Abstract
Sequence-specific gene silencing by short hairpin (sh) RNAs has recently emerged as an indispensable tool for understanding gene function and a promising avenue for drug discovery. However, a wider biomedical use of this approach is hindered by the lack of straightforward methods for achieving uniform expression of shRNAs in mammalian cell cultures. Here we report a high-efficiency and low-background (HILO) recombination-mediated cassette exchange (RMCE) technology that yields virtually homogeneous cell pools containing doxycycline-inducible shRNA elements in a matter of days and with minimal efforts. To ensure immediate utility of this approach for a wider research community, we modified 11 commonly used human (A549, HT1080, HEK293T, HeLa, HeLa-S3, and U2OS) and mouse (CAD, L929, N2a, NIH 3T3, and P19) cell lines to be compatible with the HILO-RMCE process. Because of its technical simplicity and cost efficiency, the technology will be advantageous for both low- and high-throughput shRNA experiments. We also provide evidence that HILO-RMCE will facilitate a wider range of molecular and cell biology applications by allowing one to rapidly engineer cell populations expressing essentially any transgene of interest.
The discovery of RNA interference (RNAi), sequence-specific gene silencing by double-stranded (ds) RNA molecules, has revolutionized the way we study gene function (1, 2). Two alternative molecules are routinely used to trigger RNAi in mammalian cells: (i) chemically or enzymatically generated small interfering (si) RNAs and (ii) genetically encoded short hairpin (sh) RNAs that are converted into siRNAs by the cellular RNAi machinery (2–6). The relatively low cost and the possibility of inducing sustained and tunable silencing, combined with a minimized risk of off-target effects, make the shRNA approach especially attractive for high-throughput loss-of-function screens and developing RNAi-based therapies (6–9).
However, unlike siRNAs that can be delivered into most cultured mammalian cell lines in a virtually quantitative manner, equally straightforward methods for rapidly generating homogeneous cell populations expressing shRNAs have not been described. Only few lines can be transiently transfected with plasmid DNA with efficiencies sufficient for penetrant RNAi. Similarly, a major limitation of stable expression approaches relying on plasmid- or viral vector-mediated transgenesis is the random nature of the genomic integration, which often lowers the transgene expression levels because of epigenetic silencing effects (10–14). As a consequence, obtaining shRNA-expressing cell populations normally entails time-consuming and labor-intensive enrichment or cell-cloning steps. For example, the recently published lentiviral toolkits for inducible shRNA expression generate cell populations containing substantial fractions of cells expressing shRNAs at low levels, which have to be depleted by FACS for an optimal knockdown performance (10, 15). Although feasible for experiments using just a few shRNA molecules, FACS enrichment is not practical for medium- and high-throughput shRNA screens carried out in the arrayed format (7). In addition, shRNA-encoding virus vectors require considerable efforts to prepare high-titer stocks and, importantly, are associated with biosafety concerns.
Recombination-mediated cassette exchange (RMCE) uses the activity of site-specific recombinases to integrate a donor sequence flanked by self-compatible but mutually incompatible recombination sites at a predefined acceptor locus containing a similar pair of the recombination sites (16). This method eliminates the genome position effect and allows uniform transgene expression in independent recombinant clones. RMCE has recently been used for stable shRNA expression in HeLa cells and mouse embryonic stem cells (17–20). However, the published techniques still rely on cell cloning to isolate shRNA-expressing cells from the original recombinant pools because of a background of unspecific integration events.
Results and Discussion
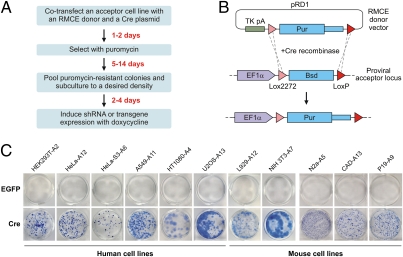
Here we developed a high-efficiency and low-background (HILO) RMCE platform that significantly accelerates engineering of mammalian cells expressing shRNAs and other genetic elements (Fig. 1A). To establish acceptor lines compatible with the HILO-RMCE procedure, we assembled a lentiviral vector-encoded cassette containing the human EF-1α promoter and a blasticidin resistance gene (Bsd) “floxed” by the mutually incompatible Cre recombinase-specific sites Lox2272 and LoxP (Fig. 1B). A single copy of this cassette was transduced into six human (HEK293T, HeLa, HeLa-S3, A549, HT1080, and U2OS) and five mouse (CAD, L929, N2a, NIH 3T3, and P19) commonly used cell lines (Fig. S1). We then constructed an RMCE donor plasmid (pRD1) containing a Lox2272- and LoxP-floxed puromycin (Pur) resistance gene (Fig. 1B). We reasoned that substituting a promoter in front of the floxed Pur gene with a strong polyadenylation signal (TK pA; derived from HSV thymidine kinase) should discourage unspecific integration events and generate puromycin-resistant colonies only in the case of correct recombination with the RMCE acceptor locus containing the strong promoter (Fig. 1B).
Fig. 1.
Establishing the HILO-RMCE acceptor cell lines. (A) Flowchart of a typical HILO-RMCE shRNA experiment. (B) Diagram of the HILO-RMCE reaction using the pRD1 donor plasmid. (C) The newly established acceptor lines were cotransfected in a 12-well (HEK293T-A2, HeLa-A12, HeLa-S3-A6, A549-A11, HT1080-A4, U2OS-A13, L929-A12, NIH 3T3-A7) or 6-well format (P19-A9, CAD-A13, N2a-A5) with a mixture containing 90% of pRD1 plasmid and 10% of a Cre-encoding plasmid (most cell lines, pCAGGS-Cre; NIH 3T3-A7, pCAGGS-nlCre) or the EGFP-encoding control plasmid pCIG. Following the puromycin selection, multiple colonies formed in the presence of Cre but not when Cre was substituted with EGFP.
The acceptor cell lines were cotransfected with a mixture of pRD1 and either the pCAGGS-Cre plasmid encoding a wild-type Cre recombinase or a control plasmid encoding EGFP (pCIG). In all cases, multiple colonies appeared 5 to 14 d following puromycin selection in the presence of Cre but not in the control wells expressing EGFP (Fig. 1C). Thus, the HILO-RMCE reaction proceeded efficiently and with a negligibly low background of Cre-independent integration, as intended.
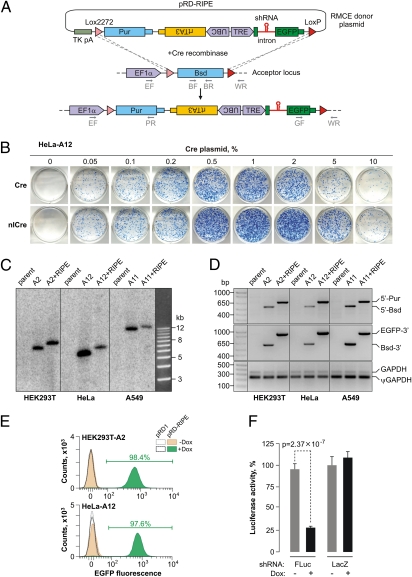
To adapt this system for inducible shRNA expression, we retrofitted pRD1 with a RIPE cassette containing a constitutively expressed reverse tetracycline transactivator gene (rtTA3) (21) and a tetracycline-inducible module that included an intron harboring a pre-miR-155 microRNA precursor-based shRNA cloning site and an EGFP expression marker (Fig. 2A). Cotransfecting the acceptor cell lines with the pRD-RIPE plasmid and pCAGGS-cre (but not the pCIG control) gave rise to multiple puromycin-resistant colonies, similar to pRD1. To further optimize the RMCE efficiency, we added an N-terminal nuclear localization signal to the Cre protein (nlCre; pCAGGS-nlCre plasmid). Consistent with earlier reports (for example, ref. 22), this modified recombinase performed better than the wild-type Cre in the tested human and mouse cell lines (Fig. 2B and Fig. S2). Genotyping the pooled recombinants suggested that the RIPE integration occurred in a precise and quantitative manner (Fig. 2 C and D). We also confirmed that the load of unspecific pRD-RIPE integration was acceptably low (Fig. S3). FACS and epifluorescent microscopy analyses showed that the RIPE-encoded EGFP gene turned on in >97% of the recombinant cells in the presence but not in the absence of the rtTA3 inducer doxycycline (Dox) (Fig. 2E and Fig. S4), an indication of tightly controlled Dox-inducible expression.
Fig. 2.
Developing the HILO-RMCE technology. (A) Diagram of the HILO-RMCE reaction using the pRD-RIPE donor plasmid. (B) HeLa-A12 cells containing the RMCE acceptor locus were cotransfected in a 12-well format with the pRD-RIPE plasmid and the indicated amounts of the pCAGGS-Cre or pCAGGS-nlCre plasmids. Note that nlCre performed better than the wild-type Cre over a wide concentration range. (C) Genomic DNA was isolated from three parental cell lines (HEK293T, HeLa, and A549; lanes labeled “parent”), the corresponding HILO-RMCE acceptor clones (“A” followed by the clone number), and pooled clones obtained by the HILO-RMCE-mediated integration of the RIPE cassette (the “A+RIPE” lanes). The DNA samples were digested with NcoI and analyzed by Southern blotting to confirm the uniform rearrangement of the acceptor locus as a result of the RMCE reaction. The results are consistent with the expected increase in the length of the acceptor locus-specific NcoI fragment by 856 bp following the RIPE integration. (D) The precision of the RMCE reaction was further confirmed by analyzing the genomic DNA samples described in C by multiplex PCR using either the 5′ (EF, BR, and PR, see A) or the 3′ junction primer mixture (GF, BF, and WR; see A). The primers were designed so that the corresponding PCR product sizes were distinct for the original acceptor (5′-Bsd and Bsd-3′) and the RIPE-targeted loci (5′-Pur and EGFP-3′). GAPDH-specific primers detecting both the bona fide gene (GAPDH) and a pseudogene (ψGAPDH) were used as a control. (E) HILO-RMCE colonies produced by cotransfecting HEK293T-A2 and HeLa-A12 cells with pCAGGS-nlCre and either pRD1 or pRD-RIPE were pooled and incubated with 2 μg/mL Dox for 48 h or left untreated. The cellular EGFP expression was then examined by FACS. Note that nearly all cells express EGFP in the Dox-treated pRD-RIPE samples. (F) HEK293T-A2 cells carrying RIPE cassettes with shRNAs against either FLuc or LacZ were induced with Dox for 36 h or left untreated. The cells were then transfected with a mixture of plasmids encoding the FLuc and RLuc luciferases and the normalized FLuc activities were assayed as described in SI Materials and Methods. Data are averaged from six transfection experiments ± SD.
To assess the utility of the RIPE cassette for RNAi, we inserted a firefly luciferase (FLuc)-specific shRNA (shFLuc) at the pre-miR-155 cloning site and integrated the RIPE-shFLuc cassette into the HEK293T-A2 cells using HILO-RMCE (Fig. S5). The cells were either preincubated with Dox or left untreated. The two populations were then transfected with a mixture of plasmids encoding FLuc and Renilla luciferase (RLuc) and the relative FLuc expression measured as a ratio between the Fluc and the Rluc activities (Fig. 2F). The FLuc expression was knocked down 3.45-fold (P = 2.37 × 10−7, t test) in the Dox-positive samples expressing shFLuc compared with the Dox-negative control. As expected, LacZ-specific shRNA control (shLacZ) had no significant effect on the FLuc expression (Fig. 2F).
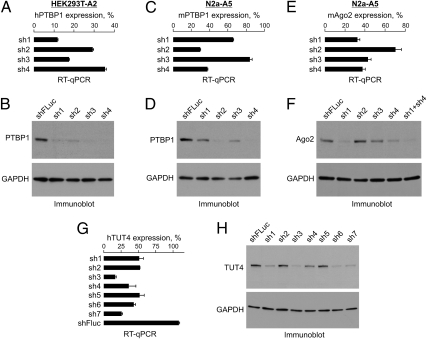
We then examined if RIPE-encoded shRNAs can be used to knock down endogenous cellular proteins. Using the above strategy, we obtained HEK293T-A2 pools expressing Dox-inducible shRNAs against human RNA-binding protein PTBP1 (23). Satisfyingly, all four computationally designed shRNAs reduced PTBP1 expression at both the mRNA and the protein levels compared with the shFLuc control (Fig. 3 A and B). Efficient knockdowns were also obtained in N2a-A5 cells using shRNAs against mouse Ptbp1 (Fig. 3 C and D) and the Argonaute protein Ago2/Eif2c2 (24, 25), a critical RNAi component and therefore potentially problematic target (Fig. 3 E and F). Interestingly, the Ago2 protein down-regulation was further improved by coexpressing the two most potent Ago2-specific shRNAs from the same RIPE cassette (lane “sh1+sh4” in Fig. 3F).
Fig. 3.
Silencing cell-encoded genes. (A and B) HEK293T-A2 cells containing four different RIPE-encoded shRNAs against human PTBP1 mRNA or the shFLuc shRNA were induced with Dox for 72 h and the efficiency of the PTBP1 knockdown analyzed by (A) RT-qPCR and (B) immunoblotting with PTBP1-specific antibodies. The RT-qPCR graph in A shows relative PTBP1 expression levels normalized to the shFLuc control. (C–F) The experiment in A and B was repeated in N2a-A5 cells using shRNAs against (C and D) mouse Ptbp1 or (E and F) Ago2 genes. Note that coexpressing the two most potent Ago2-specific shRNAs from a single RIPE cassette further improves the protein knockdown (lane “sh1+sh4” in F). (G and H) Optimization of the human TUT4/ZCCHC11 knockdown as a part of a larger-scale RNAi experiment where an shRNA library against the human TUT gene family was integrated into HEK293T-A2 cells using HILO-RMCE (see Fig. S6 for the rest of the results). (G) RT-qPCR analysis, in which the TUT4/ZCCHC11 mRNA expression levels in the Dox-treated samples were normalized to the corresponding Dox-negative controls. (H) The knockdown efficiencies were also studied by immunoblotting with an anti-TUT4 antibody. Data in A, C, E, and G are averaged from three amplifications experiments ± SD. In B, D, F, and H, a GAPDH-specific antibody was used to control lane loading.
As a test of the method performance in a larger scale RNAi experiment, we turned to the family of human terminal uridyl transferases (TUTs) implicated in various aspects of RNA metabolism including 3′-terminal modifications of microRNA and their precursors (26, 27) and designed a RIPE-encoded shRNA library containing four to seven shRNAs against each of its seven members (TUT1 to -7). Similar to the above results, efficient Dox-inducible shRNAs were identified for all of the TUT genes (Fig. 3 G and H and Fig. S6).
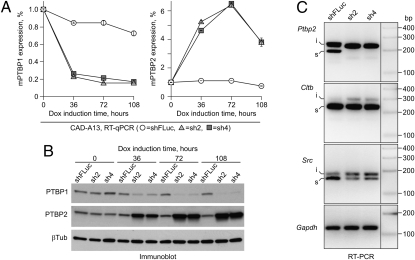
To ensure that the knockdown efficiencies afforded by the HILO-RMCE technology are sufficient to induce biological phenotypes, we established CAD-A13 cell populations encoding the optimized shRNAs against mouse Ptbp1 (sh2 and sh4) and examined the effect of these shRNAs on the expression of the Ptbp2 gene, which is known to be repressed by the Ptbp1 protein through an alternative splicing mechanism (28–30). Continued expression of either sh2 or sh4 reduced the Ptbp1 mRNA and protein levels, which was accompanied by an increase in the Ptbp2 mRNA and protein levels (Fig. 4 A and B). As expected, the inclusion of the Ptbp2 alternative exon 10 was stimulated dramatically in the Ptbp1-knockdown samples (Fig. 4C) (28–30). Moreover, Ptbp1 knockdown activated the inclusion of other alternative exons known to be regulated by this protein in CAD cells (Fig. 4C) (29).
Fig. 4.
Knocking down Ptbp1 in HILO-RMCE-generated populations modifies cellular alternative pre-mRNA splicing patterns. CAD-A13 cells containing RIPE-encoded shRNAs against mouse Ptbp1 (sh2 or sh4) or the shFLuc shRNA were induced with 2 μg/mL doxycycline and the time course of the Ptbp1 mRNA knockdown was followed for 108 h using RT-qPCR (A, Left). We also examined the time course of the Ptbp2 mRNA accumulation (A, Right), an expected outcome of the reduced Ptbp1 abundance (28–30). (B) The Ptbp1 down-regulation and the Ptbp2 up-regulation were also confirmed by immunoblotting with corresponding antibodies. (C) RT-PCR analysis of the 72-h induced samples were carried out to examine the splicing patterns of three alternative cassette exons known to be repressed by the Ptbp1 protein: exon 10 of the Ptbp2 gene, exon N1 of the Src gene, and exon 5 of the Cltb gene. Note that the inclusion of these exons is stimulated in the Ptbp1-knockdown samples compared with the shFLuc control. i, exon-included splice form; s, exon-skipped splice form. Gapdh, an RT-PCR amplification control.
Although we could successfully knock down several genes, we reasoned that the shRNA expression in our system might be subject to the provirus integration site position effect and thus could be further optimized by screening additional RMCE acceptor clones. We therefore examined the expression of the RIPE cassette in 12 independent RMCE acceptor clones of the HeLa cell line (Fig. S7 A and B). Notably, following the Dox induction, several clones, including HeLa-A5 and HeLa-A10, accumulated larger amounts of EGFP than the originally characterized HeLa-A12 clone (Fig. S7 A and B).
We then examined the performance of a RIPE-encoded human TUT4/ZCCHC11-specific shRNA (sh3) (Fig. 3 G and H) in all 12 clones using RT-qPCR (Fig S7C). As judged by this method, the EGFP expression did not always correlate with the knockdown efficiency. However, HeLa-A5 and HeLa-A10 clones did show improved RNAi compared with HeLa-A12 (Fig. S7C). TUT4 knockdown in HeLa-A5 and HeLa-A10 was virtually complete at the protein level (Fig. S7D). Interestingly, the TUT4 protein levels were also noticeably diminished in clones that did not fare well in the RT-qPCR assay (for example HeLa-A6 and HeLa-A7), thus suggesting that the RT-qPCR data were likely an underestimation of the true RNAi performance (Fig. S7 C and D).
We repeated the above experiment for three additional human lines: HEK293T, A549 and U2OS (Fig. S8). Six independent acceptor clones were examined for each cell line, and in each case we identified clones with >70% TUT4 mRNA knockdown (Fig. S8). Similar efforts using mouse Ptbp1 shRNA (sh2) (Fig. 4) have identified RNAi-proficient acceptor clones for three commonly used mouse cell lines: CAD, N2a, and L929 (Fig. S9). We concluded that RNAi efficiency can indeed be optimized by screening several acceptor clones.
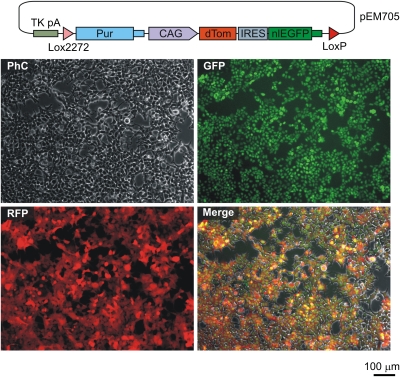
We finally explored the possibility of using the HILO-RMCE platform for rapidly generating homogeneous cell pools expressing genetic elements other than shRNAs. For this purpose, we designed three pRD1-based donor plasmids containing various transgenes under control of a constitutive promoter (CAG) and cotransfected HEK293T-A2 cells with these constructs and the pCAGGS-nlCre plasmid (Fig. 5 and Fig. S10). In all three cases, the populations of puromycin-resistant recombinant cells expressed the integrated transgenes at readily detectable levels and in a homogeneous manner (Fig. 5 and Fig. S10).
Fig. 5.
HILO-RMCE can be readily adapted for rapid engineering of transgenic cell pools. HEK293T-A2 cells were cotransfected with pCAGGS-nlCre and pRD1-based donor plasmid (pEM705) containing a CAG promoter-driven bicistronic cassette encoding dTomato (dTom) (37) and a nuclear localized EGFP proteins. Recombinant cells were selected with puromycin for 7 d, pooled and propagated for another 4 d. The fluorescent protein expression was then visualized by microscopy. Diagram of pEM705 is shown on the top of the panel.
In conclusion, HILO-RMCE transforms the shRNA experiment into a user-friendly procedure that combines the possibility of long-term RNAi with the speed and convenience of siRNA-based approaches. The cost efficiency and the technical simplicity make HILO-RMCE an ideal platform for optimizing shRNA efficiency and other types of low-throughput projects in diverse laboratory settings, as well as a useful addition to the high-throughput RNAi screening toolbox. Moreover, our data on other types of transgenic cassettes suggest that the described platform should facilitate a wider range of molecular and cell biology experiments.
Materials and Methods
Plasmids.
To construct the RMCE acceptor lentiviral vector (pEM584), we modified the pHAGE backbone (31) by inserting a human EF-1α promoter from a derivative of the pEF-BOS vector (32) followed by a Lox2272- and LoxP-floxed blasticidin resistance gene (Bsd) from the pLenti6/V5-DEST plasmid (Invitrogen) between the cpPu (Trip) and WPRE elements. The RMCE donor vector (pRD1; alternative name pEM601) was generated by subcloning the polyadenylation signal-containing HSV thymidine kinase (TK) gene from a pEasyFlox derivative (33) into the pBluescript II KS(+) backbone. Immediately downstream of the TK polyadenylation signal, we introduced a Lox2272- and LoxP-floxed promoter-less puromycin resistance (Pur) gene from the pPUR plasmid (Clontech). The pRD-RIPE plasmid (alternative name pEM791) was derived from pRD1 by removing the TK promoter and coding sequence and inserting a UBC promoter-driven reverse tetracycline transactivator gene (rtTA3) and the tet-inducible promoter TRE (both elements adapted from the pTRIPZ plasmid; Open Biosystems) between the Pur gene and the LoxP site. The TRE promoter was followed by an intron-EGFP module containing an intronic pre-miR-155–based shRNA cloning site with two BsmBI sequences (34). The pCAGGS-nlCre (alternative name pEM784) was modified from pCAGGS-Cre (35) by substituting the original sequence 5′-ACTTTACTTAAAACCATTATCTGAGTGTGAAATG-3′ in front of the Cre gene with the sequence 5′-CTAGACTCGACCATGCCCAAGAAGAAGAGGAAGGTG-3′ encoding the N-terminal nuclear localization sequence (underlined) from the Large T antigen of the SV40 virus. Complete sequences and maps of all pEM plasmids are available upon request.
Design and Cloning of pre-miR-155–Based shRNAs.
The shRNAs were designed using the miR RNAi design option of the Block-iT RNAi Designer program (https://rnaidesigner.invitrogen.com/rnaiexpress/). Wherever possible, multiple shRNAs targeting the ORF and 3′UTR were chosen for a given target gene. For each shRNA, two 64-nt long cDNA oligonucleotides were ordered from Sigma Life Science, annealed, and inserted into pRD-RIPE plasmid at the BsmBI sites (for detailed description of the pre-miR-155–based shRNA cloning strategy, see ref. 34 and https://rnaidesigner.invitrogen.com/rnaiexpress/). For example, the firefly liciferase-specific shRNA (shFLuc) was engineered by annealing the following oligonucleotides: 5′-TGCTGTATTCAGCCCATATCGTTTCAGTTTTGGCCACTGACTGACTGAAACGATGGGCTGAATA-3′ and 5′-CCTGTATTCAGCCCATCGTTTCAGTCAGTCAGTGGCCAAAACTGAAACGATATGGGCTGAATAC-3′. Information on other shRNA sequences used in this study is available on request. To improve the mAgo2 knockdown efficiency, the mAgo2 sh4 shRNA element was amplified using the RIPE-specific primers shRNAdimer_F1 and shRNAdimer_R1 (Table S1), the PCR fragment was treated with MfeI (New England Biolabs) (Table S1, underlined sequence) and inserted into the mAgo2 sh1-containing RIPE cassette at the EcoRI-EcoRV sites located downstream of the miR-155 element to create an intron-encoded sh1-sh4 tandem. Because the shRNAdimer_R1 primer restores both the EcoRI and the EcoRV sites (Table S1, italicized sequences), this procedure can theoretically be repeated to generate tandem shRNA arrays of any desired length and complexity.
Cells.
Parental cell lines used in this study were from the ATCC except for the CAD line, which was kindly provided by the authors (36). A549, HEK293T, HeLa, HeLa-S3, HT1080, L929, N2a, NIH 3T3, and U2OS were routinely propagated in DMEM/high-glucose medium (HyClone) supplemented with 10% FBS (“characterized” grade; HyClone), 1 mM sodium pyruvate (Invitrogen), and 1× penicillin-streptomycin (100 U/mL penicillin, 100 μg/mL streptomycin; Invitrogen). For passaging, adherent cells were detached using 1× trypsin-EDTA (Invitrogen). In some experiments, regular FBS was substituted with certified tetracycline-negative FBS (PAA). CAD cells were cultured in a similar medium except FBS was substituted with the FetalClone III serum (HyClone). P19 cells were cultured in αMEM medium supplemented with 2.5% FBS, 7.5% Bovine Calf Serum (BCS; HyClone) and 1× penicillin-streptomycin. When required, media were supplemented with 2.5 to 10 μg/mL blasticidin S or 1 to 16 μg/mL puromycin. To turn on the Tet-inducible expression, Dox was added to the final concentration of 2 μg/mL.
RMCE Acceptor Cell Lines.
To establish RMCE acceptor cell lines, ∼40% confluent cell cultures were incubated with serial dilutions of the pEM584 lentiviral stock (1–200 cfu per 10-cm plate) for 18 h without polybrene. The medium was then changed and the cells were incubated for another 18 h before the addition of blasticidin S to 5 to 10 μg/mL. The incubation was continued in the presence of blasticidin until noninfected cells died and visible blasticidin-resistant colonies formed. For each cell line, 12 to 18 individual colonies were picked using 200-μL pipette tips and clonally expanded. Dishes containing >50 colonies were discarded to avoid multiple integration events and colony cross-contamination. Clones with optimal RMCE performance were maintained in the presence of 2.5 to 5 μg/mL blasticidin S. For long-term storage, cells were cryopreserved in a mixture containing 90% of the appropriate culture medium and 10% DMSO.
RMCE Protocol.
RMCE acceptor cell lines were plated in 12-well plates at 1.0 to 1.5 × 105 cells per well in an antibiotic-free medium 12 to 18 h before transfection except L929-A and P19-A cells that were plated 3 h before transfection. Cells were then cotransfected with an RMCE donor plasmid (pRD, pRD-RIPE, or a derivative of pRD-RIPE containing a gene-specific shRNA sequence) blended with 0.5% to 10% (wt/wt) of a Cre-encoding plasmid (pCAGGS-Cre or pCAGGS-nlCre). To transfect one well of a 12-well plate, 0.5 μg of total DNA was mixed with 1.25 μL Lipofectamine 2000/32.5 μL Opti-MEM I (Invitrogen) following the manufacturer's protocol. Cells were incubated with the transfection mixtures overnight, the medium was changed and the incubation continued for another 24 h before adding puromycin. For most cell lines, we used a three-step selection protocol beginning with one-half of the maximal puromycin concentration for the first 24 to 48 h of selection, followed by the maximal concentration (HeLa-A and A549-A clones, 2 μg/mL; CAD-A, HeLa-S3-A, HT1080-A, N2a-A, NIH 3T3-A, and U2OS-A clones, 5 μg/mL and HEK293T-A clones, 8 μg/mL) for several days until the puromycin-sensitive cells were eliminated and then returning to the one-half maximal concentration to accelerate the proliferation of the puromycin-resistant cells. L929-A and P19-A cells were immediately exposed to the maximal puromycin concentration (L929-A, 16 μg/mL; P19-A, 5 μg/mL) followed by the one-half concentration step after the death of the puromycin-sensitive cells. The cultures were incubated until the appearance of visible puromycin-resistant colonies, which were either pooled and expanded in a medium containing one-half maximal puromycin concentration or alternatively stained with 0.1% methylene blue in 50% methanol and photographed.
Supplementary Material
Acknowledgments
We thank Guangwei Du, Richard Mulligan, I-Hsin Su, and Ken-ichi Yamamura for reagents, Geraldine Lee for the help with FACS, and Snezhka Oliferenko for comments on the manuscript. This work was supported by National Research Foundation Grant NRF-RF2008-06 (to E.V.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103532108/-/DCSupplemental.
References
- 1.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 3.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 6.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: Similarities and differences. Adv Drug Deliv Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Mohr S, Bakal C, Perrimon N. Genomic screening with RNAi: Results and challenges. Annu Rev Biochem. 2010;79:37–64. doi: 10.1146/annurev-biochem-060408-092949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimm D. Small silencing RNAs: State-of-the-art. Adv Drug Deliv Rev. 2009;61:672–703. doi: 10.1016/j.addr.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Wiznerowicz M, Szulc J, Trono D. Tuning silence: Conditional systems for RNA interference. Nat Methods. 2006;3:682–688. doi: 10.1038/nmeth914. [DOI] [PubMed] [Google Scholar]
- 10.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011;29(1):79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis J, Hotta A, Rastegar M. Retrovirus silencing by an epigenetic TRIM. Cell. 2007;131(1):13–14. doi: 10.1016/j.cell.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lund AH, Duch M, Pedersen FS. Transcriptional silencing of retroviral vectors. J Biomed Sci. 1996;3:365–378. doi: 10.1007/BF02258042. [DOI] [PubMed] [Google Scholar]
- 15.Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci USA. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirth D, et al. Road to precision: Recombinase-based targeting technologies for genome engineering. Curr Opin Biotechnol. 2007;18:411–419. doi: 10.1016/j.copbio.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Berger SM, et al. Quantitative analysis of conditional gene inactivation using rationally designed, tetracycline-controlled miRNAs. Nucleic Acids Res. 2010;38(17):e168. doi: 10.1093/nar/gkq616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito F, et al. Development and application of a stable HeLa cell line capable of site-specific transgenesis using the Cre-lox system: Establishment and application of a stable TNFRI knockdown cell line to cytotoxicity assay. Toxicol In Vitro. 2008;22:1077–1087. doi: 10.1016/j.tiv.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Weidenfeld I, et al. Inducible expression of coding and inhibitory RNAs from retargetable genomic loci. Nucleic Acids Res. 2009;37(7):e50. doi: 10.1093/nar/gkp108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Premsrirut PK, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145(1):145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das AT, et al. Viral evolution as a tool to improve the tetracycline-regulated gene expression system. J Biol Chem. 2004;279:18776–18782. doi: 10.1074/jbc.M313895200. [DOI] [PubMed] [Google Scholar]
- 22.Andreas S, Schwenk F, Küter-Luks B, Faust N, Kühn R. Enhanced efficiency through nuclear localization signal fusion on phage PhiC31-integrase: Activity comparison with Cre and FLPe recombinase in mammalian cells. Nucleic Acids Res. 2002;30:2299–2306. doi: 10.1093/nar/30.11.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: A multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 24.Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 25.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Katoh T, et al. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc Natl Acad Sci USA. 2006;103:16406–16411. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casola S. Conditional gene mutagenesis in B-lineage cells. Methods Mol Biol. 2004;271:91–109. doi: 10.1385/1-59259-796-3:091. [DOI] [PubMed] [Google Scholar]
- 34.Du G, Yonekubo J, Zeng Y, Osisami M, Frohman MA. Design of expression vectors for RNA interference based on miRNAs and RNA splicing. FEBS J. 2006;273:5421–5427. doi: 10.1111/j.1742-4658.2006.05534.x. [DOI] [PubMed] [Google Scholar]
- 35.Araki K, Araki M, Yamamura K. Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res. 1997;25:868–872. doi: 10.1093/nar/25.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi Y, Wang JK, McMillian M, Chikaraishi DM. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J Neurosci. 1997;17:1217–1225. doi: 10.1523/JNEUROSCI.17-04-01217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.