Abstract
With the development of next-generation sequencing technology, ever-expanding databases of genetic information from various organisms are available to researchers. However, our ability to study the biological meaning of genetic information and to apply our genetic knowledge to produce genetically modified crops and animals is limited, largely due to the lack of molecular tools to manipulate genomes. Recently, targeted cleavage of the genome using engineered DNA scissors called zinc finger nucleases (ZFNs) has successfully supported the precise manipulation of genetic information in various cells, animals, and plants. In this review, we will discuss the development and applications of ZFN technology for genome engineering and highlight recent reports on its use in plants.
Keywords: Zinc finger nucleases, Genome engineering, Gene targeting, Genetically modified organism
Introduction
Genetic tools to achieve specific genome modifications are essential for reverse genetics and could also provide powerful means to develop genetically modified organisms in biotechnology. Gene targeting via homology-directed recombination has been established as a primary tool for targeted genome modification in certain systems (Capecchi 2005; Smithies 2001). This process essentially involves a transfer of donor DNA (which contains homologous DNA sequences to target loci of the genome of interest) into cells and the use of this donor DNA by the cell’s homology-directed recombination machinery to exchange genetic information between the donor DNA and chromosomal target loci (Vasquez et al. 2001). Although gene targeting has been extremely useful, this process is limited to model systems in which donor DNA can be efficiently delivered and endogenous DNA repair machinery allows recombination between the donor DNA and the endogenous target loci with high frequency. Indeed, these requirements have limited gene targeting primarily to mouse and yeast systems. The frequency of homologous recombination (HR) events between foreign donor DNA and its chromosomal target can be dramatically enhanced by introducing a specific double strand break (DSB) in the chromosomal target in most research models (Johnson and Jasin 2001; Puchta et al. 1993). Although several different approaches for inducing a site-specific DSB in a cell have been suggested, zinc finger nucleases (ZFNs) have emerged as a versatile and universal tool to enable gene targeting in various organisms (Porteus and Carroll 2005).
ZFNs are chimeric DNA restriction enzymes that consist of a custom-designed zinc finger (ZF) DNA-binding domain and the nonspecific nuclease domain from the FokI restriction enzyme (Porteus and Carroll 2005). Because ZF domains can be engineered to bind to a pre-determined DNA sequence at a genomic locus of interest, ZFNs can be used to introduce DSBs at specific genomic sites in a targeted manner, thereby improving gene targeting efficiency (Bibikova et al. 2003). However, the versatility of ZFNs as a tool for the targeted manipulation of the genome goes far beyond the improvement of HR-mediated gene targeting.
In a eukaryotic cell, DSBs are mainly repaired by two evolutionarily conserved DSB repair pathways, HR and non-homologous end joining (NHEJ) (Wyman and Kanaar 2006). By exploiting these two DSB repair pathways, one can use ZFN-induced DSBs to manipulate the genome in a specific manner. With their versatility and universal nature, ZFNs have successfully supported genome engineering in cultured cells and various animal models, including C. elegans and D. melanogaster, which have long been used for genetic studies (Beumer et al. 2006; Bibikova et al. 2002; Carroll et al. 2008; Kim et al. 2009; Morton et al. 2006; Santiago et al. 2008; Urnov et al. 2005). In addition, ZFNs also have emerged as robust and reproducible tools for genome engineering in important model systems in which traditional reverse genetic methodologies are not easily applicable. These systems include rat, zebrafish, silk worm, sea urchin, model plants, and crops (Doyon et al. 2008; Geurts et al. 2009; Meng et al. 2008; Ochiai et al. 2010; Osakabe et al. 2010; Shukla et al. 2009; Takasu et al. 2010; Townsend et al. 2009; Zhang et al. 2010).
In this review, we will first describe different approaches for developing ZFNs for specific target sequences. Then we will explore how ZFNs were used to achieve genome engineering in recent studies, emphasizing their uses in plants.
Engineering ZFNs
The structure of ZFNs
ZFNs are created by combining the nuclease domain of the FokI type II S restriction enzyme with engineered ZF DNA-binding domains (Chandrasegaran and Smith 1999; Kim et al. 1996). Within this structure, ZF DNA-binding domains—composed of an array of multiple C2H2 zinc fingers—determine the DNA binding specificity, thus guiding ZFNs to their target site (Fig. 1). The nuclease domain provides the enzymatic activity that cleaves DNA strands (Durai et al. 2005; Porteus and Carroll 2005). Because one ZF module recognizes a 3-base pair (bp) DNA subsite, a ZFN with an array of 3 or 4 ZFs recognizes and binds to a sequence of 9 or 12 nucleotides. Since the cleavage activity of the FokI nuclease domain requires dimerization, a pair of ZFNs forms a site-specific endonuclease enzyme. Thus, a target site of a pair of three-finger ZFNs consists of two 9-bp recognition sequences flanking a spacer 5 or 6 bp in length. Thus, the target site of a ZFN pair is expected to be very rare—if not unique—in a genome, enabling ZFNs to introduce site-specific DSBs.
Fig. 1.
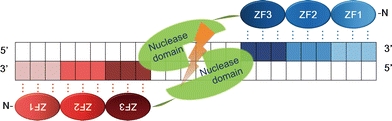
Structure of ZFNs. Schematic representation of ZFNs bound to their target sites. These nucleases are created by fusing an engineered ZF DNA binding domain to the DNA cleavage domain of the FokI type IIS restriction enzyme. An array of 3 ZFs (shaded ovals) for each ZFN protein will recognize a target sequence on DNA (shaded boxes). The binding of two ZFNs on the target DNA with a spacer (5/6 bp) in the middle will trigger the dimerization of the ZFN nuclease domains and the cleavage of the target DNA
The specificity of this system has recently been increased further through the development of an improved ZFN architecture (Miller et al. 2007; Szczepek et al. 2007). When a pair of ZFNs is expressed together in a cell with the aim of digesting a target site, homodimerization of each ZFN could result in possible off-target site cleavages. To overcome this caveat, two groups have independently developed FokI nuclease domain variants that are engineered to show cleavage activity only as a heterodimer. The use of such obligatory heterodimeric FokI nuclease variants has greatly improved the specificity and reduced the cellular toxicity of ZFNs.
ZF binding domains and different methods of building custom-designed ZF arrays
ZF DNA-binding domains are among the most abundant DNA-binding domains encoded in higher eukaryotic genomes (Klug 2010). Such domains are composed of a tandem array of multiple ZF modules in which each ZF recognizes a 3 bp DNA subsite (Wolfe et al. 2000). Importantly, individual ZF modules retain their DNA-binding specificities in the context of artificially constructed multi-finger ZF proteins. This observation provoked the idea that one could use a directory of ZF modules, each recognizing one of the 64 possible 3-bp DNA subsites, to build an artificial ZF DNA binding domain that could bind to any DNA sequence (Pabo et al. 2001). To this end, in the last 15 years many research groups have undertaken efforts to isolate naturally occurring ZF modules with various sequence specificities, as well as to use rational, combinatorial approaches to engineer ZF modules with altered DNA binding specificities. Such efforts have established a directory of ZFs that recognize many of the 64 possible 3-bp DNA subsites (Bae et al. 2003; Lee et al. 2003). These ZFs have been successfully used to build custom-designed DNA-binding domains for artificial transcription factors that can regulate biologically and clinically important genes in various organisms (Jamieson et al. 2003; Park et al. 2003, 2005). During the course of this work, several different methods of building new ZF DNA binding domains using ZF modules have been devised. Currently, three different methods are used to construct novel ZFNs: modular assembly, cell-based selection, and a proprietary method used by Sangamo Biosciences (Carroll et al. 2006; Kim et al. 2010a; Maeder et al. 2009; Urnov et al. 2010; Wright et al. 2006).
The traditional modular-assembly method involves a simple directory-based assembly of preselected ZFs as building blocks to construct multi-finger ZF arrays via standard recombinant DNA technology (Carroll et al. 2006). There are several in silico tools (Kim et al. 2009; Mandell and Barbas 2006; Sander et al. 2007) and sources of ZF modules (Bae et al. 2003; Segal et al. 1999; Wright et al. 2006) to facilitate the modular assembly of ZFNs. Modularly assembled ZF arrays can be tested for their DNA-binding activities using standard assays that analyze DNA–protein interactions. Alternatively, the arrays can be assembled to form ZFNs that can be directly tested for their ability to induce DSBs at their target sites in the genome or a reporter construct. Reported success rates of modularly assembled ZFNs vary greatly, ranging from 6 to 30% depending on the study (Kim et al. 2009, 2010a; Lee et al. 2010; Ramirez et al. 2008). It should be noted that we have found that the careful selection and use of reliable modules consistently improves the success rates of modularly assembled ZFNs (Lee et al. 2010) (Kim et al. 2010b). Modularly assembled ZFNs have been successfully used for genome engineering in various systems, including cultured mammalian cells, C. elegans, D. melanogaster, tobacco, and Arabidopsis (Beumer et al. 2006; Bibikova et al. 2002; Kim et al. 2009; Morton et al. 2006; Osakabe et al. 2010; Siekmann et al. 2009; Townsend et al. 2009). With its simple procedure, coupled with well-established assays to test the activity of ZFNs, the modular-assembly method provides an easily accessible and reliable approach for rapidly generating and screening many ZFN pairs to target a specific sequence. Commercial services for the modular assembly and activity validation of ZFNs are also available from ToolGen, Inc. (http://www.toolgen.com).
A proprietary method used by Sangamo Biosciences is based on a principle similar to that of simple modular assembly, except that an archive of validated two-finger modules is used to assemble longer ZF arrays (Moore et al. 2001). Although ZFNs made by this approach have been successfully used to engineer the genomes of cultured mammalian cells, animals, and plants (Cost et al. 2010; Doyon et al. 2008; Geurts et al. 2009; Liu et al. 2010; Moehle et al. 2007; Orlando et al. 2010; Santiago et al. 2008; Shukla et al. 2009; Urnov et al. 2005), detailed information about the two-finger archive is not publically accessible. Instead, ZFNs developed by this approach are commercially available from Sigma-Aldrich (http://www.sigma.com), which licensed this technology from Sangamo; however, they are not available for use in plants because of an exclusive licensing agreement between Sangamo and Dow Agrosciences.
More recently, a cell-based selection approach for building site-specific ZFNs, termed the Oligomerized Pool ENgineering (OPEN) method, was developed (Maeder et al. 2008; Maeder et al. 2009). In this approach, pools of ZFs that are selected to bind different 3-bp DNA subsites are established from a library of ZFs with random mutations at their DNA-contacting residues using an E. coli two-hybrid selection system. Using a web-based analysis tool, a small number of candidate ZFN sites within a given target sequence are selected and a library of three-finger arrays for each target half site is constructed (Sander et al. 2010). Then, using an E. coli two-hybrid selection system, three-finger arrays that bind to the 9-bp target sequence are selected and further analyzed. Selected three-finger arrays are assembled to form ZFNs, and their ability to introduce DSBs at the target site is determined. ZFNs created using the OPEN approach have been successfully used for genome engineering in cultured mammalian cells, zebrafish, tobacco, and Arabidopsis (Foley et al. 2009; Maeder et al. 2008; Townsend et al. 2009; Zou et al. 2009). This method, however, is both labor intensive and time consuming, because a few rounds of E. coli-based selection steps are required before making ZFNs. Furthermore, this method is limited to the production of three-finger ZFNs but not four-finger ZFNs; the complexity of four-finger arrays is about 100 times greater than that of three-finger arrays, and cannot be covered by the cell-based method. Four-finger ZFNs, in general, are more specific and reliable than are three-finger ZFNs (Lee et al. 2010).
Targeted genome engineering using ZFNs
DSBs pose a uniquely high threat to the integrity of the genetic information of a cell. Not only can the unfaithful repair of a DSB result in unwanted modifications of genetic information at the site of the DSB, but a failure to repair such a break could also lead to the loss of larger chromosomal fragments (Agarwal et al. 2006). Thus, eukaryotic cells have evolved elaborate mechanisms to repair DSBs faithfully and efficiently. Two major mechanisms of DSB repair are known, NHEJ and HR. A site-specific DSB induced by ZFNs will trigger the repair process by NHEJ or HR, both of which can be exploited to achieve targeted genome engineering such as gene knockout and gene addition.
Targeted genome engineering via NHEJ
NHEJ is a rapid and efficient DSB repair mechanism that involves a simple ligation of the two DNA ends that result from a DSB (Lieber 2010). The protein machinery of the canonical NHEJ pathway provides a mechanism to hold the two DNA ends in close physical proximity, trim the DNA ends to prepare them for subsequent ligation, and enzymatically ligate the two DNA ends to complete the repair process. Overall, the NHEJ mechanism is highly accurate and efficient in the repair of DSBs. The innate nature of NHEJ that lacks the use of an intact copy (a homologous sequence) to restore genetic information at DSBs, however, result in the formation of sporadic unfaithful repair products with the signature of small nucleotide deletions or insertions (indels).
Thus, if the ZFN target site is within a protein-coding region, small indels that arise from an unfaithful repair of ZFN-induced DSBs will likely result in a frameshift mutation in the target gene and thus a truncation of the target protein (Porteus and Carroll 2005). NHEJ-mediated knockout of a target gene by ZFNs has some advantages over the HR-mediated gene knockout process by ZFNs. First, the frequency of NHEJ-mediated repair of DSBs is generally much higher than that of HR-mediated DSB repair. This is because NHEJ is a preferred DSB repair mechanism in higher eukaryotic cells. Additionally, the faithful repair of ZFN-induced DSBs will result in the restoration of the ZFN target site. In contrast, small indels created by unfaithful NHEJ will destroy the ZFN target site. Thus, while a cell is expressing ZFNs, the ZFN target site is likely to experience multiple rounds of digestion and repair, which will substantially increase the occurrence of NHEJ-mediated small indels. Another important advantage of the NHEJ-mediated gene knockout procedure is that, unlike the HR-mediated process, it does not require the simultaneous delivery of donor DNA with ZFNs into the cell.
NHEJ-mediated gene knockout using ZFNs has been extremely successful in animal models and cultured mammalian cells. In particular, the direct injection of mRNA encoding ZFNs into one-cell embryos has provided an efficient and rapid approach to establish gene knockout animal models (Remy et al. 2010). Initially reported in Drosophila, this method has recently been successfully used in higher metazoan systems like zebrafish and rat, two important animal models for human diseases (Beumer et al. 2008; Carbery et al. 2010; Doyon et al. 2008; Foley et al. 2009; Geurts et al. 2009; Mashimo et al. 2010; Meng et al. 2008). Using this approach, the efficient delivery of ZFNs is ensured. In addition, because ZFNs are delivered to a one-cell embryo from which a whole organism originates, one can expect a high rate of target gene knockout among somatic and germ line cells of direct offspring, expediting the process of establishing knockout animals, as compared to traditional gene targeting methods in mouse embryonic cell lines.
NHEJ-mediated gene knockout in cultured mammalian cells has been achieved by the transient expression of ZFNs via various gene delivery methods. In cells of a variety of species and origins, chemical transfection, electroporation, and integrase-deficient lentivirus-mediated delivery of DNA/RNA for ZFN expression have supported efficient gene knockout via NHEJ (Kim et al. 2009; Lombardo et al. 2007; Maeder et al. 2009; Santiago et al. 2008). The frequency of gene knockout using this approach ranged widely, from 1 to 50%. This high frequency of gene knockout by transient ZFN expression allowed researchers to isolate single cell-derived knockout clones without using any drug-mediated selection process. Indeed, this process was so efficient that a research group was able to knockout three genes in Chinese hamster ovary (CHO) cells by simple repetition of ZFN expression and single-clone isolation processes (Liu et al. 2010).
In addition to the creation of simple gene knockouts, NHEJ-mediated repair of ZFN-induced DSBs can be exploited to achieve more complex and elaborate genome engineering (Fig. 2). First, the repair of two DSBs—each specifically induced by ZFNs via NHEJ—produces a subset of cells with chromosomal variations that involve the loss of genetic information between the two DSB sites (if the two DSB sites are on the same chromosome) and a translocation between two chromosomes (if the two DSB sites are on different chromosomes) (Brunet et al. 2009; Lee et al. 2010; Liu et al. 2010; Petolino et al. 2010; Sollu et al. 2010). Second, small DNA fragments with appropriate overhang structures that are codelivered into cells with ZFNs can be incorporated into the DSB site with relatively high frequency (Orlando et al. 2010) (Fig. 2a). Thus, the targeted addition of genetic information into the genome is possible using ZFNs via NHEJ. Yet to be applied in broader research models, these additional applications in targeted genome engineering suggest that ZFNs can be used to achieve a range of genomic variations, beyond simple gene knockouts, that have been found in comparative genomics research. In addition, ZFN-mediated large chromosomal deletions can allow the removal of a cluster of homologous and redundant genes in a single step.
Fig. 2.
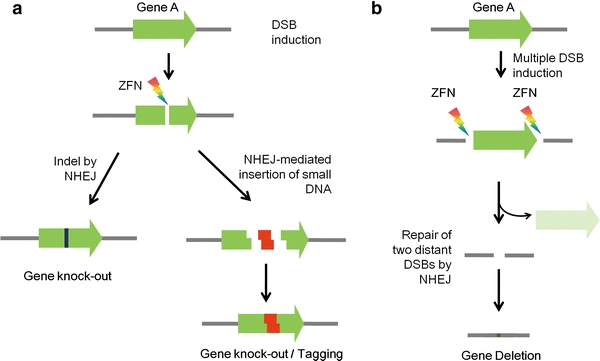
ZFN-mediated targeted genome engineering via NHEJ. a Gene knockout and DNA insertion. A DSB created by a pair of ZFNs stimulates the error-prone nonhomologous end joining (NHEJ) pathway, and the target gene can be disrupted by small indels induced by NHEJ repair (left pathway). Small DNA fragments delivered into the cell can be captured into the target site of a DSB induced by ZFNs via NHEJ (right pathway). b Gene deletion. The NHEJ repair of two concurrent DSBs flanking a target gene is induced by two ZFN pairs, which can give rise to targeted deletions of genes between the two sites
HR-mediated targeted genome engineering by ZFNs
Homology-directed DSB repair involves the use of template DNA with homology to a DSB locus to restore genetic information in an error-free manner (San Filippo et al. 2008). Even without inducing site-specific DSBs, foreign DNA transferred by researchers into cells can trigger the HR process between target and donor DNA to achieve gene targeting. This process, however, is extremely inefficient in most systems, with the exception of mouse embryonic stem cells and yeast. Site-specific DSBs at the gene targeting site have been proven to enhance the frequency of gene targeting greatly (100- to 10,000-fold) in various cells (Johnson and Jasin 2001). Thus, ZFNs can be used to enable HR-mediated genome engineering when they are delivered into cells together with appropriately designed donor DNA (Porteus and Carroll 2005) (Fig. 3). Like classical gene targeting, HR-mediated gene knockout using ZFNs enables researchers to use a drug resistance mediated selection scheme for isolating the small number of cells that have undergone the desired gene-targeting event. The efficient addition of up to 8 kb of genetic information from donor DNA into the ZFN target site via HR has also been reported (Moehle et al. 2007). Additionally, because HR involves the faithful transfer of genetic information from donor DNA to the DSB site, researchers can achieve gene correction and gene editing, which are very difficult, if not impossible, with the NHEJ-mediated process. In fact, the first application of ZFNs at endogenous target loci in cultured human cells was to use ZFNs to correct inherited mutations in the human IL2RG gene that causes severe immune deficiency (Urnov et al. 2005). More recently, ZFNs have supported efficient gene targeting and gene insertion at various target loci in human embryonic stem cells and induced pluripotent stem cells (Hockemeyer et al. 2009; Zou et al. 2009). Together, these studies suggest that ZFNs could be used for gene and cell therapy for inherited monogenic disorders.
Fig. 3.
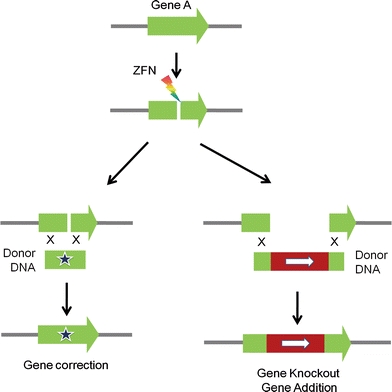
ZFN-mediated targeted genome engineering via HR. The repair of ZFN-induced DSBs via the HR pathway can be exploited to induce gene correction, gene knockout, and gene addition using properly designed donor DNA
Targeted genome engineering using ZFNs in plants
Initial reports on the feasibility of ZFN-mediated genome engineering in plants used a previously characterized ZFN whose target sequence was artificially incorporated into a plant genome. In the first report, through the use of Agrobacterium-mediated gene delivery, Arabidopsis was engineered to carry a ZFN with a QQR zinc finger domain under the regulation of a heat-inducible promoter as well as a target site for the QQR ZFN homodimer (Lloyd et al. 2005). The integrity of the QQR ZFN target site was analyzed in seven of these transgenic Arabidopsis lines, which were induced to express ZFN by heatshock. This analysis showed that 1.7–19.6% of the target sites contained ZFN-induced mutations. Among the mutagenized sites, 78% showed simple deletions of 1–52 bp, 13% showed simple insertions of 1–4 bp, and the rest showed more complex rearrangements, with both deletions and insertions. This study was followed by a report on successful HR-mediated reporter gene correction using ZFNs in tobacco (Wright et al. 2005). To test if the ZFNs could enhance HR events in this species, the authors first carefully established a transgenic tobacco line carrying a defective version of a reporter gene that consisted of a fusion between sequences encoding b-glucuronidase (GUS) and neomycin phosphotransferase (NPTII), which confers resistance to kanamycin. This fusion reporter had a deletion of 600 bp at the fusion site and an insertion of a Zif268 ZFN binding site. When protoplasts from this transgenic tobacco line were transformed with donor DNA to restore the activity of the GUS:NTPII reporter gene, no GUS+ and kanamycin-resistant colonies were obtained. However, when the same donor DNA was transformed with Zif268 ZFN expression plasmids, double-positive colonies were recovered at a frequency of ~1 × 10−4. Further analysis estimated that 20% of these GUS+/Kan-resistant cells contained a legitimate target structure that resulted solely from the intended HR event, whereas the other cells contained additional accompanying DNA rearrangements. Together, these two studies demonstrated the possibility of ZFN-mediated genome engineering via NHEJ and HR in plants.
Since these reports, many research groups have developed methods to construct and deliver ZFNs and reporter systems to test the activity of ZFNs in plants (Cai et al. 2009; de Pater et al. 2009; Tovkach et al. 2009). However, the real breakthrough in the application of ZFNs to plant research occurred in 2009, when endogenous genomic loci in tobacco and maize were successfully engineered using ZFNs (Maeder et al. 2008; Shukla et al. 2009; Townsend et al. 2009) (Table 1).
Table 1.
ZFN-mediated engineering of endogenous genes in plants
| Plant | Target gene | ZFN delivery | ZFN development | Genome engineering | Reference |
|---|---|---|---|---|---|
| Tobacco | SurA and B | Electroporation | OPEN | NHEJ (gene knockout) | Maeder et al. (2009) |
| Tobacco | SurA and B | Electroporation | OPEN, modular assembly | HR (gene correction) | Townsend et al. (2009) |
| Zea mays | IPK1 | Whisker-mediated transformation | Two-finger modules | HR (gene knockout by selection marker addition) | Shukla et al. (2009) |
| Tobacco | CHN50 | Agrobacterium | Two-finger modules | HR (gene knockout by selection marker addition) | Cai et al. (2009) |
| Arabidopsis | ABI4, NHEJ | Agrobacterium | Modular assembly | NHEJ (gene knockout) | Osakabe et al. (2010) |
| Arabidopsis | ADH1, TT4 | Agrobacterium | OPEN | NHEJ (gene knockout) | Zhang et al. (2010) |
Successful ZFN-mediated gene knockouts in tobacco were first achieved via NHEJ on the acetolactate synthase genes SuRA and SuRB (Maeder et al. 2008). Among 66 transgenic plants generated from protoplasts that stably expressed a pair of ZFNs targeting both the SuRA and SuRB genes, three plants contained mutations in SuRA, and one plant was a homozygous mutant. No mutant SuRB allele was detected. In a subsequent study, the endogenous tobacco SuRA and SuRB genes were also efficiently engineered using ZFNs via an HR-mediated mechanism (Townsend et al. 2009). For this purpose, additional ZFN pairs targeting the SuRA gene were developed using the modular assembly and OPEN methods and validated in a yeast-based reporter assay. The abilities of functional SuRA ZFN pairs to induce site-specific small indels via NHEJ upon expression in tobacco protoplasts were compared by pyrosequencing of the ZFN target site. Although site-specific mutagenesis by transient expression of ZFNs in protoplasts occurred at very low efficiency, this approach has enabled the comparison of ZFN activities on their endogenous targets in plant cells. The authors then constructed gene-correction donor DNA containing known missense mutations in the SuRA gene that confer herbicide resistance to tobacco. When this donor DNA was transformed together with ZFN expression plasmids into tobacco protoplasts, herbicide-resistant cells induced by an HR event between the target loci and donor DNA were obtained at a frequency of 0.2~4%. HR-mediated gene addition was also reported for tobacco (Cai et al. 2009). In this study, ZFNs targeting the CHN50 gene were developed and used to knock in a herbicide-tolerance gene (PAT). Both the ZFN expression vectors and the donor DNA were delivered into tobacco BY2 suspension culture cells using Agrobacterium. 5–10% of the selected plants were found to have undergone ZFN-induced targeted gene insertion at the CHN50 locus. Because the CHN50 gene displays robust expression in tobacco cells in suspension culture at the stationary phase, this locus could provide a safe harbor for gene addition for the high and stable expression of recombinant proteins under these conditions.
Plants in which ZFNs can be used for HR-mediated gene modifications include species beyond the model plant systems. Recently, this approach was used to produce maize plants with IPK1 mutations. The IPK1 gene encodes inositol-1,3,4,5,6-pentakisphosphate 2-kinase, which catalyzes the final step in phytate biosynthesis. Knockout of IPK1 is expected to render an agriculturally and environmentally beneficial phenotype in maize (Shukla et al. 2009). Multiple ZFN pairs were constructed to target the IPK1 gene. Appropriate HR donor DNA for the targeted insertion of a herbicide-tolerance gene (PAT), with or without an autonomous promoter, were also constructed for each ZFN pair. The delivery of IPK1-targeted ZFNs together with the corresponding donor DNA successfully resulted in the production of herbicide-tolerant calli with site-specific insertion of the PAT gene. The use of autonomous donor DNAs for the targeted insertion of the PAT gene resulted in more herbicide-tolerant calli than the use of nonautonomous donor DNA. The ratio of calli with a site-specific insertion of PAT at the IPK1 locus was much higher, however, when nonautonomous donor DNA was used. Importantly, insertion of the PAT gene into the IPK1 ZFN target site will result in the knockout of the IPK1 gene. The authors successfully produced an IPK1 knockout maize strain; furthermore, they showed decreased phytate and increased inorganic phosphate levels, as expected from the loss of the IPK1 enzyme.
More recently, efficient ZFN-mediated gene knockouts via NHEJ in Arabidopsis were reported (Table 1). In one study, ZFNs targeting the ADH1 and TT4 genes were developed and constructed in an estrogen-inducible expression system (Zhang et al. 2010). ADH1- and TT4-specific ZFN pairs were introduced into wild-type Arabidopsis plants by the Agrobacterium floral dip method. Transgenic plants containing ZFN constructs were selected; ZFN expression was induced with estradiol for ten days during germination. Subsequent analysis of somatic mutations in T1 seedlings revealed that the ZFNs efficiently induced mutations in the ADH1 and TT4 target sites with estimated rates of 16 and 7%, respectively. Importantly, these mutant T1 seedlings also showed a high frequency of germinal transmission of the ADH1 and TT4 mutations. In an independent study, ZFNs targeting the ABA-INSENSITIVE4 (ABI4) gene were developed and transferred to Arabidopsis via Agrobacterium-mediated transformation (Osakabe et al. 2010). Expression of the transferred ABI4 ZFNs was under the control of the Arabidopsis heat-shock protein HSP18.2 gene promoter. When ABI4 ZFN expression was induced by heat shock in nine transgenic plants, mutations at the ABI4 ZFN target site were induced at various rates ranging from 0.26 to 2.86%. In subsequent analysis, the authors successfully generated homozygous ABI4 knockout progenies from these T1 plants and showed that these plants exhibited the expected phenotypes of ABA and glucose insensitivity. These two studies establish ZFN-mediated gene knockout via NHEJ as an efficient and reliable approach in Arabidopsis.
Conclusion
With recent successes in the application of ZFN technology to model plants and crops, ZFN could represent a reliable and robust method for targeted genome modification, which has been an elusive goal for plant researchers so far. The successful implementation of ZFN technology generally requires reliable sources of target-specific ZFNs, efficient methods of delivering and expressing ZFNs, sensitive methods of validating the activity of ZFNs, and robust strategies to isolate engineered organisms. Fortunately, many of the above requirements have already been established in previous efforts to use ZFNs in cultured cells and animals, and they are easily applicable to plants. More efforts to adapt ZFN technology for various well-established gene delivery and expression systems in plants, as described in a recent report on the efficient delivery of ZFNs using tobacco rattle virus (TRV), may greatly broaden the spectrum of plants to which ZFN-mediated genome engineering can be efficiently applied (Marton et al. 2010).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Seokjoong Kim, FAX: +82-2-8891568, Email: sjkim.toolgen@gmail.com.
Jin-Soo Kim, FAX: +82-2-8747455, Email: jskim01@snu.ac.kr.
References
- Agarwal S, Tafel AA, Kanaar R. DNA double-strand break repair and chromosome translocations. DNA Repair (Amst) 2006;5:1075–1081. doi: 10.1016/j.dnarep.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, Kwon HS, Kim HW, Yeh BI, Lee HW, Sohn SH, Yoon J, Seol W, Kim JS. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D (2006) Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics 172:2391–2403 [DOI] [PMC free article] [PubMed]
- Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, Carroll D (2008) Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci USA 105:19821–19826 [DOI] [PMC free article] [PubMed]
- Bibikova M, Golic M, Golic KG, Carroll D (2002) Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161:1169–1175 [DOI] [PMC free article] [PubMed]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci USA. 2009;106:10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, Rock JM, Lee YL, Garrison R, Schulenberg L, Blue R, Worden A, Baker L, Faraji F, Zhang L, Holmes MC, Rebar EJ, Collingwood TN, Rubin-Wilson B, Gregory PD, Urnov FD, Petolino JF. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol. 2009;69:699–709. doi: 10.1007/s11103-008-9449-7. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui X. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- Carroll D, Beumer KJ, Morton JJ, Bozas A, Trautman JK (2008) Gene targeting in Drosophila and Caenorhabditis elegans with zinc-finger nucleases. Methods Mol Biol 435:63–77 [DOI] [PubMed]
- Chandrasegaran S, Smith J. Chimeric restriction enzymes: what is next? Biol Chem. 1999;380:841–848. doi: 10.1515/BC.1999.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Freyvert Y, Vafiadis A, Santiago Y, Miller JC, Rebar E, Collingwood TN, Snowden A, Gregory PD. BAK and BAX deletion using zinc-finger nucleases yields apoptosis-resistant CHO cells. Biotechnol Bioeng. 2010;105:330–340. doi: 10.1002/bit.22541. [DOI] [PubMed] [Google Scholar]
- de Pater S, Neuteboom LW, Pinas JE, Hooykaas PJ, van der Zaal BJ (2009) ZFN-induced mutagenesis and gene-targeting in Arabidopsis through Agrobacterium-mediated floral dip transformation. Plant Biotechnol J 7:821–835 [DOI] [PubMed]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS One. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Menoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, Meng X, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat Rev Drug Discov. 2003;2:361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem Soc Trans. 2001;29:196–201. doi: 10.1042/BST0290196. [DOI] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Lee HJ, Carroll D (2010a) Genome editing with modularly assembled zinc-finger nucleases. Nat Methods 7:91 (author reply 91–92) [DOI] [PMC free article] [PubMed]
- Kim S, Lee MJ, Kim H, Kang M, Kim JS (2010b) Pre-assembled zinc finger arrays for rapid construction of zinc finger nucleases. Nat Methods (in press) [DOI] [PubMed]
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Lee DK, Seol W, Kim JS. Custom DNA-binding proteins and artificial transcription factors. Curr Top Med Chem. 2003;3:645–657. doi: 10.2174/1568026033452384. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PQ, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, Collingwood TN, Gregory PD. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol Bioeng. 2010;106:97–105. doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- Lloyd A, Plaisier CL, Carroll D, Drews GN (2005) Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci USA 102:2232–2237 [DOI] [PMC free article] [PubMed]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Muller-Lerch F, Fu F, Pearlberg J, Gobel C, Dassie JP, Pruett-Miller SM, Porteus MH, Sgroi DC, Iafrate AJ, Dobbs D, McCray PB, Jr, Cathomen T, Voytas DF, Joung JK. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK (2009) Oligomerized pool engineering (OPEN): an “open-source” protocol for making customized zinc-finger arrays. Nat Protoc 4:1471–1501 [DOI] [PMC free article] [PubMed]
- Mandell JG, Barbas CF 3rd (2006) Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res 34:W516–W523 [DOI] [PMC free article] [PubMed]
- Marton I, Zuker A, Shklarman E, Zeevi V, Tovkach A, Roffe S, Ovadis M, Tzfira T, Vainstein A. Nontransgenic genome modification in plant cells. Plant Physiol. 2010;154:1079–1087. doi: 10.1104/pp.110.164806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, Serikawa T. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS One. 2010;5:e8870. doi: 10.1371/journal.pone.0008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Klug A, Choo Y. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc Natl Acad Sci USA. 2001;98:1437–1441. doi: 10.1073/pnas.98.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci USA. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai H, Fujita K, Suzuki K, Nishikawa M, Shibata T, Sakamoto N, Yamamoto T. Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells. 2010;15:875–885. doi: 10.1111/j.1365-2443.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- Orlando SJ, Santiago Y, DeKelver RC, Freyvert Y, Boydston EA, Moehle EA, Choi VM, Gopalan SM, Lou JF, Li J, Miller JC, Holmes MC, Gregory PD, Urnov FD, Cost GJ. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Osakabe Y, Toki S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc Natl Acad Sci USA. 2010;107:12034–12039. doi: 10.1073/pnas.1000234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- Park KS, Lee DK, Lee H, Lee Y, Jang YS, Kim YH, Yang HY, Lee SI, Seol W, Kim JS. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat Biotechnol. 2003;21:1208–1214. doi: 10.1038/nbt868. [DOI] [PubMed] [Google Scholar]
- Park KS, Jang YS, Lee H, Kim JS. Phenotypic alteration and target gene identification using combinatorial libraries of zinc finger proteins in prokaryotic cells. J Bacteriol. 2005;187:5496–5499. doi: 10.1128/JB.187.15.5496-5499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petolino JF, Worden A, Curlee K, Connell J, Strange Moynahan TL, Larsen C, Russell S. Zinc finger nuclease-mediated transgene deletion. Plant Mol Biol. 2010;73:617–628. doi: 10.1007/s11103-010-9641-4. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993;21:5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, Cathomen T, Voytas DF, Joung JK. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy S, Tesson L, Menoret S, Usal C, Scharenberg AM, Anegon I. Zinc-finger nucleases: a powerful tool for genetic engineering of animals. Transgenic Res. 2010;19:363–371. doi: 10.1007/s11248-009-9323-7. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–W605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38(Suppl):W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459:437–441 [DOI] [PubMed]
- Siekmann AF, Standley C, Fogarty KE, Wolfe SA, Lawson ND. Chemokine signaling guides regional patterning of the first embryonic artery. Genes Dev. 2009;23:2272–2277. doi: 10.1101/gad.1813509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O. Forty years with homologous recombination. Nat Med. 2001;7:1083–1086. doi: 10.1038/nm1001-1083. [DOI] [PubMed] [Google Scholar]
- Sollu C, Pars K, Cornu TI, Thibodeau-Beganny S, Maeder ML, Joung JK, Heilbronn R, Cathomen T (2010) Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res. doi:10.1093/nar/gkq720 [DOI] [PMC free article] [PubMed]
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Takasu Y, Kobayashi I, Beumer K, Uchino K, Sezutsu H, Sajwan S, Carroll D, Tamura T, Zurovec M. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem Mol Biol. 2010;40:759–765. doi: 10.1016/j.ibmb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Tovkach A, Zeevi V, Tzfira T. A toolbox and procedural notes for characterizing novel zinc finger nucleases for genome editing in plant cells. Plant J. 2009;57:747–757. doi: 10.1111/j.1365-313X.2008.03718.x. [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc Natl Acad Sci USA. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 2005;44:693–705. doi: 10.1111/j.1365-313X.2005.02551.x. [DOI] [PubMed] [Google Scholar]
- Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, Joung JK. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, Joung JK, Voytas DF (2010) High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci USA 107:12028–12033 [DOI] [PMC free article] [PubMed]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, Porteus MH, Joung JK, Cheng L. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]


