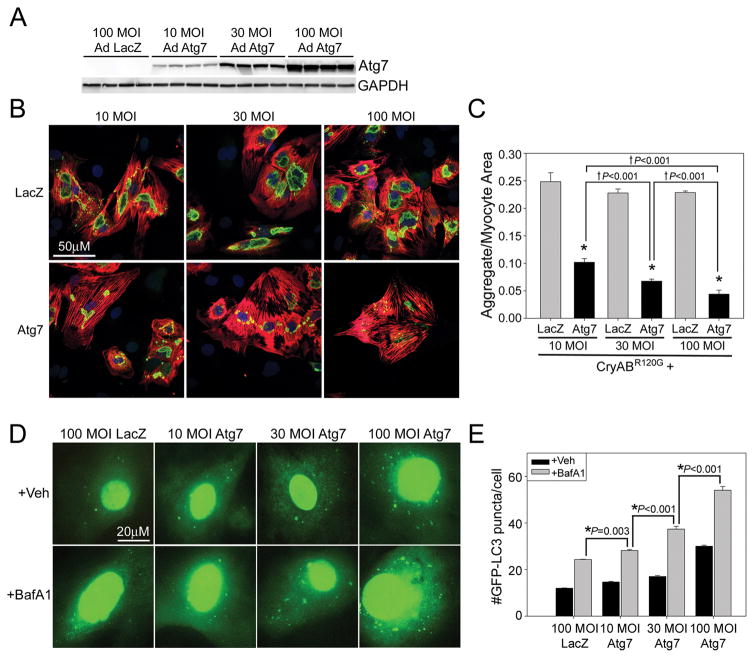
To better define the effective dose of Atg7 we tested the effects of increasing MOI of AdAtg7 relative to AdLacZ. Increasing MOI of AdAtg7 results in a dose-dependent increase in Atg7 protein levels (Figure 4A). Increasing MOI of Atg7 has no effect on cytotoxicity (Supplementary Figure 4). Increasing the MOI of AdAtg7 with a constant MOI of AdCryABR120G resulted in a dose-dependent reduction in CryABR120G aggregate content (Figure 4B and 4C). To determine whether increasing doses of AdAtg7 resulted in increasing levels of autophagy, autophagic flux assays with GFP-LC3 were performed (Figure 4D). Autophagic flux was significantly increased in a dose-dependent manner from 100 MOI AdLacZ to 10, 30, and 100MOI of AdAtg7 (Figure 4E).
Figure 4. Increasing Atg7 expression causes a dose-dependent reduction in aggregate content.
A, RNC’s were infected with 100 MOI AdLacZ, or 10, 30, or 100 MOI AdAtg7 to show that Atg7 protein levels increase in a dose-dependent manner (n=4/group). B, RNCs were co-infected with 10 MOI of AdCryABR120G and 10, 30, or 100 MOI of AdLacZ or AdAtg7, respectively. Five days post-infection cells were fixed and immunostained with CryAB (green, aggregates), TnI (red, cardiomyocytes) and TO-PRO-3 (blue, nuclei). C, Aggregate area relative to the cardiomyocyte area (n=8/treatment). *P<0.001, significant difference between Atg7 versus LacZ. †P<0.1, significant difference between 10 versus 30 versus 100 MOI of Atg7. D, Autophagic flux was evaluated by co-infecting Ad GFP-LC3 with Ad LacZ (100MOI) or Ad Atg7 (10, 30 or 100 MOI) in the presence or absence of BafA1. E, Autophagic flux is significantly increased in a dose-dependent manner with increasing MOI of AdAtg7 (n=4 wells/group). *P<0.001, significant difference between BafA1 treated versus BafA1 treated with additional Atg7.
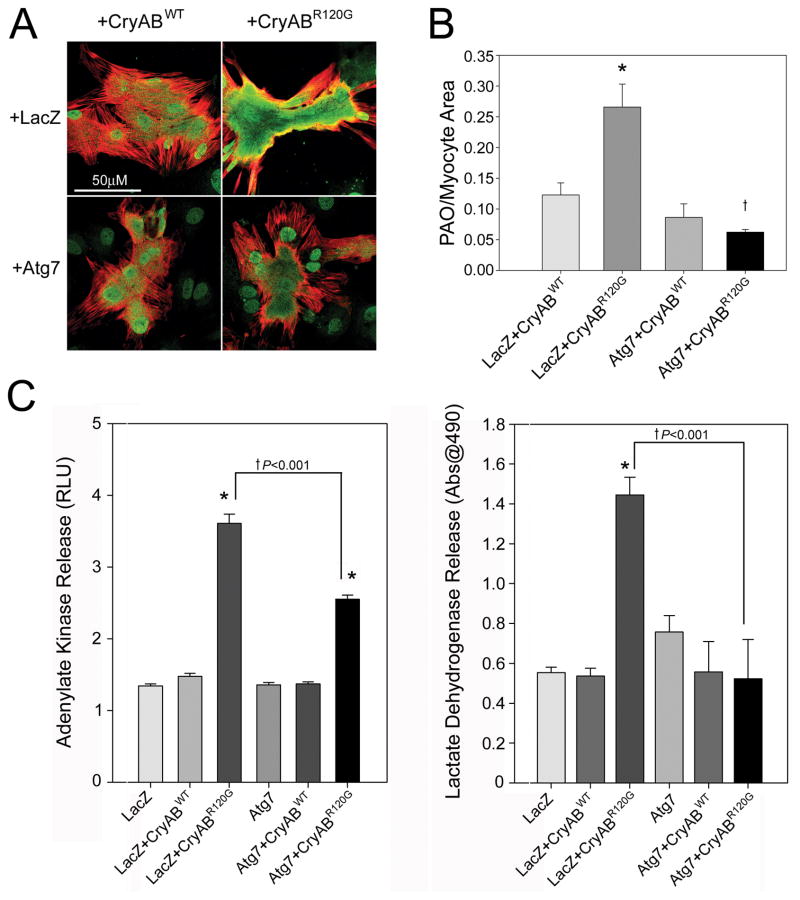
Previous data from the CryABR120G model showed that PAO accumulation is associated with cytotoxicity and heart failure.25,27,28 To test the hypothesis that Atg7 induction of autophagy could reduce PAO content we immunostained transfected RNCs with a conformation-specific antibody29–31 that uniquely stains PAOs and, under these culture conditions, nuclei (Figure 5A). The non-nuclear staining represents toxic oligomer accumulation.25,27,28 We observed a significant, 4.2-fold reduction in non-nuclear PAO staining when AdAtg7 was co-expressed with AdCryABR120G (Figure 5B). To quantify non-nuclear PAO reduction, the area of cytoplasmic PAO staining (green) was measured relative to cardiomyocyte area as visualized using TnI staining (red). Consistent with PAO’s cytotoxicity, we observed that Atg7 expression also significantly reduced CryABR120G cytotoxicity as measured by adenylate kinase and lactate dehydrogenase release assays (Figure 5C).
Figure 5. Atg7 reduces CryABR120G PAO content and cytotoxicity.
A, Immunofluorescent staining shows decreased pre-amyloid oligomer PAO (green) content when Atg7 is co-expressed with CryABR120G. Cardiomyocytes counterstained by a TnI antibody (red). B, PAO area relative to the cardiomyocyte area (n=4 wells/treatment). *P<0.001, significant difference between CryABR120G versus CryABWT. †P<0.05, significant difference between CryABR120G+Atg7 versus CryABR120G+LacZ (P<0.001). C, CryABR120G adenylate kinase and lactate dehydrogenase release are reduced by Atg7 co-expression at 5 days post-infection (n=4/treatment). *P<0.001, significant difference between CryABR120G versus CryABWT. †P<0.001, significant difference between CryABR120G+Atg7 versus CryABR120G+LacZ.
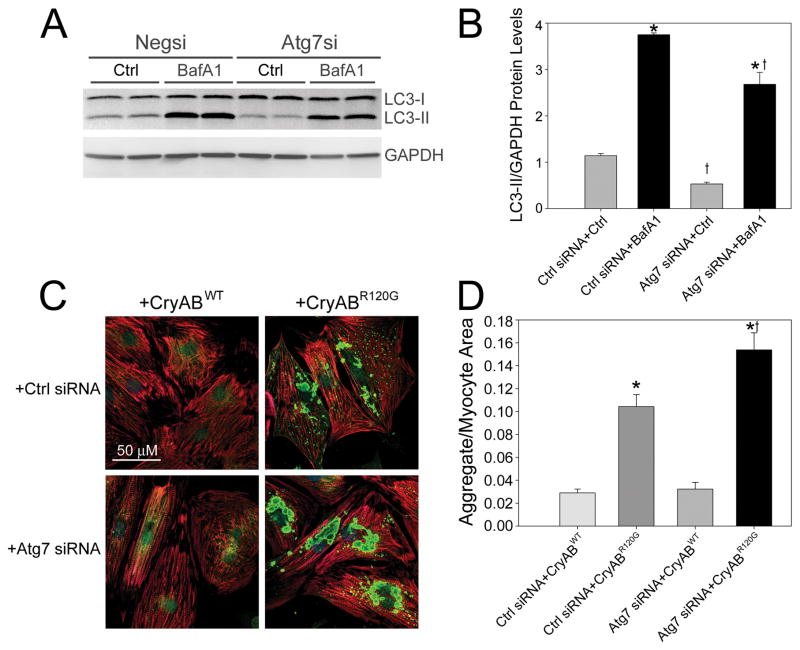
Silencing Atg7 can reduce autophagy.12,32 However, the question of whether autophagic deficiency leads directly to increased aggregate content is relatively unexplored. To test this hypothesis we used an Atg7 silencing siRNA (Supplementary Figure 3A-C) with AdCryABR120G-infected RNCs to see if aggregate content increased as a result of autophagic silencing. To confirm that Atg7 siRNA successfully inhibited autophagy we measured autophagic flux (Figure 6A). The data showed that, much like CryABR120G expression, decreased Atg7 levels significantly impaired LC3 synthesis (Figure 6B). Silencing Atg7 expression while overexpressing CryABR120G resulted in a marked 47% increase in aggregate content per cardiomyocyte (Figure 6C and 6D).
Figure 6. Atg7 silencing inhibits autophagy and increases aggregate content of CryABR120G.
A, Representative blot showing autophagic flux assay with reduced LC3-II levels from Atg7 knockdown with BafA1 (n=4/treatment). B, Decreased LC3-II protein levels with Atg7 siRNA. *P<0.05, significant difference between Veh versus BafA1. †P<0.05, significant difference Atg7 siRNA versus Ctrl siRNA. C, Increased CryAB aggregate content (green) when Atg7 siRNA is coupled with CryABR120G. Cardiomyocytes are counterstained with a TnI antibody (red). D, Aggregate area relative to the cardiomyocyte area (n=4/treatment) *P<0.001, significant difference CryABWT versus CryABR120G. †P<0.00 1, significant difference CryABR120G+Atg7 siRNA versus CryABR120G+Ctrl siRNA.
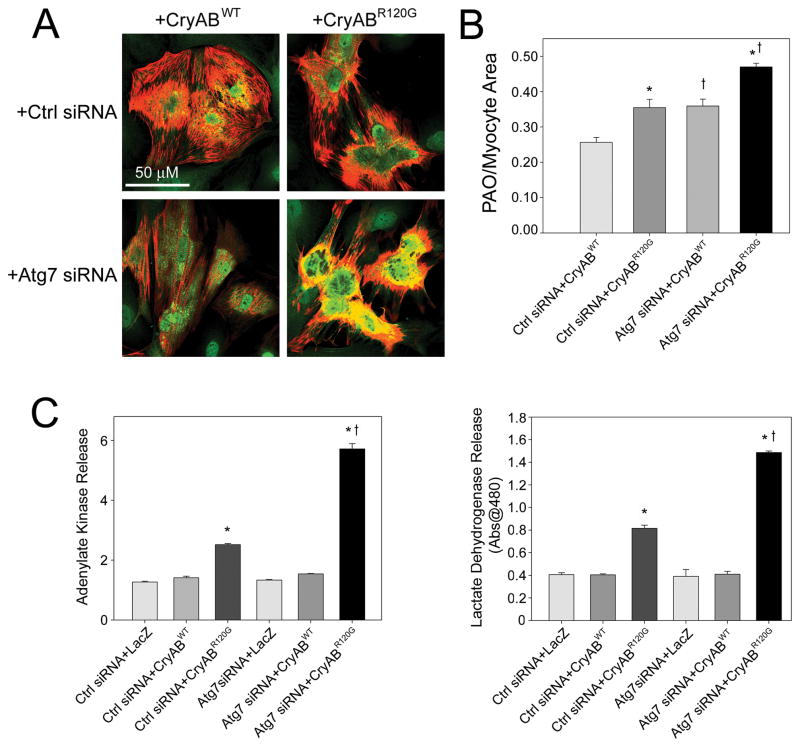
Impaired autophagic function has not previously been associated with PAO content. We therefore tested whether Atg7 silencing with CryABR120G expression could increase PAO content in cardiomyocytes. Immunocytochemistry showed that knockdown of Atg7 exacerbated the degree of PAO staining in CryABR120G expressing cardiomyocytes (Figure 7A). By quantifying PAO area (green) relative to cardiomyocyte are (red), we found that Atg7 silencing significantly increased PAO/cardiomyocyte area (Figure 7B). Atg7 siRNA doubled the magnitude increase in PAO staining between CryABWT and CryABR120G. Similarly, knocking down Atg7 in CryABR120G expressing cardiomyocytes resulted in increased cytotoxicity as measured by adenylate kinase and lactate dehydrogenase release assays (Figure 7C and 7D).
Figure 7. Atg7 siRNA increases CryABR120G PAO content and cytotoxicity.
A, PAO content (green) is increased when Atg7 siRNA is co-expressed with CryABR120G. Cardiomyocytes are counterstained with a TnI antibody (red). B, PAO area relative to the cardiomyocyte area (n=4/treatment). *P<0.05, significant difference CryABWT versus CryABR120G. †P<0.05, significant difference between Atg7 siRNA versus Ctrl siRNA. C, CryABR120G adenylate kinase and lactate dehydrogenase release are increased by Atg7 siRNA co-expression at 5 days post-infection (n=4/treatment). *P<0.001, significant difference between CryABR120G versus CryABWT. †P<0.001, significant difference between CryABR120G+Atg7 siRNA versus CryABR120G+Ctrl siRNA.
Discussion
Our data demonstrate that enhanced levels of Atg7 are sufficient to induce autophagy. Furthermore, Atg7’s ability to impact on autophagy did not require preconditioning by cell starvation, meaning that Atg7 can, unlike Beclin1, increase basal autophagy, not just starvation-induced autophagy.14 The capacity to induce autophagy without starvation makes Atg7 a unique tool for autophagic studies. Numerous studies have concluded that elevated autophagy is detrimental, after observing an increase in autophagic markers in association with a deleterious context.14,33 Our work demonstrates that Atg7 overexpression clearly increases autophagic flux (autophagic function) without increasing cytotoxicity. In fact when Atg7 was co-expressed with cytotoxic CryABR120G expression it reduced overall cytotoxicity. These data show that autophagy upregulation is not universally detrimental but can in fact be beneficial. Whether Atg7 induction represents a widely used pathway for beneficial autophagy remains to be determined.
Because of the necessary roles of Atg7 in processing LC3-I to active LC3-II and in the conjugation of Atg5 to Atg12, we hypothesized that upregulating what might be a rate limiting enzyme, Atg7, would result in more available LC3-II, and increase autophagosomal content. However, we observed no depletion in LC3-I with Atg7 overexpression, suggesting that Atg7 upregulates the process of autophagy overall rather than simply impacting a single, rate-limiting step.
Increasing evidence suggests that misfolded proteins, PAOs and aggregates can all cause or functionally participate in the development of cardiotoxicity and heart failure,27,28,34,35 and pharmacological inhibition of autophagy has been associated with CryABR120G aggregate accumulation.36 While our lab has studied the CryABR120G model extensively, we have not defined a single, that is, necessary and sufficient mechanism for PAOs and aggregate accumulation.25,26,34,37 The autophagic flux assays for CryABR120G expressing cardiomyocytes clearly demonstrate that CryABR120G expression causes an autophagic deficiency. Atg7 silencing causes a similar autophagic deficiency, such that when we combined the autophagic deficiency of CryABR120G with Atg7 knockdown, we observed increased accumulation of PAOs and aggregates. Additional PAO and aggregate accumulation led to increased cytotoxicity. These data show that conditions of autophagic deficiency exacerbate the accumulation of misfolded proteins and aggregates, contributing to the proteotoxicity phenotype in the cardiomyocyte.
If autophagic insufficiency plays a role in the CryABR120G phenotype of PAO and aggregate accumulation, then overexpression of Atg7 to induce autophagy should attenuate the pathology. Indeed, Atg7 expression decreased CryABR120G aggregate and PAO content in cardiomyocytes. Consistent with the hypothesis that impaired autophagy with PAO and aggregate accumulation induces a proteotoxic stress, Atg7 stimulation of autophagy also reduced CryABR120G-induced cardiomyocyte toxicity. Additionally, increasing levels of Atg7 expression led to a dose-dependent reduction in CryABR120G aggregate content, suggesting that further elevating autophagic function increased clearance misfolded protein structures.
While it is logical to think that upregulation of autophagy, which functions to degrade misfolded proteins and aggregates, enhances their clearance, few if any models have shown the sufficiency of autophagy for this process.22 Our data clearly show that inducing autophagy by Atg7 overexpression is sufficient to reduce CryABR120G PAO and aggregate content, with a concomitant decrease in cytotoxicity. Thus Atg7 rescues the autophagic deficiency caused by CryABR120G expression. Future studies will be aimed at testing the effects of Atg7 overexpression in vivo and its capacity to prevent aggregate formation and heart disease as well as its potential for rescuing pre-existing disease.
Our understanding of the role of autophagy is still in its infancy. It is currently unclear how prevalent autophagic deficiency is in models of heart disease or neurodegeneration, as most publications report associative studies with autophagic markers and disease. Several genetic models of heart disease, including Tg-Mst1, truncated MYBPC3, DesΔ7, PQ83 and CryABR120G, result in the accumulation of intracellular aggregates.27,28,34,38,39 However, recent evidence suggests that autophagic induction may not be generally effective for aggregate diseases as, in an expanded polyglutamine model of aggregate formation, autophagosomes failed to recognize and engulf the polyglutamine aggregates.40 It remains to be seen whether upregulation of autophagy is beneficial in other models of protein aggregation, PAO accumulation and heart disease. However, if other aggregate-inducing models of heart failure are due to insufficient autophagy, inducing autophagy may provide a mechanistic rescue for those models of heart disease.
A limitation in many studies of autophagy is the exclusive reliance on measuring levels of autophagic markers such as LC3 either by blotting or by puncta accumulation. Upregulation of LC3 levels can be observed under both conditions of increased autophagic synthesis and decreased lysosomal function.20 Actual, autophagic function, determined with autophagic flux assays, which compares LC3 synthesis versus degradation through use of a lysosomal inhibitor,21 is relatively rare in the literature. As assays of autophagic function come into widespread use, the relationship between autophagic function and disease will be better defined.
One of the biggest hurdles to our understanding of the processes’ implications is the lack of genetic tools by which autophagy can be regulated. Our data describe a new genetic target for the upregulation of autophagy, Atg7. Additional genetic reagents will also need to be developed to better define the pathways that induce and repress autophagic function in order to more fully understand autophagy’s role in cardiac disease.
Supplementary Material
Novelty and Significance.
What is known?
Atg7 is necessary for autophagic function.
Impaired autophagy results in aggregate accumulation.
Mutant αB-crystallin expression causes the accumulation of misfolded pre-amyloid oligomers and aggregates.
What New Information Does This Article Contribute?
Atg7 expression is sufficient to induce autophagic function in cardiac myocytes without nutrient deprivation.
CryABR120G expression inhibits autophagy.
Atg7-induced autophagy rescues the impaired autophagy of CryABR120G expression, as evidenced by dose-dependent reduction in aggregate and pre-amyloid oligomer content per cardiomyocyte, demonstrating that autophagy upregulation can be beneficial for proteotoxic disease.
Only a few genes have been demonstrated to regulate autophagy. Here, we show that Atg7 expression can upregulate autophagic flux in cardiac myocytes. A model of desmin-related myopathy, brought about through expression of a mutated αB-crystallin (CryABR120G) that causes the human disease, forms protein aggregates and pre-amyloid oligomers in cardiomyocytes. We found that CryABR120G expression resulted in impaired autophagy, which may contribute to the accumulation of aggregates and pre-amyloid oligomers, characteristic of desmin-related myopathy. To test whether inducing autophagy could reduce CryABR120G pathology we co-expressed Atg7 with CryABR120G, which resulted in reduced aggregate and pre-amyloid oligomer content per cardiomyocyte with a corresponding reduction in cytotoxicity. Conversely, silencing Atg7 inhibited autophagy, which exacerbated the aggregate and pre-amyloid accumulation with CryABR120G expression. Our findings suggest a novel role for Atg7 in inducing a beneficial form of autophagy, which may be a potential therapeutic avenue for the treatment of desminopathy pathology.
Acknowledgments
Sources of Funding
This work was supported by NIH grants P01HL69799, P50HL074728, P50HL077101, P01HL059408, and R01HL087862 (to J.R.). National Institutes of Health fellowship awards T32 HL07752 and F32 HL087478; and an American Heart Association Postdoctoral Fellowship (to JSP).
Non-standard abbreviations
- PAO
pre-amyloid oligomer
- CryABR120G
the αB-crystallin R120G mutation
- BafA1
Bafilomycin A1
Footnotes
Disclosures
None.
References
- 1.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 2.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 3.Roberts EA, Deretic V. Autophagic proteolysis of long-lived proteins in nonliver cells. Methods Mol Biol. 2008;445:111–117. doi: 10.1007/978-1-59745-157-4_6. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 6.Goswami SK, Das DK. Autophagy in the myocardium: Dying for survival? Exp Clin Cardiol. 2006;11:183–188. [PMC free article] [PubMed] [Google Scholar]
- 7.Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- 8.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circ Res. 2008;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell Death Differ. 2009;16:31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- 10.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 12.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arsov I, Li X, Matthews G, Coradin J, Hartmann B, Simon AK, Sealfon SC, Yue Z. BAC-mediated transgenic expression of fluorescent autophagic protein Beclin 1 reveals a role for Beclin 1 in lymphocyte development. Cell Death Differ. 2008;15:1385–1395. doi: 10.1038/cdd.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circ Res. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komatsu M, Tanida I, Ueno T, Ohsumi M, Ohsumi Y, Kominami E. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1–E2 complex formation. J Biol Chem. 2001;276:9846–9854. doi: 10.1074/jbc.M007737200. [DOI] [PubMed] [Google Scholar]
- 18.Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, Klionsky DJ. In search of an “autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 22.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, Rubinsztein DC. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 23.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 24.Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, Rubinsztein DC. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 25.Maloyan A, Gulick J, Glabe CG, Kayed R, Robbins J. Exercise reverses preamyloid oligomer and prolongs survival in alphaB-crystallin-based desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2007;104:5995–6000. doi: 10.1073/pnas.0609202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maloyan A, Sanbe A, Osinska H, Westfall M, Robinson D, Imahashi K, Murphy E, Robbins J. Mitochondrial dysfunction and apoptosis underlie the pathogenic process in alpha-B-crystallin desmin-related cardiomyopathy. Circulation. 2005;112:3451–3461. doi: 10.1161/CIRCULATIONAHA.105.572552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, Robbins J. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanbe A, Osinska H, Villa C, Gulick J, Klevitsky R, Glabe CG, Kayed R, Robbins J. Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2005;102:13592–13597. doi: 10.1073/pnas.0503324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glabe CG, Kayed R. Common structure and toxic function of amyloid oligomers implies a common mechanism of pathogenesis. Neurology. 2006;66:S74–78. doi: 10.1212/01.wnl.0000192103.24796.42. [DOI] [PubMed] [Google Scholar]
- 30.Kayed R, Glabe CG. Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol. 2006;413:326–344. doi: 10.1016/S0076-6879(06)13017-7. [DOI] [PubMed] [Google Scholar]
- 31.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 32.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 33.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 34.Pattison JS, Sanbe A, Maloyan A, Osinska H, Klevitsky R, Robbins J. Cardiomyocyte expression of a polyglutamine preamyloid oligomer causes heart failure. Circulation. 2008;117:2743–2751. doi: 10.1161/CIRCULATIONAHA.107.750232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ, Jr, Rothermel BA, Hill JA. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B, Rothermel BA, Hill JA. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maloyan A, Sayegh J, Osinska H, Chua BH, Robbins J. Manipulation of death pathways in desmin-related cardiomyopathy. Circ Res. 106:1524–1532. doi: 10.1161/CIRCRESAHA.109.212639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Osinska H, Dorn GW, 2nd, Nieman M, Lorenz JN, Gerdes AM, Witt S, Kimball T, Gulick J, Robbins J. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 39.Sarikas A, Carrier L, Schenke C, Doll D, Flavigny J, Lindenberg KS, Eschenhagen T, Zolk O. Impairment of the ubiquitin-proteasome system by truncated cardiac myosin binding protein C mutants. Cardiovasc Res. 2005;66:33–44. doi: 10.1016/j.cardiores.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.