Abstract
Aspergillus niger FETL FT3, a local extracellular tannase producer strain that was isolated from one of dumping sites of tannin-rich barks of Rhizophora apiculata in Perak, Malaysia. This fungus was cultivated in 250 mL Erlenmeyer flask under submerged fermentation system. Various physical parameters were studied in order to maximize the tannase production. Maximal yield of tannase production, that is, 2.81 U per mL was obtained on the fourth day of cultivation when the submerged fermentation was carried out using liquid Czapek-Dox medium containing (percent; weight per volume) 0.25% NaNO3, 0.1% KH2PO4, 0.05% MgSO4 ·7H2O, 0.05% KCl, and 1.0% tannic acid. The physical parameters used initial medium pH of 6.0, incubation temperature of 30°C, agitation speed of 200 rpm and inoculums size of 6 × 106 spores/ ml. This research has showed that physical parameters were influenced the tannase production by the fungus with 156.4 percent increment.
1. Introduction
Tannin acyl hydrolase (E.C.3.11.20) or commonly known as tannase is an inducible enzyme that catalyzes the hydrolysis of ester and depside bonds in hydrolysable tannin to produce glucose and gallic acid [1]. Tannase has wide applications especially in food, beverage, and pharmaceutical industries [2, 3]. It involves in the manufacturing of instant tea, coffee flavored, soft drinks, beer and wine where tannase is used to eliminate the water soluble precipitates [4–6]. Tannase is also used in debittering of fruit juices [7] and in improving vegetable fermentations [8]. Another important application of tannase is the production of gallic acid and propyl gallate [9, 10]. Gallic acid is used in the pharmaceutical industry for the synthesis of antibacterial drugs and also in the food industry as a substrate for the chemical synthesis of food preservatives, whereas propyl gallate is an important food additive to prevent food oxidation.
Tannase is produced by a number of microorganisms and microbial. Tannase finds immense application in various industries due to its high stability and availability. Among the microbial tannase producers, filamentous fungi like Aspergillus [11–13], Rhizophs [14], Paecilomyces [15], and Penicillium [16] were studied extensively. Besides fungi, yeasts [17] and bacteria [18–22] have also been reported to produce tannase, but yeast was not extensively studied.
In our study, we are reporting the production of tannase by Aspergillus niger FETL FT3 which we had successfully isolated from the dumping site of Rhizophora apiculata barks at the mangrove area in Perak, Malaysia, This fungal strain has an ability to produce extracellular tannase significantly. Therefore, in order to increase the tannase production by this fungus in submerged fermentation system, improvement through physical parameter has to be carried out in this research.
2. Materials and Methods
2.1. Microorganism, Culture Maintenance and Inoculums Preparation
Aspergillus niger FETL FT3 were isolated from the dumping sites of tannin-rich barks of Rhizophora apiculata in Perak, Malaysia was used throughout this study. The fungal culture was maintained on 2 percent (weight per volume) malt extract agar slant supplemented with 0.01 percent (weight per volume) tannic acid at 30°C for 5 days aerobically until sporulate, before storing them at 4°C until further used. The subculturing was performed every three weeks to assure its viability.
The inoculum was prepared by adding 5.0 mL of sterile distilled water to the agar slants and shaking vigorously. The spore suspension that obtained was adjusted to 1 × 107 spores per mL using a haemocytometer slide chamber (Neubauer, Germany) and used as the inoculums.
2.2. Cultivation Method for Enzyme Production
The fermentation was carried out in a 250 mL Erlenmeyer flask containing 50 mL of Czapek-Dox liquid medium [23] containing (percent; weight per volume) 0.25% NaNO3, 0.1% KH2PO4, 0.05% MgSo4·7H2O, 0.05% KCl, and 1.0% tannic acid (as a sole carbon source). The initial experiment was conducted in order to determine the optimal incubation or cultivation time that produced highest tannase production. Therefore, an experiment using Czapek-Dox medium (pH 5) with the addition of 1 percent (volume per volume; 1 × 107 spores per mL) inoculum, agitated at 200 rpm, and incubated at 30°C was carried-out.
2.3. Improvement of Cultural Conditions
Various optimal physical parameters required for maximal tannase production by Aspergillus niger FETL FT3 in a shake flask system were studied. These included various initial medium pH (4, 5, 6, 7, and 8), temperature (25, 30, 35, 40, 45, and 50°C), agitation speed (0, 50, 100, 150, 200, and 250 rpm) and inoculums sizes (1 × 102, 1 × 103, 1 × 104, 1 × 105, 1 × 106, 1 × 107, and 1 × 108). All experiments were carriedout in triplicate and the values were reported as standard deviations
2.4. Enzyme Extraction
The cultivation medium was filtered through GF/A glass fiber filter (Whatman) to separate supernatant from mycelial mass. The cell-free culture filtrate was then filtered again using a membrane filter with a pore size of 0.22 μm (Millipore) and was regards as a source of tannase.
2.5. Tannase Activity Determination
Extracellular tannase activity was determined according to method described by Deschamps et al. [1]. 1 mL of the culture filtrate was incubated with 1 mL of tannic acid solution (1% w/v tannic acid in 0.2 M acetate buffer, pH 5.0) at 40°C for 30 min in a water bath. The enzymatic reaction was then stopped by adding 4 mL of standard protein solution (1.0 mg/mL bovine serum albumin at room temperature (30 ± 2°C). The gallic acid produced was not precipitated by the protein solution in this method due to its low molecular weight. A reference tube with heat inactivated enzyme was treated in the same manner except that the tannic acid was precipitated immediately without incubation. The tubes were then centrifuged at 3000 rpm (Chermle Z 320) for 20 min and the absorbance of 1 : 80-fold diluted supernatant (containing liberated gallic acid by action of tannase on tannic acid) against distilled water as blank in a UV spectrophotometer (UV-120-01 Shimadzu, Japan). The amount of gallic acid produced in the reaction mixture was then estimated from a reference curve standardized with commercial gallic acid (Fluka AG) with concentration ranging from 0 to 30 μmol/mL. Enzyme activity as expressed in U per mL and one unit of tannase was defined as the amount of enzyme required to release 1.0 μmol of gallic acid per minute per mL of fermentation broth under standard assay conditions.
2.6. Growth Determination
Growth was determined based on the dry weight of the fungal mycelia after filtration using GF/A glass fiber filter, followed by drying at 65°C until constant weight obtained.
3. Results and Discussion
Studies on the improvement of incubation time required for the production of tannase under submerged fermentation conditions by the newly isolated Aspergillus niger FETL FT3 was carried out in Czapek Dox medium containing 1 percent (weight per volume) tannic acid, whereby the growth, production of tannase, and pH of the medium were monitored at an interval of 24 h for 14 days. Before the improvement of physical parameters was carried out, the study on the incubation time was performed first. It was necessary to determine the optimal day of cultivation which produced maximal tannase. Therefore the particular incubation time can be used to improve the other parameters involved in this study. Figure 1 shows the results obtained for the incubation time that was carried-out for 14 days. The results showed that the maximal tannase production of 1.096 U per mL was achieved on day 4 of cultivation time. After that, the amount of tannase decreased considerably with further increase in the cultivation time. Rajakumar and Nandy [16] and Gautam et al. [24] have indicated that the reason for the initial increase in tannase activity and followed by a decrease may be due to the inhibition or denaturation of the enzyme, while Sabu et al. [25] and Rodrigues et al. [26] suggested that reduced nutrient level of medium affected the enzyme synthesis. Besides the possibility of substrate limitation, Kar et al. [27] pointed out that the initial rise followed by a decrease in tannase was also possibly due to the secretion of toxic substances which could cause cell lysis. However, Deschamps et al. [1] and Bradoo et al. [28] suggested that the decline in enzyme production may be due to gallic acid production as the present enzyme shows strict end-product repression. In addition, to obtain maximum extracellular tannase on 4th day of cultivation, the fungus was also observed to attain its highest growth on the 6th day of cultivation. This indicated that tannase formation by this fungus was not directly growth related as the highest fungal biomass was achieved approximately 2 days later than tannase production. The results also showed that tannase was produced during a primary phase of growth. Similarly, Deschamps et al. [1] also observed that maximal tannase production was produced by bacteria at their active phase of growth. Additionally, the production of tannase during the primary phase of growth has also been reported in Penicillium chrysogenum [16], Aspergillus aculeatus [13, 29], Aspergillus flavus [30], and Bacillus licheniformis [31, 32]. Furthermore, a rapid breakdown of tannin allows cell multiplication by the assimilation of the degradation products [1], presumably glucose being metabolized faster than gallic acid [31, 33]. However, the fungal growth showed that the biomass did not increase continuously but experienced a deceleration of growth on the 6th day of cultivation, followed by a growth cessation. The reason for this may be due to limited supply of the degradation products to the fungus (i.e., glucose and gallic acid) at time point onwards. During the enzyme production and fungal growth, the pH of the medium was also observed to decrease sharply (became more acidic) with a lowest pH of 3.97 attained on the 5th day of cultivation and then increased slowly. Mondal and Pati [31] indicated that this may be due to the assimilation of glucose and the accumulation of gallic acid in the medium at the early stage of fungal growth and subsequent utilization of gallic acid in the latter stage of its growth. Consequently, the stability of tannic acid may have been affected by these changes in pH values during cultivation [26], whereby pH value was low during cultivation is resulted in decreased tannase synthesis. Besides, enzymes are also very sensitive to change in pH of medium, as reported by Ramirez-Coronel et al. [12], that tannase activity from Aspergillus niger was stable only at pH values between 3.5 and 7.0.
Figure 1.
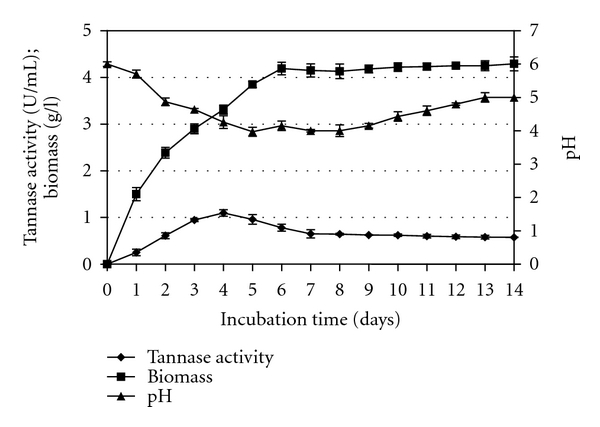
Effect of incubation period on tannase production by Aspergillus niger FETL FT3 in a shake-flask system.
An uninoculated reference flask (data not shown) was also used to measure any spontaneous tannin hydrolysis and showed only slight degradation of about 5% hydrolysis devoid of tannase activity after 14 days of incubation. This indicated that hydrolysis of tannic acid observed in the above experiment was purely due to the extracellular tannase produced by the fungus. Therefore, based on the results obtained, incubation time of 4 days was used to improve other physical parameters.
3.1. Effect of Incubation Temperature on Tannase Production
Temperature is one of the factors that affect various cell metabolic processes and among them are protein denaturation, enzymatic inhibition, promotion or inhibition on the production of a particular metabolite and also cell death. Therefore, the effect of incubation temperature on tannase production needs to be studied.
Figure 2 shows that the maximal tannase production of 1.121 U per mL was achieved at the incubation temperature of 30°C, where the fungal growth also achieved its maximal level. However, the amount of tannase production and also the fungal growth were decreased considerably at higher temperatures above 30°C. Lekha and Lonsane [10] reported that temperature between 30–35°C was optimal for most of the fungal strains to produce maximal tannase activity, whereby Barthomeuf et al. [11] reported 33°C as optimal for tannase production by Aspergillus niger LCF 8, while Yamada et al. [30] and Rajakumar and Nandy [16] reported an optimal temperature of 30°C and 28°C for tannase production by Aspergillus flavus and Penicillium chrysogenum, respectively. Similarly, Bradoo et al. [28] reported that Aspergillus japonicus produced maximal tannase at 30°C. Obviously, the optimal temperature for most of the fungal tannase production is near 30°C and thus is comparable with this study. However, Frost and Moss [34] specified that the optimal temperature for a synthesis of any particular enzyme may differ from the optimal temperature for growth, suggesting that the optimal temperature for maximal enzyme production and growth recorded at 30°C in this study was merely a coincidence. The rapid growth of the fungal culture observed at temperatures between 30 and 35°C also suggested that the fungus is mesophilic, and this could be explained by the fact that this fungus was isolated from a mangrove environment. Therefore, temperatures of 40°C and above did not enhance its growth, as the growth of the microorganism may have adversely been affected by heat [23], while temperatures below 30°C may have resulted in freezing of the protoplasmic membrane which causes inactivation of solute transport systems in the cells [35]. Another possible reason for initial rise in tannase activity followed by a decrease, thereafter, is because of the atoms in the enzyme molecule that have greater energy and a greater tendency to move with increased temperature and after certain temperature they acquire sufficient energy to overcome the weak interactions holding the globular protein structure together and deactivation follows. Besides, the variation in enzyme production with changes in temperature has also been reported to be due to denaturation of some of the heat sensitive biochemical products produced during fermentation [23] and variation of dissolved oxygen tension in the medium with increasing temperatures [35].
Figure 2.
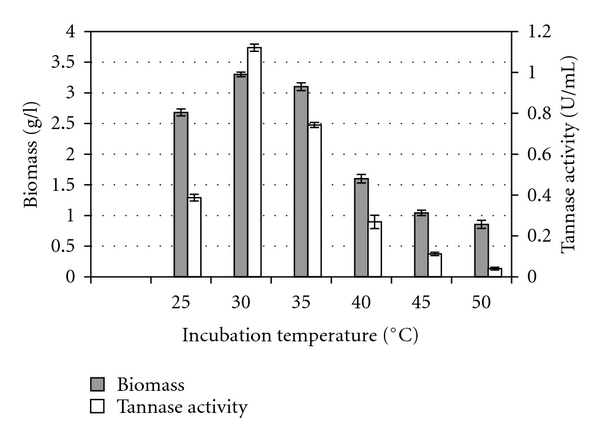
Effect of incubation temperature on biomass and tannase production by Aspergillus niger FETL FT3.
3.2. Effect of pH on Tannase Production
To observe the effect of initial medium pH on tannase production, the medium was adjusted to different pH levels, and the enzyme production was observed. Figure 3 shows that the optimal pH for tannase production by A. niger FETL FT3 was 6. However, the highest fungal growth was achieved at pH 7. The same pattern of findings was reported by Bradoo et al. [28], using Aspergillus japonicus where tannase production was maximal between pH 6 and 7 and optimal growth was observed at the pH between 4 and 7. These findings indicated that the enzyme production was not growth related. Similarly, these studies also showed that pH affects tannase production in the fermentation broth but has minimal effect on the growth of the fungus. The optimal extracellular tannase production observed at pH 6 in this experiment, which was somewhere near the growth pH of the fungus, could also be explained by the fact that this fungus was isolated from the decayed Rhizophora apiculata barks, which is also quite acidic in nature. Furthermore, the optimal initial medium pH for tannase production by submerged fermentation has also been reported in the acidic range of 4.5 [11] to 6.0 [30]. Accordingly, Pourrat et al. [36] found no significant tannase activity in the culture medium where the initial medium pH was less than 4. Barthomeuf et al. [11] reported that the tannase enzyme produced by their Aspergillus niger at pH value below 3.5 was unstable, while at pH values above 5.0, substrate hydrolysis and diffusion of enzyme into the medium occurred. In addition to that, Lekha and Lonsane [10] indicated that with increase in pH, the enzyme production decreased as tannase is acidic proteins with an optimal pH around 5.5.
Figure 3.
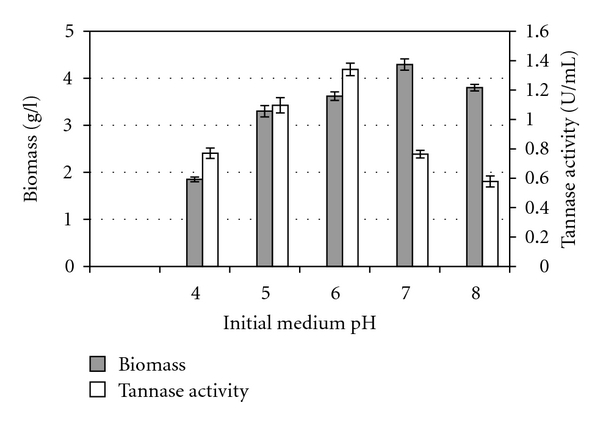
Effect of initial medium pH on biomass and tannase production by Aspergillus niger FETL FT3.
3.3. Effect of Agitation Speed on Tannase Production
In the submerged cultures, agitation speed was found to be a critical factor influencing both the fungal mycelial biomass and tannase production. Figure 4 shows that the enzyme activity and the fungal growth increased with the increasing of agitation speed up to 200 rpm, where it achieved its maximal tannase activity of 1.410 U per mL and fungal biomass of 3.75 grams per liter. Lower or higher agitation speed than 200 rpm produced lower enzyme activity and fungal growth. Similarly, Seth and Chand [37] also observed a drastic fall in enzyme activity when the agitation speed was increased beyond 400 rpm in a 3 liter mechanically agitated bioreactor. Seth and Chand [37] explained that at lower agitation speed, the inadequate mixing of the broth towards the later stages of growth affected the enzyme synthesis by accumulation of gallic acid on the surface of the mycelium, while the drastic fall in enzyme activity at higher agitation rates, was largely due to shearing effect on the mycelium. In the same way, Barthomeuf et al. [11] and Pourrat et al. [9, 36] found that excessive aeration favored oxidation of tannins and thus had inhibiting effects on the biosynthesis of the enzyme. Additionally, changes in the morphology of Aspergillus niger caused by different agitation speeds [35, 38–42] were also found to influence tannase production and growth of the organism.
Figure 4.
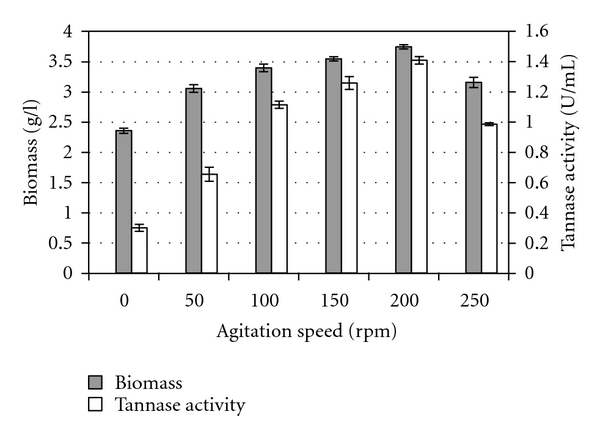
Effect of agitation rate on biomass and tannase production by Aspergillus niger FETL FT3.
3.4. Effects of Inoculums Size on Tannase Production
Inoculum size is another important parameter for tannase production and the size of inoculum has been reported to play a significant role in the production of metabolites [20]. In the present study, fungal spores were used as the inoculum and different inoculum's sizes, that is, 1 percent (volume per volume) of about 1 × 102 to 1 × 108 spore per mL were studied to enhance the utilization of tannic acid and thereby improving tannase activity (Figure 5). Aspergillus niger FETL FT3 pellets were observed at all the different level of spore inoculum studied, however, studies on the effect of inoculum's size on tannase production showed an increase in tannase production with increasing inoculum's sizes up to spore concentrations in the range of 1 × 105 to 1 × 106 spore/mL (Figure 5(a)). In order to determine the actual inoculums size that gave maximal tannase production, the spore concentration was narrow down to 2 × 106–10 × 106 spores/mL (Figure 5(b)). The results revealed that the maximal tannase production of 1.650 U per mL was obtained at the spore concentrations of about 6 × 106 spores/mL (Figure 5(b)). However, there was not much different in tannase production at the inoculums size between 6 × 106 to 10 × 106 spores per mL.
Figure 5.
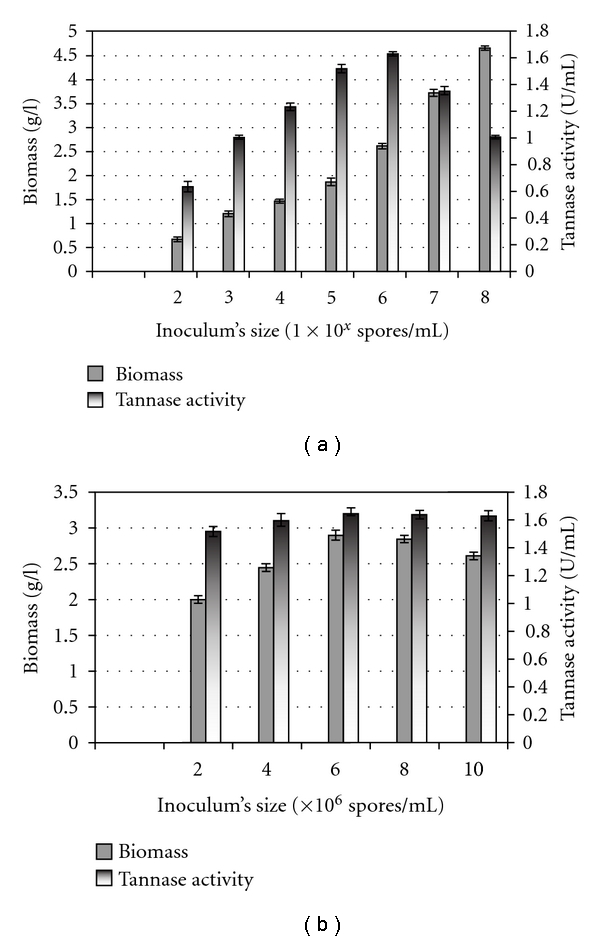
Effect of inoculum's size on biomass and tannase production by cells of Aspergillus niger FETL FT3 in a shake-flask system. (a) Wide range, (b) narrow range.
The results obtained suggested that lower concentration of inoculums size may have not been sufficient for initiating growth and enzyme synthesis [20, 25, 43], and, therefore, an increase in the number of spores ensured a rapid production of biomass and enzyme synthesis [44]. However, the decreased in enzyme production after an inoculum's size of about 6 × 106 spores per mL was presumably because of the depletion of nutrients due to enhanced biomass production [20, 25].
3.5. Enzyme Production Using Improved Physical Parameters
After the improvement of the various physical parameters, a time-course study was conducted to see the cumulative effect of various parameters. The experiment was conducted by incorporating all the improved physical parameters, and samples were taken at 24-hour intervals. The results of the time-course study after and before improvements are shown in Figure 6. The maximal incubation period for the enzyme production was 4 days for both conditions, that is, maximal incubation period for tannase production was retained on 4th day upon improvement. Additionally, a comparison of enzyme production shows that improvement resulted in an increase of 156.4 percent (1.096 U per mL before and 2.81 U per mL after improvements).
Figure 6.
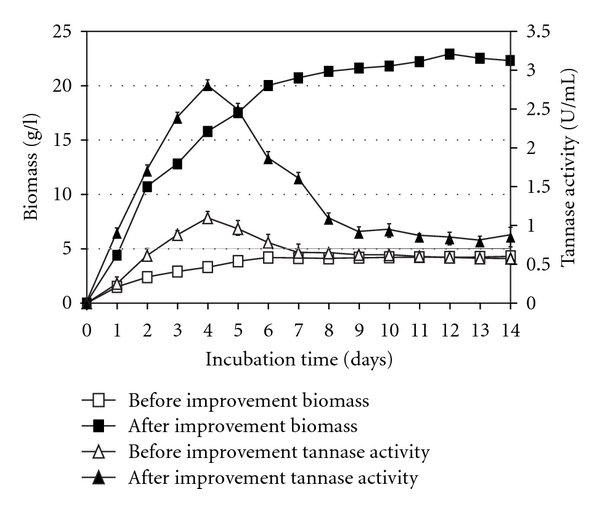
Tannase production after and before improvement of physical parameter in a shake-flask system by Aspergillus niger FETL FT3.
4. Conclusion
This study revealed that a locally isolated fungal strain, A. niger FETL FT3 was a potential fungal tannase producer. The physical parameters studied did influence its tannase production in a submerged fermentation using a shake-flask system. Nevertheless, a scaling-up of tannase production in a fermenter with a bigger volume of cultivation medium will be reported elsewhere.
References
- 1.Deschamps AM, Otuk G, Lebeault J. Production of tannase and degradation of chestnut tannin by bacteria. Journal of Fermentation Technology. 1983;61(1):55–59. [Google Scholar]
- 2.Boadi DK, Neufeld RJ. Encapsulation of tannase for the hydrolysis of tea tannins. Enzyme and Microbial Technology. 2001;28(7-8):590–595. doi: 10.1016/s0141-0229(01)00295-2. [DOI] [PubMed] [Google Scholar]
- 3.Aguilar CN, Rodriguez R, Gutierrez-Sanchez G, et al. Microbial tannases: advances and perspectives. Applied Microbiology and Biotechnology. 2007;76(1):47–59. doi: 10.1007/s00253-007-1000-2. [DOI] [PubMed] [Google Scholar]
- 4.Sanderson GW, Coggon P. Tech. Rep. 3812266. Arlington, Va, USA: US Patent; 1974. Green tea conversion using tannase and natural tea leaves. [Google Scholar]
- 5.Aguilar CN, Gutierrez-Sanches G. Review: sources, properties, applications and potential uses of tannin acyl hydrolase. Food Science and Technology International. 2001;7(5):373–382. [Google Scholar]
- 6.Vaquero I, Marcobal A, Munoz R. Tannase activity by lactic acid bacteria isolated from grape must and wine. International Journal of Food Microbiology. 2004;96(2):199–204. doi: 10.1016/j.ijfoodmicro.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Rout S, Banerjee R. Production of tannase under mSSF and its application in fruit juice debittering. Indian Journal of Biotechnology. 2006;5(3):346–350. [Google Scholar]
- 8.Ayed L, Hamdi M. Culture conditions of tannase production by Lactobacillus plantarum. Biotechnology Letters. 2002;24(21):1763–1765. [Google Scholar]
- 9.Pourrat H, Regerat F, Morvan P, Pourrat A. Microbiological production of gallic acid from Rhus coriaria L. Biotechnology Letters. 1987;9(10):731–734. [Google Scholar]
- 10.Lekha PK, Lonsane BK. Production and application of tannin acyl hydrolsase: state of the art. Advances in Applied Microbiology. 1997;44:215–260. doi: 10.1016/s0065-2164(08)70463-5. [DOI] [PubMed] [Google Scholar]
- 11.Barthomeuf C, Regerat F, Pourrat H. Production, purification and characterization of a tannase from Aspergillus niger LCF 8. Journal of Fermentation and Bioengineering. 1994;77(3):320–323. [Google Scholar]
- 12.Ramirez-Coronel MA, Viniegra-Gonzalez G, Darvill A, Augur C. A novel tannase from Aspergillus niger with β-glucosidase activity. Microbiology. 2003;149(10):2941–2946. doi: 10.1099/mic.0.26346-0. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee D, Mondal KC, Pati BR. Production and characterization of extracellular and intracellular tannase from newly isolated Aspergillus aculeatus DBF 9. Journal of Basic Microbiology. 2001;41(6):313–318. doi: 10.1002/1521-4028(200112)41:6<313::AID-JOBM313>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 14.Hadi TA, Banerjee R, Bhattacharyya BC. Optimization of tannase biosynthesis by a newly isolated Rhizopus oryzae. Bioprocess Engineering. 1994;11(6):239–243. [Google Scholar]
- 15.Mahendran B, Raman N, Kim DJ. Purification and characterization of tannase from Paecilomyces variotii: hydrolysis of tannic acid using immobilized tannase. Applied Microbiology and Biotechnology. 2006;70(4):444–450. doi: 10.1007/s00253-005-0082-y. [DOI] [PubMed] [Google Scholar]
- 16.Rajakumar GS, Nandy SC. Decomposition of tannic acid and gallic acid by Penicillium chrysogenum. Leather Science. 1985;32:278–280. [Google Scholar]
- 17.Aoki K, Shinka R, Nishira H. Purification and some properties of yeast tannase. Agriculture Biology and Chemistry. 1976;40(1):79–85. [Google Scholar]
- 18.Mondal KC, Banerjee D, Banerjee R, Pati BR. Production and characterization of tannase from Bacillus cereus KBR9. Journal of General and Applied Microbiology. 2001;47(5):263–267. doi: 10.2323/jgam.47.263. [DOI] [PubMed] [Google Scholar]
- 19.Nishitani Y, Sasaki E, Fujisawa T, Osawa R. Genotypic analyses of lactobacilli with a range of tannase activities isolated from human feces and fermented food. Systematic and Applied Microbiology. 2004;27(1):109–117. doi: 10.1078/0723-2020-00262. [DOI] [PubMed] [Google Scholar]
- 20.Sabu A, Augur C, Swati C, Pandey A. Tannase production by Lactobacillus sp. ASR-S1 under solid-state fermentation. Process Biochemistry. 2006;41(3):575–580. [Google Scholar]
- 21.Osawa R, Kuroiso K, Goto S, Shimizu A. Isolation of tannin-degrading Lactobacilli from humans and fermented foods. Applied and Environmental Microbiology. 2000;66(7):3093–3097. doi: 10.1128/aem.66.7.3093-3097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natarajan K, Rajendran A. Effect of fermentation parameters on extra cellular tannase production by Lactobacillus plantarum MTCC 1407. E-Journal of Chemistry. 2009;6(4):979–984. [Google Scholar]
- 23.Kar B, Banerjee R. Biosynthesis of tannin acyl hydrolase from tannin-rich forest residue under different fermentation conditions. Journal of Industrial Microbiology and Biotechnology. 2000;25(1):29–38. [Google Scholar]
- 24.Gautam P, Sabu A, Pandey A, Szakacs G, Soccol CR. Microbial production of extra-cellular phytase using polystyrene as inert solid support. Bioresource Technology. 2002;83(3):229–233. doi: 10.1016/s0960-8524(01)00215-2. [DOI] [PubMed] [Google Scholar]
- 25.Sabu A, Pandey A, Daud MJ, Szakacs G. Tamarind seed powder and palm kernel cake: two novel agro residues for the production of tannase under solid state fermentation by Aspergillus niger ATCC 16620. Bioresource Technology. 2005;96(11):1223–1228. doi: 10.1016/j.biortech.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigues THS, Dantas MAA, Pinto GAS, Goncalves LRB. Tannase production by solid state fermentation of cashew apples baggase. Aplied Biochemistry and Biotechnology. 2007;137–140(1–12):675–688. doi: 10.1007/s12010-007-9088-5. [DOI] [PubMed] [Google Scholar]
- 27.Kar B, Banerjee R, Bhattacharyya BC. Microbial production of gallic acid by modified solid state fermentation. Journal of Industrial Microbiology and Biotechnology. 1999;23(3):173–177. [Google Scholar]
- 28.Bradoo S, Gupta R, Saxena RK. Parametric optimization and biochemical regulation of extracellular tannase from Aspergillus japonicus. Process Biochemistry. 1997;32(2):135–139. [Google Scholar]
- 29.Banerjee D, Mondal KC, Pati BR. Tannase production by Aspergillus aculeatus DBF9 through solid-state fermentation. Acta Microbiologica et Immunologica Hungarica. 2007;54(2):159–166. doi: 10.1556/AMicr.54.2007.2.6. [DOI] [PubMed] [Google Scholar]
- 30.Yamada H, Adachi O, Watanbe M, Sato N. Studies on fungal tannase. Part I: formation, purification and catalytic properties of tannase of Aspergillus flavus. Agricultural and Biological Chemistry. 1968;32:1070–1078. [Google Scholar]
- 31.Mondal KC, Pati BR. Studies on the extracellular tannase from newly isolated Bacillus licheniformis KBR 6. Journal of Basic Microbiology. 2000;40(4):223–232. doi: 10.1002/1521-4028(200008)40:4<223::AID-JOBM223>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 32.Mondal KC, Banerjee R, Pati BR. Tannase production by Bacillus licheniformis. Biotechnology Letters. 2000;22(9):767–769. [Google Scholar]
- 33.Van de Lagemaat J, Pyle DL. Modelling the uptake and growth kinetics of Penicillium glabrum in a tannic acid-containing solid-state fermentation for tannase production. Process Biochemistry. 2005;40(5):1773–1782. [Google Scholar]
- 34.G. M. Frost, Moss DA. Production of enzymes by fermentation. In: Kennedy JF, editor. Biotechnology Vol. 7a. London, UK: Elsevier Applied Science; 1987. p. 134. [Google Scholar]
- 35.Papagianni M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnology Advances. 2004;22(3):189–259. doi: 10.1016/j.biotechadv.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Pourrat H, Regerat F, Pourrat A. Production of gallic acid from Tara by a strain of Aspergillus niger. Journal of Fermentation Technology. 1985;63:401–403. [Google Scholar]
- 37.Seth M, Chand S. Biosynthesis of tannase and hydrolysis of tannins to gallic acid by Aspergillus awamori—optimisation of process parameters. Process Biochemistry. 2000;36(1-2):39–44. [Google Scholar]
- 38.Taguchi H, Yoshida T, Tomita Y, Teramoto S. The effects of agitation on disruption of the mycelia pellets in stirred fermenters. Journal of Fermentaion Technology. 1986;46:814–822. [Google Scholar]
- 39.Smith JJ, Lilly MD, Fox RI. The effect of agitation on the morphology and penicillin production of Penicillium chrysogenum. Biotechnology and Bioengineering. 1990;35(10):1011–1023. doi: 10.1002/bit.260351009. [DOI] [PubMed] [Google Scholar]
- 40.Van de Lagemaat J, Pyle DL. Solid-state fermentation and bioremediation: development of a continuous process for the production of fungal tannase. Chemical Engineering Journal. 2001;84(2):115–123. [Google Scholar]
- 41.Park JP, Kim YM, Kim SW, et al. Effect of agitation intensity on the exo-biopolymer production and mycelial morphology in Cordyceps militaris. Letters in Applied Microbiology. 2002;34(6):433–438. doi: 10.1046/j.1472-765x.2002.01126.x. [DOI] [PubMed] [Google Scholar]
- 42.Kim SW, Hwang HJ, Xu CP, Choi JW, Yun JW. Effect of aeration and agitation on the production of mycelial biomass and exopolysaccharides in an enthomopathogenic fungus Paecilomyces sinclairii. Letters in Applied Microbiology. 2003;36(5):321–326. doi: 10.1046/j.1472-765x.2003.01318.x. [DOI] [PubMed] [Google Scholar]
- 43.Mukherjee G, Banerjee R. Biosynthesis of tannase and gallic acid from tannin rich substrates by Rhizopus oryzae and Aspergillus foetidus. Journal of Basic Microbiology. 2004;44(1):42–48. doi: 10.1002/jobm.200310317. [DOI] [PubMed] [Google Scholar]
- 44.Kashyap P, Sabu A, Pandey A, Szakacs G, Soccol CR. Extra-cellular L-glutaminase production by Zygosaccharomyces rouxii under solid-state fermentation. Process Biochemistry. 2002;38(3):307–312. [Google Scholar]


