Abstract
Cernunnos/XLF is a core protein of the nonhomologous DNA end-joining (NHEJ) pathway that processes the majority of DNA double-strand breaks in mammals. Cernunnos stimulates the final ligation step catalyzed by the complex between DNA ligase IV and Xrcc4 (X4). Here we present the crystal structure of the X41–157-Cernunnos1–224 complex at 5.5-Å resolution and identify the relative positions of the two factors and their binding sites. The X-ray structure reveals a filament arrangement for X41–157 and Cernunnos1–224 homodimers mediated by repeated interactions through their N-terminal head domains. A filament arrangement of the X4–Cernunnos complex was confirmed by transmission electron microscopy analyses both with truncated and full-length proteins. We further modeled the interface and used structure-based site-directed mutagenesis and calorimetry to characterize the roles of various residues at the X4–Cernunnos interface. We identified four X4 residues (Glu55, Asp58, Met61, and Phe106) essential for the interaction with Cernunnos. These findings provide new insights into the molecular bases for stimulatory and bridging roles of Cernunnos in the final DNA ligation step.
Keywords: nonhomologous end-joining, protein–protein interaction, DNA repair, V(D)J recombination, immunodeficiency
DNA double-strand breaks (DSBs) are the most toxic DNA lesions in the genome, and unrepaired DSBs can cause large-scale losses of genetic information through chromosome rearrangement (1, 2). These DNA damages result from exposure to exogenous damaging agents, such as ionizing radiation, radiomimetic compounds, and topoisomerase inhibitors. DSBs are also obligate intermediates in several recombination processes in vertebrates, including antigen receptor gene rearrangement, V(D)J recombination (3, 4). In higher eukaryotes, DSBs are repaired by several mechanisms, among which nonhomologous end-joining (NHEJ) represents the major pathway, particularly when sister chromatids are not available (5). Deficiency in the NHEJ machinery results in sensitivity to ionizing radiation and severe combined immune deficiencies in humans and mice due to abortive V(D)J recombination (6). NHEJ is orchestrated by at least seven proteins. The Ku70/Ku80 heterodimer adopts a preformed ring-shaped structure that recognizes and encircles the duplex DNA ends at the DSB (7). Ku70/Ku80 recruits a 469-kDa serine/threonine protein kinase, the DNA-PK catalytic subunit (DNA-PKcs), via a direct interaction and shifts about 10 bp inward so that DNA-PKcs acquires a position at the terminus through its large open-ring cradle structure (8). Upon association with Ku and DNA, DNA-PKcs is activated and phosphorylates several proteins including itself and Ku70/Ku80. The DNA-PK holoenzyme, constituted by Ku and DNA-PKcs, plays a central role in NHEJ. Among other functions, DNA-PK mediates the end-bridging of the DSB extremities (9), regulates access to the DNA ends by processing enzymes such as the DNA-PKcs-associated Artemis nuclease (10, 11), and recruits the Xrcc4–ligase IV complex to DNA ends for the ligation step (12). The Xrcc4–ligase IV complex carries out the final joining of synapsed DNA ends in association with a third ligation factor, called Cernunnos/XLF. Cernunnos was identified through complementation of a fibroblast cell line obtained from a human patient with immunodeficiency and microcephaly (13). The same factor, referred to as XLF for Xrcc4-like factor, was identified through a yeast two-hybrid screen for proteins that interact with Xrcc4 (14) (for simplicity, we will subsequently refer to ligase IV, Xrcc4 and Cernunnos/XLF as L4, X4 and Cernunnos, respectively). Sequence analyses revealed that Cernunnos and X4 belong to the same structural family, which also includes Nej1, the yeast ortholog of Cernunnos (15), and Nej1 was shown to interact with Lif1, the yeast ortholog of X4 (16).
The L4–X4 interaction is extremely tight in vivo and is resistant to high ionic strength in vitro (17, 18). Cells deficient for X4 present no stable L4 (19), whereas L4-deficient cells present a reduced level of X4 (20). Structural studies of the L4–X4 complex revealed a stoichiometry of one L4 to one X4 homodimer and extensive protein–protein interactions mediated by the coiled-coil tails of X4 and the carboxy-terminal tandem BRCT repeat of L4 (21, 22). Cernunnos and the L4–X4 complex coprecipitate from crude extract (14, 15) through an interaction sensitive to ionic strength (20). The association between Cernunnos and L4–X4 complex is mediated primarily through an interaction with X4 (14), although a weak interaction with L4 has also been reported (16). Cernunnos stimulates the ligase activity of the L4–X4 complex (20, 21, 23–25) and seems particularly important for the ligation of mismatched or noncohesive DNA ends (25, 26). Like X4, Cernunnos is a homodimer (27, 28). It presents important structural similarities with X4, which include its N-terminal globular head domain (amino acids 1–127) and a central coiled-coil (amino acids 128–169). The Cernunnos coiled-coil is 60 Å shorter than that of X4 and folds back toward the N terminus. Both X4 and Cernunnos have C-terminal regions that are predicted to be unstructured.
Two-hybrid mapping and mutagenesis studies showed that X4 and Cernunnos interact through their N-terminal head domains (16, 27, 29). In particular, single mutations at any of three Cernunnos residues (R64, L65, and L115) disrupt its interaction with X4 and abrogate Cernunnos function in several DNA repair assays, despite correct Cernunnos expression and localization (29). Low-resolution models of an X4–Cernunnos complex obtained by small angle X-ray scattering (SAXS) were recently reported (30). The results of these experiments supported a head-to-head interaction model between X4 and Cernunnos and predicted a mixture of oligomers, from tetramers to octamers. While this study suggested that X4 and Cernunnos can form filaments, no direct observation of such high-order arrangement has been reported.
Here we provide insights into the molecular interactions of Cernunnos during the DSB ligation step. We solved the crystal structure of the X41–157-Cernunnos1–224 complex at 5.5-Å resolution and used seleno-methionine (SeMet) labeling to unambiguously determine the positions of both factors. We observed that X4 and Cernunnos produced filament arrangements in the crystal through repetition of the same interface. The capacity of the X4–Cernunnos complex to form filaments was further confirmed by transmission electron microscopy with truncated and full-length proteins. Finally, the individual contribution of interface residues to the stability of the complex was analyzed by mutagenesis and calorimetry.
Results
Crystal Structure of the X41–157-Cernunnos1–224 Complex Demonstrates an Interaction Through the N-Terminal Head Domains.
To crystallize the X4–Cernunnos complex, one Cernunnos construct, 1–224, and two X4 constructs, 1–203 and 1–157, were purified separately (see Materials and Methods and Fig. S1A). The shorter X41–157 construct corresponded to a protein with a central coiled-coil that contained approximately 12 fewer helical turns (46 amino acids) than X41–203 construct. X41–157 did not contain the X4 residues involved in the L4 interaction (154–188) (21). Calorimetry measurements (Table 1) showed that X41–157 interacted with Cernunnos with a dissociation constant (Kd) of 4.1 ± 0.2 μM, which was similar to that obtained previously in studies using the longer construct X41–203 (Kd = 3.9 ± 0.2 μM) (29). We obtained crystals only with the X41–157-Cernunnos1–224 complex. These crystals diffracted to 5.5-Å resolution on the synchrotron beamline PX1 (SOLEIL, France). They presented a large cell parameter of 856 Å with a hexagonal symmetry, P6522. Molecular replacement allowed positioning two X4 homodimers (named X4-A and -B) and two Cernunnos homodimers (named Cernunnos-A and -B) in the asymmetric unit, using the individual crystal structures of X4 and Cernunnos (21, 27) (see Table S1 for the data collection and refinement statistics).
Table 1.
Analysis of X4-Cernunnos interactions by calorimetry
| Cernunnos * | Xrcc4 * | Kd (μM) | Commentary | Reference |
| WT1–224 | WT1–203 | 3.9 ± 0.2 | 29 | |
| WT1–224 | WT1–157 | 4.1 ± 0.2 | X4(1-157) central coiled-coil 60 Å shorter than X4(1–203) | this study |
| WT1–224 | E55R1–157 | no interaction | in helix α1, salt bridge with R64C | this study |
| WT1–224 | D58R1–157 | no interaction | in helix α1, close to E55C and R64C | this study |
| WT1–224 | M61R1–157 | no interaction | in loop α1–α2, in contact with P116C and L115C | this study |
| WT1–224 | E62R1–157 | 3.8 ± 0.2 | in helix α2, salt bridge with K65X | this study |
| WT1–224 | K65E1–157 | > 40 | in helix α2, salt bridge with E111C and E62X | this study |
| WT1–224 | E69R1–157 | 5.0 ± 0.2 | in helix α2, salt bridge with K99X and hydrogen bond with Y84X | this study |
| WT1–224 | F106E1–157 | no interaction | in strand β7, in contact with L65C, A67C, F72C, and L112C | this study |
| WT1–224 | F106A1–157 | > 40 | see above | this study |
| WT1–224 | F106Y1–157 | > 40 | see above | this study |
| E111K1–224 | WT1–157 | 6.9 ± 0.2 | in strand β6, salt bridge with K65X | this study |
| R64E1–224 | WT1–203 | no interaction | in loop α2–α3, salt-bridge with E55X and close to D58X | 29 |
| L65D1–224 | WT1–203 | > 80 | in loop α2–α3, in contact with F106X | 29 |
| L115D1-224 | WT1–203 | no interaction | in strand β7, in contact with F106XM61X, M59X and L108X | 29 |
*The limits of the constructs used for X4 and Cernunnos are indicated in superscript.
The electron density of the X4 and Cernunnos main chains was clearly visible in the composite omit annealed electron density map (Fig. S2 A and B). The individual crystal structures of X4 and Cernunnos showed that these proteins share structural homology in their N-terminal head domains but presented key differences that allowed them to be identified in the electron density at 5.5-Å resolution. The first major difference concerned the C-terminal helical domain of the proteins, which is a single protruding helix in X4 and consists of three helices in Cernunnos. In particular, the Cernunnos helices α5 and α6 fold back toward the N terminus. A second major difference is that when the coiled-coil domains of the Cernunnos and the X4 coiled-coil tails are superimposed, the trajectories of their N-terminal domains differed by 45°. This is due to the presence of the α6 helix between the head domain and the coiled-coil of Cernunnos. The electron densities of the head domains, of the coiled-coils, and of the Cernunnos helices α5 and α6 allowed us to unambiguously discriminate both proteins. Nonetheless, to further validate this structure, data were collected on crystals of the X4–Cernunnos complex using SeMet-Cernunnos molecules. Electron density maps obtained from these crystals confirmed the solution obtained by molecular replacement and exhibited peaks between 3.7 σ and 5.9 σ levels in the anomalous difference Fourier maps on eight out of the nine Cernunnos methionines (Fig. S2C).
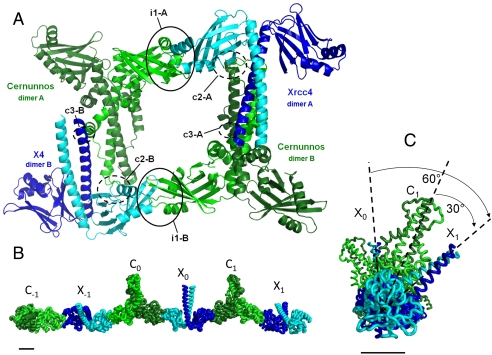
The X41–157 and Cernunnos1–224 homodimers present in the asymmetric unit contact each other at three sites (named “i1,” “c2,” and “c3”), with each one occurring twice (designated as A and B) within the asymmetric unit (Fig. 1A). The distal regions of the X4 and Cernunnos N-terminal head domains form the main interface, i1, which in X4 involves amino acids 55–69 in helix α1, loop α1-α2, helix α2, plus amino acids 99–108 in strand β6, loop β6–β7 and strand β7. The Cernunnos contribution to the i1 interface involves two regions: amino acids 64–72 located in loop α2–α3 and helix α3, and amino acids 109–118 in strands β6, loop β6–β7 and strand β7 (see X4 and Cernunnos multiple sequence alignment in Fig. S3A). The total area buried by X4 and Cernunnos in the i1 interface is approximately 1,300 Å2. The complex has a pseudosymmetric character because the interface involves the mutually corresponding loops and secondary structures of the Cernunnos and X4 N-terminal head domains. The second and third contact regions, c2 and c3, are likely crystal contacts because they present buried areas far smaller than that of i1 (approximately 600 Å2 for c2 and approximately 200 Å2 for c3) and were not observed in the X4–Cernunnos(SeMet) crystal form (see Fig. S2D for detailed view of c2 contact region).
Fig. 1.
Crystal structure of X4–Cernunnos/XLF complex. (A) The asymmetric unit of the crystal contains two complexes consisting of a X4 homodimer (dark and light blue) and a Cernunnos homodimer (dark and light green). These two complexes interact through a main interface “i1” and two contact regions “c2” and “c3” that are likely crystal contacts. Each of these interfaces and contact regions are found twice (A and B). Interfaces i1-A and i1-B (black circles) are mediated by the head domains. (B) Filament arrangement of the Cernunnos–X4 complex in the crystal. Three Cernunnos homodimers (dark and light green, named C-1, C0, C1) and three X4 homodimers (dark and light blue, named X-1, X0, X1) are shown in a ribbon representation. Each N-terminal head of the X4 and Cernunnos dimers interacts with the same i1 interface all along the c axis of the crystal. (C) The same representation as in B with a rotation of 90° around the vertical axis and a higher zoom. This view shows that X4 and Cernunnos coiled-coils are tilted at an angle of 30° with respect to each other (dashed lines). The coiled-coil tails of two consecutive Cernunnos or two consecutive X4 are thus tilted by 60°, in agreement with the crystal symmetry 65 element. Bars represent 2 nm.
These results are consistent with an analysis of the domains required for the X4–Cernunnos interaction (16) and with previously reported site-directed mutagenesis studies (27, 29), all of which identified the i1 interface, involving the X4 and Cernunnos N-terminal head domains, as the primary interface.
The X41–157-Cernunnos1–224 Complex Presents a Filament Arrangement in the Crystal.
Analysis of the crystal packing revealed that the second head domain of each X4 and Cernunnos homodimer present in the asymmetric unit interacts with a second Cernunnos head domain and a second X4 head domain, respectively. This is generated by the 65 crystal symmetry element that corresponds to a rotation of -60° and a translation of one-sixth of the unit cell c along this axis (Fig. 1 B and C and Fig. S2E). These interfaces with symmetrical molecules superimpose well with the i1 interface observed in the asymmetric unit (rmsd of 0.7 Å over 1,048 X4 and Cernunnos main-chain atoms). All X4–Cernunnos complexes present in the crystal are organized as parallel filaments, wrapping along the c axis of the unit cell (Fig. S2G). The crystal form obtained with the SeMet–Cernunnos protein belongs to a related spacegroup (P6422, a = b = 105 Å, c = 427 Å), with an asymmetric unit of half the size of the native complex and only one X4–Cernunnos complex. The overall arrangement is very similar with parallel filaments (Fig. S2F). The change in spacegroup arises from a small reorientation of the X4 and Cernunnos molecules at the interface (translation of 1.5 Å and rotation of 1 °).
Electron Microscopy Analyses Further Confirm the Filament Arrangement of X41–157-Cernunnos1-224 Complex and Reveal a Filament Arrangement for the Full-Length X4 and Cernunnos Proteins.
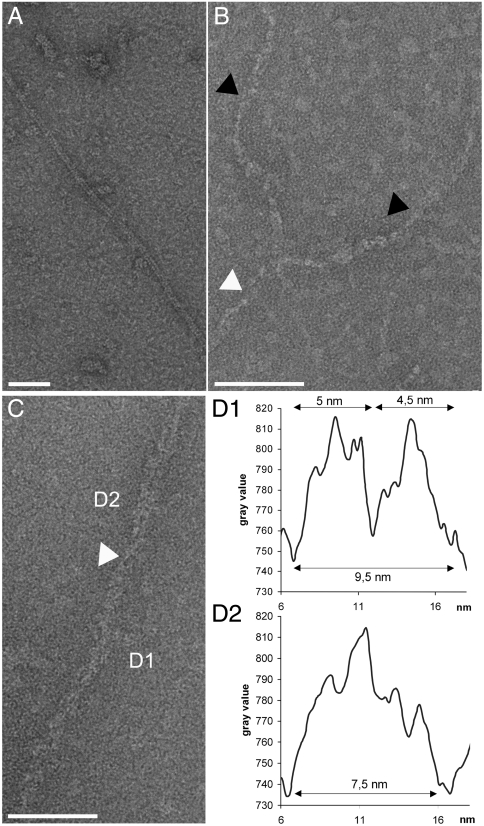
We next used transmission electron microscopy (TEM) to further explore the ability of X4, Cernunnos, and the X4–Cernunnos complex to make filaments. We reproducibly observed that the X4–Cernunnos complex formed filaments of various thicknesses and with lengths ranging from 0.5 μm to several μm. Most filaments were approximately 10 nm thick (Fig. 2A and Fig. S5A). These 10-nm filaments were homogenous in diameter, curvilinear, and followed variable trajectories. TEM micrographs showed that most prevalent 10-nm filaments were composed of two thinner filaments (Fig. 2 B and C and a schematic representation in Fig. S5D), which were wrapped around each other in a helical manner (Fig. 2B, black arrow). Cross-sectional measurements of the 10-nm filaments confirmed that they were made of two thinner filaments of about 5 nm in diameter (two peaks, Fig. 2 C and D). The loss of two peaks on the profile suggested that the 10-nm filament had rotated on itself. In some micrographs, the 10-nm filaments appeared to cross each other with a helical arrangement (Fig. 2B, white arrow). We also observed filaments having a thickness of approximately 20 nm and a more regular structure (Fig. S5E). Long filaments were specific to the X4–Cernunnos complex as neither the X4 nor the Cernunnos proteins alone adopted this conformation, although we did observe short oligomers with the X4 protein alone (maximum particle length approximately 50 nm) (Fig. S5 B and C).
Fig. 2.
Electron micrographs of Xrcc4–Cernunnos complexes and of the Xrcc4 and Cernunnos proteins alone. (A) The X4–Cernunnos complex reproducibly forms filaments with a homogeneous thickness (approximately 10 nm) and with lengths ranging from 0.5 μm to several μm (28,980× magnification). (B) The most prevalent 10-nm filaments (white arrow) are composed of two thin filaments (black arrows) wrapped in a helical shape in some micrographs (63,000× magnification). The individual thin filaments also adopt a helical configuration as illustrated by the variation of their apparent diameter. (C and D) Gray level profiles of a cross section of a filament integrated over a length of 50 nm (D1 and D2). In the D1 section, two peaks were observed on the graph and show that two 5-nm filaments constitute the 10-nm filament. The white arrow marks a cross between two 5-nm filaments. The gray level profile of D2 presents a single signal suggesting that the filament is rotated on itself (63,000× magnification).
We then purified X4 and Cernunnos full-length (FL) proteins to evaluate the influence of the C-terminal regions of these proteins on their direct interaction and on their propensity to form filaments alone or in complex. Using size-exclusion chromatography, we detected a direct interaction between X4FL and CernunnosFL (Fig. S1B). By TEM, we reproducibly observed that X4FL/CernunnosFL complexes formed filaments approximately 20–25 nm thick, which had a tight braid-like structure (Fig. S5F). These particular filaments were specific to X4FL-CernunnosFL complexes. X4FL alone made some filaments, though with different geometries, whereas CernunnosFL alone made only a very few straight and regular filaments, which appeared rarely in some micrographs.
In summary, the crystal structure and TEM micrographs of X41–157-Cernunnos1–224 complexes present two independent direct visualizations of X4–Cernunnos filaments. Analysis of the full-length proteins by TEM suggested that complexes of these X4 and Cernunnos proteins, which included their respective C-terminal regions, also formed filaments.
Insight into the X41-157-Cernunnos1-224 Interface Through Molecular Modeling.
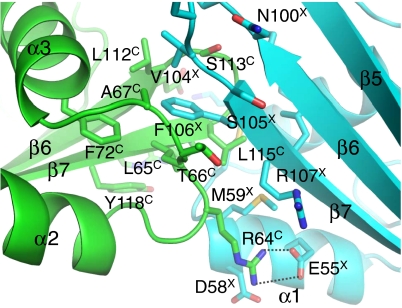
The 5.5-Å resolution electron density maps allowed the X4 and Cernunnos main-chain traces to be positioned but were insufficient to determine the side-chain conformations. We next modeled the complex between the Cernunnos and X4 head domains using a module from the Rosetta software that allows modeling within the constraints of electron density maps (31). The model superimposed with an rmsd of 1.4 Å over 188 X4 and Cernunnos main-chain atoms of residues at the interface. The X4–Cernunnos model obtained with Rosetta involved 15 X4 residues and 13 Cernunnos residues (Fig. 3). The center of the interface consisted of hydrophobic residues from X4 (M59, M61, L101, V104, and F106) and from Cernunnos (L65, A67, F72, L112, L115, P116, and Y118). In the Rosetta model, the interaction involved six hydrogen bonds and two salt bridges (between E55X and R64C and between K65X and E111C) at the periphery (Fig. 3 and Fig. S3B).
Fig. 3.
Interface of the X41–157-Cernunnos1–224 complex formed by the distal part of their N-terminal head domains. Residues involved in the interface between X4 (cyan) and Cernunnos (green) are shown in a stick representation. Residues are labeled with “X” and “C” superscripts for X4 and Cernunnos, respectively. Dashed lines indicate inter- and intra-molecular hydrogen bonds and salt bridges between side-chain atoms proposed from Rosetta modeling.
Energetic Characterization of the Individual Contribution of X41–157 and Cernunnos1-224 Interface Residues by Calorimetry.
In order to characterize the energetic contribution of the residues at the X4–Cernunnos interface, we purified nine X4 mutants and measured the impact of the corresponding mutations on the interaction using isothermal titration calorimetry (ITC). Based on our structural data, we targeted three groups of residues for mutation. The first group consisted of the hydrophobic X4 residues, M61X and F106X, which stack against Cernunnos residues L65C, A67C, L112C, P116C, and L115C. These residues were mutated into charged residues, M61RX and F106EX, and F106 was also mutated into alanine and tyrosine. The second group included the charged residues E55X and K65X, which are involved in salt bridges with Cernunnos residues R64C and E111C, respectively. This group also included residue D58X, located near the salt bridge between E55X and R64C (Fig. 3). We reversed the charges of these residues by making the substitutions E55RX, K65EX, and D58RX. We also measured the contribution made by Cernunnos residue E111, which makes a salt bridge with K65X in the Rosetta model, by testing an E111KC mutant. The third group included charged residues present at the edges of the interfaces. We thus analyzed X4 mutants E62RX and E69RX.
All the X41–157 and Cernunnos1–224 variants were purified to homogeneity and their overall structural integrity was verified by analytical size-exclusion chromatography (Fig. S1C). ITC measurements were performed in duplicate. X4 WT or variant proteins in the cell were titrated by WT or variant Cernunnos proteins (Table 1 and Fig. S3 for the thermograms). Within the first group of mutated residues, mutations at the hydrophobic positions, M61RX and F106EX, led to a complete loss of interaction between X4 and Cernunnos. This result is in good agreement with the central position of these residues in the X4–Cernunnos interface of the Rosetta model. In the second group, the modification of two charged residues by the E55RX and K65EX substitutions also impaired the X4–Cernunnos interaction. The X4 substitution E55RX, like the Cernunnos variant R64EC (29), completely disrupted interaction, which is in agreement with the salt bridge between E55X and R64C proposed by the Rosetta model. The K65EX variant reduced interaction by a factor at least ten compared to WT X4, agreeing with its potential role in a salt bridge with residue E111C. Mutation of the other member of this salt bridge pair, represented by the E111KC variant, did not diminish interaction. However, analysis of the interface suggested that introduction of a lysine at Cernunnos position 111 could result in a salt bridge with position E62X and thus compensate for the loss of interaction with residue K65X (Fig. S3B). The last substitution of the second group, D58RX, also disrupted the X4–Cernunnos interaction. Position D58X is located close to the aforementioned E55X/R64C salt bridge (Fig. 3). Introduction of a positive charge close to these two residues could indirectly alter the strength of their salt bridge. Another possibility is that R64C could interact with both of the negatively charged residues, D58X and E55X, due to their proximity. Finally, in the third group of mutations, the two variants harboring substitutions near the edge of the putative interface (E62RX and E69RX) interacted with Cernunnos at near WT levels. With these calorimetry experiments, the four X4 residues (E55X, D58X, M61X, and F106X) that we found to be essential for interaction with Cernunnos were in good agreement with the X4–Cernunnos Rosetta model.
Discussion
The low-resolution crystal structure presented here allowed us to define an unambiguous head-to-head interaction between X41–157 and Cernunnos1-224. This structure is consistent with the mutagenesis data presented by Andres et al., which identified roles for the X4 residues K65X and K99X and for the Cernunnos residue L115C (27). In our Rosetta model, Cernunnos position L115C is buried in the i1 interface and is in contact with four X4 hydrophobic residues (M59X, M61X, F106X, and L108X). X4 position K65X makes an intramolecular salt bridge with E62X and an intermolecular salt bridge with Cernunnos E111C. From our calorimetry results on Cernunnos variant E111KC, we propose that the K65(E/Q)X mutants affect the X4–Cernunnos interaction through local rearrangement of the X4 interaction site. Position K99X makes an intramolecular salt bridge with E69X and is in van der Waals contact with Cernunnos main-chain residues S113C-G114C in the β6–β7 loop that is central in the interaction. Our results suggest that the variant K99EX may affect the X4 interaction site structure and perturb the contacts made with Cernunnos near loop β6–β7. The complex presented here is also compatible with yeast two-hybrid and coprecipitation studies suggesting that X4 and Cernunnos interact through their globular heads (16). Intriguingly, this last study also showed that the Saccharomyces cerevisiae Lif1 and Nej1 proteins, the respective homologs of X4 and Cernunnos, interact with each other through the head domain of Lif1 but through the C-terminal region of Nej1 (16).
Our crystal structure and electron microscopy analyses clearly showed that X41–157-Cernunnos1-224 complexes can adopt a filamentous organization and that complexes formed by the full-length proteins also form filaments detectable by TEM. This property arises from the distal positions of the interaction sites on both protein head domains and from the homodimeric nature of both X4 and Cernunnos. This observation agrees with low-resolution models obtained by SAXS using X41–140 and Cernunnos1–248 constructs (30). In this study, the authors obtained the best fit to their experimental data by mixing tetramer, hexamer, and octamer assemblies of X4–Cernunnos. The best fitting model obtained by SAXS included a tilt angle of approximately 45° between the X41–140 and Cernunnos1–248 coiled-coil regions, which is close to the approximately 30° angle observed in our crystal structure with the X41–157-Cernunnos1-224 complex.
Most of the filaments observed in the electron micrographs of X4–Cernunnos complexes were 10 nm in diameter and the TEM micrographs indicated that these filaments were composed of two thinner filaments having diameters of 5 nm. The maximum dimensions of the X41–157 and Cernunnos1-224 molecules, deduced from their crystal structures, suggest that the 5-nm filaments correspond to linear chains having the width of a single X41–157-Cernunnos1–224 complex as shown in Fig. 1B. The preponderance of the 10-nm filaments on the electron micrographs suggests that individual 5-nm filaments are prone to “dimerize” through additional interactions mediated either by X4/X4, Cernunnos/Cernunnos or X4/Cernunnos contacts.
Do these in vitro observed filaments have any physiological relevance in vivo? Further cellular microscopy analyses will help to evaluate this issue. It would be informative to evaluate the stability and the structure of these filaments in the presence of L4 and/or DNA, either or both of which might somehow modify this organization. A superimposition of the structure of the complex between X41–203 and the C-terminal domain of L4654–911 (21) with the structure of the X41–157-Cernunnos1–224 complex suggests that X4 can bind simultaneously Cernunnos1–224 through the i1 interface and L4(CTD)654–911 (Fig. S4A). The low-resolution structure of the L4FL-X4FL complex, recently determined by cryoelectron microscopy (32), suggests that the large catalytic domain of L4 could be located near the i1 X41–157-Cernunnos1–224 head-to-head interaction site that we’ve described here. The crystal structure of the L4–X4–Cernunnos complex with the full-length proteins should help to clarify the binding mode of the complete ligation complex. DSB repair studies in the presence or absence of Cernunnos have shown that Cernunnos promotes DNA end ligation by the L4–X4 complex (25, 33). From our observation, we propose that the oligomerization of the X4–Cernunnos complex could be a mechanism to facilitate the recruitment of two or more L4–X4 complexes to DNA DSB repair points (Fig. S4B).
A recent structural study reported that the centriolar homodimeric protein, SAS-6, which belongs to the X4-fold family, forms oligomeric rings which have ninefold symmetry (34). Interestingly, these SAS-6 oligomeric rings are mediated by its N-terminal domain and by loops equivalent to the ones involved in forming the X4 and Cernunnos in the filaments presented here. The results with SAS-6 suggest that the X4-fold may represent a favorable structural template for forming higher order oligomeric structures in different cellular contexts.
Material and Methods
Sample Preparation, Crystallization and Structure Determination.
X41–157 and Cernunnos1–224 were produced as previously described (29). The X41–336 and Cernunnos1–299 full-length proteins were purified as described in SI Text. Production of Cernunnos1–224 labeled with seleno-methionine was performed with the same protocol, except that the cellular cultures were performed in minimal medium supplemented with most amino-acids and seleno-methionine (see SI Text for the detailed protocol). Crystals of the X4–Cernunnos complex were grown by vapor diffusion from a solution containing both proteins at a stoichiometry of 1∶1. We performed the crystallization screens at the HTX platform (EMBL, Grenoble). The optimum condition was obtained by mixing 2 μL of the X4–Cernunnos complex (5.3 mg/mL) with 2 μL of a reservoir solution containing 9% v/v MPD (methylpentanediol), 50 mM MgSO4 and 0.1 M sodium cacodylate buffer at pH 6.5. All X-ray diffraction data were collected on the Proxima 1 beamline (SOLEIL Synchrotron, France). The native crystals diffracted to a resolution of 5.5 Å. The crystals belonged to the spacegroup P6522 with a large cell parameter c of 856 Å (see Table S1). Due to a small crystal mosaicity (measured by XDS between 0.1 and 0.2 °), and to the alignment of the c axis along the spindle axis, there were no significant overlap problems. Incompleteness arose in the highest resolution shell due to the strong anisotropy of diffraction. The mean intensity of the Bragg reflections in the ab direction falls off quickly after 8 Å, whereas in the c direction diffraction can be clearly seen beyond 4 Å. The structure was determined by molecular replacement using MolRep (35) and the individual crystal structures of Cernunnos homodimer (PDB ID code 2R9A) (27) and of X4 homodimer (PDB ID code 3II6) (21). A rigid body refinement was obtained with the Buster program (36). A full composite torsion annealed omit electron density maps was calculated with the CNS program version 1.3 using strong NCS restraints (37). The crystals of the X4–(SeMet)Cernunnos complex were obtained in similar conditions. These crystals diffracted to a resolution of 6.6 Å and belonged to the spacegroup P6422 with a cell parameter c of 427 Å. The crystal structure was determined by molecular replacement. The anomalous difference Fourier maps were calculated using the program Coot (38), and phases were obtained after constrained refinement with the Buster program. Figures were prepared with PyMOL (PyMOL Molecular Graphics System, Schrödinger, LLC).
Transmission Electron Microscopy (TEM).
For negative staining, the stock solutions are diluted in salt buffers (Tris 10 mM, pH 7.5, plus 50 or 150 mM NaCl). We selected the dilution conditions to observe the X4–Cernunnos complex on the grid support with homogeneous distributions. 5 μL of solution were adsorbed onto a 300-mesh copper grid coated with a collodion film covered by a thin carbon film, and activated by glow-discharge. After 1 min, grids were washed with aqueous 2% w/vol uranyl acetate (Merck) and then dried with ashless filter paper (VWR). TEM observations were carried out with a Zeiss 912AB transmission electron microscope in filtered zero loss mode. Electron micrographs, profiles of gray level and measurements were obtained using a ProScan 1024 HSC digital camera and Soft Imaging Software system.
Isothermal Titration Calorimetry (ITC).
The calorimetric titration experiments were performed as previously described (29).
Rosetta Modeling.
Molecular modeling with Rosetta was initiated with the crystal structure refined to 5.5-Å resolution with Buster (for details see Fig. S2)
Supplementary Material
Acknowledgments.
We thank S. Marcand, B. Gilquin, and S. Zinn-Justin for critical comments and helpful discussions; C. Tellier, M. Gondry (Commissariat à l'Energie Atomique et aux Energies Alternatives, Institut de Biologie et de Technologies de Saclay), and G. Stier (European Molecular Biology Laboratory, Heildelberg) for gifts of expression plasmids; and R. Thai (Commissariat à l'Energie Atomique et aux Energies Alternatives, Institut de Biologie et de Technologies de Saclay) for mass spectrometry analyses. This work was supported by institutional grants from the Institut National du Cancer/Cancéropôle Ile-de-France. V.R. is financed by Institut National du Cancer. We thank the European infrastructure P-CUBE for access to the HTX platform. We thank synchrotron European Synchrotron Radiation Facility (Grenoble, France) for access to beamlines ID23-1, ID23-2, and ID29 at the beginning of the project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and the structure factors of the X4–Cernunnos complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3Q4F).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100758108/-/DCSupplemental.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 4.de Villartay JP. V(D)J recombination deficiencies. Adv Exp Med Biol. 2009;650:46–58. doi: 10.1007/978-1-4419-0296-2_4. [DOI] [PubMed] [Google Scholar]
- 5.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Revy P, Buck D, le Deist F, de Villartay JP. The repair of DNA damages/modifications during the maturation of the immune system: Lessons from human primary immunodeficiency disorders and animal models. Adv Immunol. 2005;87:237–295. doi: 10.1016/S0065-2776(05)87007-5. [DOI] [PubMed] [Google Scholar]
- 7.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 8.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2009;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moshous D, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 11.Goodarzi AA, et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calsou P, Delteil C, Frit P, Drouet J, Salles B. Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment. J Mol Biol. 2003;326:93–103. doi: 10.1016/s0022-2836(02)01328-1. [DOI] [PubMed] [Google Scholar]
- 13.Buck D, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 14.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 15.Callebaut I, et al. Cernunnos interacts with the XRCC4 x DNA–ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J Biol Chem. 2006;281:13857–13860. doi: 10.1074/jbc.C500473200. [DOI] [PubMed] [Google Scholar]
- 16.Deshpande RA, Wilson TE. Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4. DNA Repair (Amst) 2007;6:1507–1516. doi: 10.1016/j.dnarep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grawunder U, et al. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 18.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 19.Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: Evidence for stabilization by XRCC4. Mutat Res. 1999;433:53–8. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- 20.Riballo E, et al. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu PY, et al. Structural and functional interaction between the human DNA repair proteins DNA Ligase IVand XRCC4. Mol Cell Biol. 2009;29:3163–3172. doi: 10.1128/MCB.01895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sibanda BL, et al. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- 23.Hentges P, et al. Evolutionary and functional conservation of the DNA non-homologous end-joining protein, XLF/ Cernunnos. J Biol Chem. 2006;281:37517–37526. doi: 10.1074/jbc.M608727200. [DOI] [PubMed] [Google Scholar]
- 24.Lu H, Pannicke U, Schwarz K, Lieber MR. Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. J Biol Chem. 2007;282:11155–11162. doi: 10.1074/jbc.M609904200. [DOI] [PubMed] [Google Scholar]
- 25.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci USA. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu J, et al. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal structure of human XLF: A twist in nonhomologous DNA end-joining. Mol Cell. 2007;28:1093–1101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, et al. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. EMBO J. 2008;27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malivert L, et al. Delineation of the Xrcc4-interacting region in the globular head domain of cernunnos/XLF. J Biol Chem. 2010;285:26475–26483. doi: 10.1074/jbc.M110.138156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammel M, Yu Y, Fang S, Lees-Miller SP, Tainer JA. XLF regulates filament architecture of the XRCC4. ligase IV complex. Structure. 2010;18:1431–1442. doi: 10.1016/j.str.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using Rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recuero-Checa MA, et al. Electron microscopy of Xrcc4 and the DNA ligase IV–Xrcc4 DNA repair complex. DNA Repair (Amst) 2009;8:1380–1389. doi: 10.1016/j.dnarep.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4–DNA ligase IV complex: Influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitagawa D, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 36.Bricogne G, et al. BUSTER version 2.9. Cambridge, UK: Global Phasing Ltd; 2010. [Google Scholar]
- 37.Brunger AT, Adams PD, Rice LM. New applications of simulated annealing in X-ray crystallography and solution NMR. Structure. 1997;5:325–336. doi: 10.1016/s0969-2126(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.