Abstract
SNARE proteins play a critical role in intracellular membrane fusion by forming tight complexes that bring two membranes together and involve sequences called SNARE motifs. These motifs have a high tendency to form amphipathic coiled-coils that assemble into four-helix bundles, and often precede transmembrane regions. NMR studies in dodecylphosphocholine (DPC) micelles suggested that the N-terminal half of the SNARE motif from the neuronal SNARE synaptobrevin binds to membranes, which appeared to contradict previous biophysical studies of synaptobrevin in liposomes. NMR analyses of synaptobrevin reconstituted into nanodiscs and into liposomes now show that most of its SNARE motif, except for the basic C terminus, is highly flexible, exhibiting cross-peak patterns and transverse relaxation rates that are very similar to those observed in solution. Considering the proximity to the bilayer imposed by membrane anchoring, our data show that most of the synaptobrevin SNARE motif has a remarkable reluctance to bind membranes. This conclusion is further supported by NMR experiments showing that the soluble synaptobrevin SNARE motif does not bind to liposomes, even though it does bind to DPC micelles. These results show that nanodiscs provide a much better membrane model than DPC micelles in this system, and that most of the SNARE motif of membrane-anchored synaptobrevin is accessible for SNARE complex formation. We propose that the charge and hydrophobicity of SNARE motifs is optimized to enable formation of highly stable SNARE complexes while at the same time avoiding membrane binding, which could hinder SNARE complex assembly.
Keywords: neurotransmitter release, membrane proteins, membrane traffic
Traffic at most eukaryotic membrane compartments is governed by members of conserved protein families that underlie a universal mechanism of intracellular membrane fusion (1). Particularly important among these proteins are the members of the SNARE family, which are characterized by sequences of about 60–70 amino acid residues that are known as SNARE motifs and often precede C-terminal transmembrane (TM) regions (2–5). Through these motifs, SNAREs from two apposed membranes form a tight four-helix bundle called the SNARE complex (6, 7), which brings the two membranes together and was proposed to provide the energy for membrane fusion (8). Although the exact fusion mechanism is still unclear (9), and fusion depends critically on other universal factors such as Sec1/Munc18 proteins (10–12) and sometimes on specialized proteins such as synaptotagmin-1 in the case of synaptic vesicle exocytosis (13–15), there is little doubt that the SNAREs play a central role in membrane fusion.
Crystal structures of the SNARE four-helix bundle that represents the postfusion state have been determined without or with the adjacent TM regions (e.g., refs. 7 and 16), but detailed structural information on the isolated SNARE motifs attached to their adjacent TM regions is more limited. Early studies of synaptobrevin, syntaxin-1, and SNAP-25, the SNAREs that mediate synaptic vesicle fusion (3–5), and of the yeast plasma membrane SNAREs, showed that soluble fragments spanning the SNARE motif are generally unstructured (17–21), although the syntaxin-1 SNARE motif can be unstructured or helical (due to oligomerization) depending on the conditions (17, 19, 22, 23). Analyses of the synaptobrevin SNARE motif in the context of the full-length protein have yielded conflicting results. On one hand, CD experiments in detergents (24) and EPR data on synaptobrevin reconstituted into lipid vesicles (25) indicated that most of the synaptobrevin SNARE motif is unstructured except for a short region at its C-terminal end, which forms an α-helix together with part of the linker joining the SNARE motif and the TM sequence (below referred to as the juxtamembrane region; see Fig. 1A). This region associates with the membrane (25) because of the abundance of basic and aromatic residues in its sequence and already has a tendency to form helical structure in solution, as observed by NMR spectroscopy (18). On the other hand, NMR studies in dodecylphosphocholine (DPC) micelles indicated that much of the N-terminal half of the synaptobrevin SNARE motif (residues 36–54) also binds to the micelles, forming an α-helix (26).
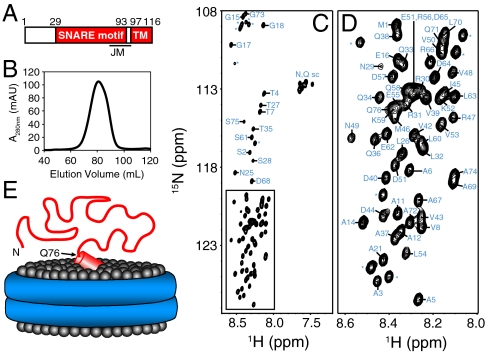
Fig. 1.
NMR analysis of synaptobrevin in nanodiscs. (A) Domain structure of synaptobrevin. Residue numbers above the bar indicate the sequence spanning the SNARE motif and the TM region. The approximate position of the juxtamembrane region (JM), which includes the C terminus of the SNARE motif and the linker joining it to the TM region, is indicated below. (B) Gel filtration on a Superdex200 column of the nanodiscs containing synaptobrevin after detergent removal and before concentrating for NMR analysis. (C, D) 1H-15N HSQC spectrum of synaptobrevin incorporated into nanodiscs. The expansion shown in D corresponds to the box of C. Cross-peak assignments are indicated (* indicates those from N-terminal residues arising from the expression vector). (E) Cartoon representing the overall structure of synaptobrevin (red) in nanodiscs with the lipid headgroups shown as gray spheres and the ApoA1 scaffold shown as a double blue ring. The diagram is meant to illustrate that residues 1–76 of synaptobrevin are highly flexible. The juxtamembrane region is represented by a tilted cylinder that represents a helix and is bound on the surface of the nanodiscs in a tilted orientation based on EPR data (25).
It is important to note that the amphipathic nature and necessary proximity of SNARE motifs to membranes could strongly favor membrane binding. Because the N-terminal halves of SNARE motifs are critical to initiate SNARE complex assembly (27, 28), membrane binding might compete with and thus hinder SNARE complex assembly. Therefore, characterizing in detail the conformational behavior of the membrane-anchored synaptobrevin SNARE motif and of membrane-anchored SNARE motifs in general is critical to understand the intrinsic anatomy of SNARE proteins and their biochemical properties, which is fundamental to elucidate the mechanism of intracellular membrane fusion. With this purpose, we have performed an NMR analysis of synaptobrevin reconstituted into phospholipid vesicles or into nanodiscs, which are disc-like phospholipid bilayers surrounded by a scaffolding protein (29). Our data conclusively show that most of the SNARE motif of reconstituted synaptobrevin is unstructured and does not interact with phospholipid bilayers under a variety of conditions, thus remaining highly accessible for SNARE complex assembly. These results suggest that the N-terminal sequences of SNARE motifs are optimized to avoid membrane interactions, which enhances their availability for binding to their cognate SNAREs or other components of the fusion machinery. Moreover, our data illustrate how using detergents as membrane mimics can sometimes yield misleading results, whereas nanodics offer a more faithful model to recapitulate the behavior of membrane proteins in their native environment.
Results
Flexibility of the SNARE Motif Synaptobrevin on Nanodiscs.
Application of solution NMR methods to study the structure of membrane proteins reconstituted into phosphoplipid vesicles is hindered by the very large size of the vesicles, which effectively ranges from tens of megadaltons to gigadaltons depending on their radii and leads to broadening beyond detection of resonances from structured parts of the proteins. This problem can be alleviated by incorporation of membrane proteins into nanodiscs (30–32), which leads to effective molecular weights in the 150–400 kDa range but still requires protein perdeuteration to obtain high-quality NMR data for structured regions. In the current study, we made no attempt to observe resonances from structured regions of reconstituted synaptobrevin and focused on determining which regions of its sequence remain flexible upon anchoring to a phospholipid bilayer. The rationale behind these experiments is that the resonances of a flexible polypeptide attached to a large macromolecular species should be observable as long as sufficient fast internal motions remain and there are no or minimal interactions with the large species. For instance, relatively sharp signals can still be observed even upon anchoring a peptide to 50-μm cross-linked polystyrene beads (33).
We first analyzed full-length synaptobrevin (residues 1–116) incorporated into nanodiscs with a lipid composition consisting of 85% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 15% 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS), which has been widely used in reconstitutions of SNARE proteins (34). Homogeneous preparations as judged by gel filtration (Fig. 1B) were obtained by carefully adjusting the ratio of lipids to synaptobrevin and scaffolding protein. The resulting nanodiscs could readily be concentrated to 90 μM (or higher), which was sufficient to obtain high-quality triple resonance data for assignment of backbone resonances from flexible regions. The 1H-15N heteronuclear single quantum coherence (HSQC) spectrum and corresponding cross-peak assignments (Fig. 1 C and D) reveal that resonances are observable for the sequence spanning residues 1 to 76, but not for the rest of the protein. These results are consistent with a model whereby the juxtamembrane and TM sequences of synaptobrevin are structured and bound to the nanodiscs, whereas residues 1–76, which include most of the SNARE motif, are unstructured and flexible (Fig. 1E).
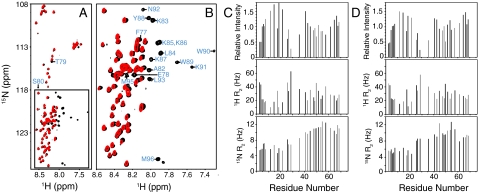
Our conclusions were further supported by comparison of the 1H-15N HSQC spectrum of full-length synaptobrevin in nanodiscs with that of the synaptobrevin cytoplasmic region (residues 1–96) in solution, for which resonance assignments were already available (18). The major differences between the two spectra correspond to the cross-peaks from residues 77–96 of the soluble fragment, which are not observed for synaptobrevin in the nanodiscs (Fig. 2 A and B). Importantly, the cross-peaks from residues 1–76 are practically in the same positions in both spectra. As shown earlier (18), this sequence is unstructured in the soluble synaptobrevin(1–96) fragment. Considering the high sensitivity of amide chemical shifts to even slight changes in chemical environment, our data indicate that residues 1–76 are similarly unstructured in the nanodisc-anchored synaptobrevin and are visiting similar conformational ensembles as in the soluble fragment. Moreover, the patterns of cross-peak intensities and transverse relaxation rates of the 1H and 15N backbone atoms of residues 1–76 are very similar for the soluble synaptobrevin (1–96) fragment and the nanodisc-anchored full-length synaptobrevin (Fig. 2 C and D). All these observations show that the segment spanning residues 1–76 does not interact with the nanodisc membranes, and that its conformational behavior is largely uncoupled from the juxtamembrane region regardless of whether this region is free or bound to a membrane.
Fig. 2.
Residues 1–74 of synaptobrevin have a similar conformational behavior in solution and on nanodiscs. (A, B) Superposition of 1H-15N HSQC spectra of the soluble synaptobrevin(1–96) fragment (black contours) and full-length synaptobrevin in nanodiscs composed of POPC∶DOPS 85∶15 (red contours). The expansion shown in B corresponds to the box of A. Cross-peak assignments are indicated for residues from the soluble synaptobrevin(1–96) fragment that are not observed on nanodiscs. (C, D) Relative cross-peak intensities (Top), and transverse relaxation rates (R2) of the 1H (Middle) and 15N (Bottom) backbone nuclei of residues 1–74 of the soluble synaptobrevin(1–96) fragment (C) or of full-length synaptobrevin in nanodiscs (D). Data were quantified only for well-resolved cross-peaks. Relative intensities were calculated by dividing the observed intensity of a cross-peak by the average of all cross-peak intensities (46).
Reluctance of the SNARE Motif of Reconstituted Synaptobrevin to Membrane Binding.
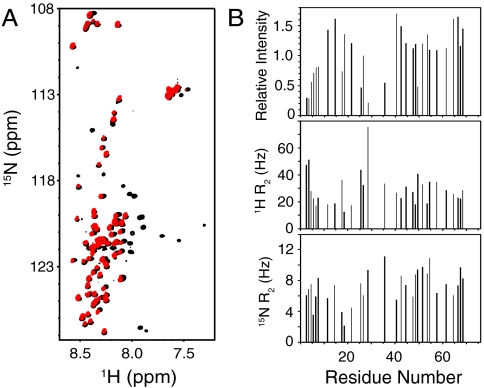
Although nanodiscs are believed to offer faithful models of phospholipid bilayers, there is only limited data demonstrating this notion. In addition, it is plausible that cis interactions of the N-terminal half of the synaptobrevin SNARE motif with the nanodisc surface might have been hindered by its limited size [10-nm diameter for the nanodiscs we prepared (29)]. To study synaptobrevin in a much more extensively used model of a phosphoilipid bilayer, we reconstituted full-length synaptobrevin into preformed 100-nm vesicles composed of POPC∶DOPS 85∶15 by detergent-assisted insertion (35), a method that yields highly homogeneous proteoliposomes (36). In these experiments, we used a 1∶500 protein-to-lipid ratio, which is well below that found in synaptic vesicles (37), to limit the likelihood that molecular crowding might hinder cis interactions of the SNARE motif with the vesicle surface. Control experiments showed that the reconstituted synaptobrevin readily formed SNARE complexes with the syntaxin-1 and SNAP-25 SNARE motifs (Fig. S1). Even though the synaptobrevin concentration was only 9 μM, we still were able to obtain high-quality 1H-15N HSQC spectra exhibiting cross-peaks for residues 1–74 (Fig. 3A, red contours). Moreover, the positions of the cross-peaks, the patterns of cross-peak intensities, as well as the 1H and 15N transverse relaxation rates of vesicle-anchored synaptobrevin (Fig. 3), are very similar to those observed for the soluble synaptobrevin(1–96) fragment and for synaptobrevin in nanodiscs (Figs. 1, 2, and 3A). These results show that residues 1–74 of vesicle-anchored synaptobrevin are highly flexible and unstructured, as they are in the nanodiscs or in solution.
Fig. 3.
Residues 1–74 of synaptobrevin have a similar conformational behavior in solution and on liposomes. (A) Superposition of 1H-15N HSQC spectra of the soluble synaptobrevin(1–96) fragment (black contours) and full-length synaptobrevin reconstituted into liposomes composed of POPC∶DOPS 85∶15 (molar ratio) (red contours). (B) Relative cross-peak intensities (Top), and transverse relaxation rates (R2) of the 1H (Middle) and 15N (Bottom) backbone nuclei of residues 1–74 of full-length synaptobrevin in liposomes (same sample as A). The data were analyzed as in Fig. 2 C and D.
In the above experiments, we included DOPS because the cytoplasmic leaflets of synaptic membranes are negatively charged. Because the synaptobrevin SNARE motif has abundant negative charges, we investigated the possibility that binding to membranes might be hindered by repulsion with the negatively charged head groups of DOPS by acquiring additional 1H-15N HSQC spectra of synaptobrevin reconstituted in POPC∶DOPS 85∶15 vesicles in the presence of 1 mM Mg2+, and of synaptobrevin reconstituted into POPC vesicles lacking DOPS. The spectra obtained under both conditions were very similar to those obtained in POPC∶DOPS 85∶15 in the absence of Mg2+ (Fig. S2 A and B). Analogous 1H-15N HSQC spectra were also obtained with synaptobrevin reconstituted into vesicles formed with POPC:1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine:DOPS:cholesterol 45∶20∶15∶20, a lipid composition that approximates that of synaptic vesicles (37) (Fig. S2C). All these results show that the lipid composition does not influence markedly the conformational behavior of residues 1–74 of reconstituted synaptobrevin.
Our results agree with previous CD and EPR studies of reconstituted synaptobrevin (24, 25), but contrast with those of an NMR study in DPC micelles, which concluded that residues 36–54 in the N-terminal half of the SNARE motif bind to membranes (26). In fact, our data suggest that this region does not have any significant affinity for membranes. Thus, if residues 36–54 of even a small population of reconstituted synaptobrevin molecules were bound to the membrane because of a weak membrane affinity, the transverse relaxation in these molecules would be extremely fast (orders of magnitude faster than those of unbound states) because of the very large size of the vesicles (effectively larger than 100 MDa). Such fast relaxation would be transferred to the unbound molecules by chemical exchange and would lead to substantial increases in the observed relaxation rates and decreases in the observed resonance intensities, even if the population of bound molecules were very small [for instance, binding to the 800-kDa GroEL tetradecamer causes strong resonance broadening on a peptide even at a 1∶400 GroEL:peptide ratio (38)]. However, the resonances of residues 36–54 of reconstituted synaptobrevin did not exhibit any overt increase in relaxation or decrease in intensity (Figs. 2 C and D and 3B). The same argument applies to the entire segment encompassing residues 1–74 of synaptobrevin. Because the proximity to the bilayer imposed by membrane anchoring should favor membrane binding by dramatically increasing the local concentration, our results show that residues 1–74 of synaptobrevin actually have a remarkable reluctance to membrane binding.
Soluble Synaptobrevin SNARE Motif Binds to DPC Micelles but not to Liposomes.
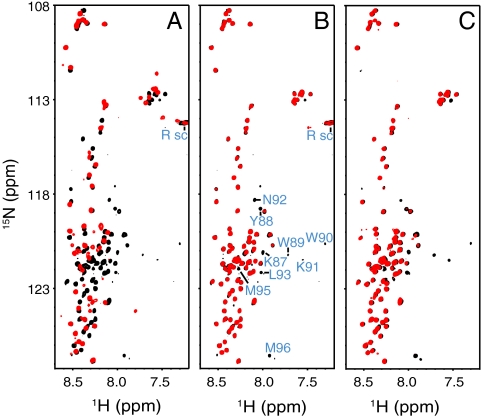
Given the contrast of our conclusions with those drawn from the results obtained in DPC micelles (26), we decided to test whether we could verify the latter results in our hands. Indeed, a 1H-15N HSQC spectrum of the soluble synaptobrevin(1–96) fragment in the presence of DPC under similar conditions to those used in ref. 26 revealed dramatic changes, including disappearance of the cross-peaks of residues 36–54 and of C-terminal residues from unstructured synaptobrevin(1–96), and the appearance of new cross-peaks corresponding to the micelle bound protein (Fig. 4A). However, 1H-15N HSQC spectra of synaptobrevin(1–96) in the presence of POPC vesicles (50 mM lipid) at the same pH (6.0) or at neutral pH exhibited much smaller perturbations, leading to broadening of only a few C-terminal residues (Fig. 4 B and C). These results further confirm the conclusion that the N-terminal half of the synaptobrevin SNARE motif does not bind to membranes, and show that DPC micelles do not constitute a good membrane model for this system.
Fig. 4.
The N-terminal half of the SNARE motif of soluble synaptobrevin binds to DPC micelles but not to liposomes. (A–C) Superposition of 1H-15N HSQC spectra of the 10 μM soluble synaptobrevin(1–96) fragment (black contours) alone or in the presence (red contours) of 300 mM DPC at pH 6.0 (A), POPC liposomes (50 mM lipids) at pH 6.0 (B), or POPC liposomes (50 mM lipids) at pH 7.0 (C). In B, cross-peaks that disappear upon addition of liposomes are labeled; cross-peaks from Arg side chains, which are observable at pH 6.0 but not at pH 7.0, are labeled Rsc. Note that in A there are less observable cross-peaks in DPC than in ref. 26 because we used a much lower protein concentration, but the data are fully consistent.
Discussion
SNARE proteins play a central role in intracellular membrane fusion by forming SNARE complexes that bring the two membranes into close proximity (8). To perform this function, the basic architecture of most SNARE proteins is relatively simple, consisting of a SNARE motif that is followed by a short linker and a C-terminal TM region [apart from the existence in some cases of regulatory N-terminal domains like the syntaxin-1 Habc domain (39)]. A potential problem with this architecture is that the SNARE motif must be able to form an amphipathic helix to assemble into a SNARE complex, and the necessary membrane proximity of the SNARE motif should favor binding of its hydrophobic side to the membrane. Although it has been suggested that such binding could facilitate SNARE complex formation by nucleating helical structure (26), the benefit of such nucleation is unclear, because helix-to-coil transitions can occur in the nanosecond time scale; in fact, it seems more likely that membrane binding would sequester the hydrophobic face of the SNARE motif, thus hindering interactions with other SNAREs or with other components of the fusion apparatus. This issue is particularly important for the N-terminal halves of the SNARE motifs, because they are believed to be critical for initiation of SNARE complex assembly (40), and they could also play important roles in binding to proteins that may form scaffolds for SNARE complex formation such as Munc13s (41–43). The results presented here show that most of the SNARE motif of reconstituted synaptobrevin does not bind to membranes and in fact appears to have a remarkable reluctance to membrane binding. It is tempting to speculate that reluctance to membrane binding is a general property of SNARE motifs that may be an intrinsic aspect of their anatomy and may be fundamental for their function in intracellular membrane fusion.
Except for the basic C terminus, the SNARE motif of synaptobrevin has abundant negative charges. This feature, which is shared by many SNARE motifs including that of syntaxin-1 (44), should lead to repulsion with the negatively charged membrane surface and is therefore very likely to contribute to the reluctance of most of the synaptobrevin SNARE motif to membrane binding. Note, however, that the very negative nature of SNARE motifs imposes an additional energy barrier to bring the membranes together, and hence may play an important role in the mechanisms of membrane fusion and its regulation. For instance, it is plausible that the repulsion between the membranes and the negative sequences from the SNARE motifs may add to the repulsion between the two membranes themselves, and that these repulsive forces are compensated by the energy of SNARE complex formation and by the interactions of the positively charged C termini of the SNARE motifs with the membranes. This combination of repulsive and attractive forces exerted by the SNARE motifs may help to generate leverage on the membranes to bend them and initiate membrane fusion. This notion has been supported by electrostatic potential calculations, and it seems likely that diverse proteins that bind to different regions of the SNARE complex (45–48) may also help to overcome the repulsive forces, at least some of them by binding in addition to one or both membranes (13, 47, 49, 50).
Regardless of the potential functional importance of negative charges in SNARE motifs, our results show that most of the synaptobrevin SNARE motif does not bind to membranes even when they do not contain negative charges (Figs. S2B and 4 B and C). Therefore, the reluctance to membrane binding does not arise only from electrostatic repulsion and may be due to a more fundamental, intrinsic property. We speculate that, perhaps because SNARE motifs might have been selected by evolution to form four-helix bundles rather than two- or three-helix bundles, the ratio between hydrophobic and polar residues may be relatively low, and the hydrophobic interactions that can be established between the SNARE motif hydrophobic surface and the interior of the membrane may not be sufficient to overcome the energy required for insertion into the lipid bilayer. Thus, the composition of SNARE sequences may be optimized to have sufficient hydrophobicity to form stable SNARE complexes while at the same time avoiding membrane insertion.
It is interesting that the N-terminal half of the synaptobrevin SNARE motif does bind to DPC micelles (26) (Fig. 4A), despite its inability to bind to membranes. The basis for this difference is unclear, but it seems plausible that micelles may be more dynamic and adaptable than phospholipid bilayers, which may facilitate interactions with sequences of limited hydrophobicity. In any case, it now seems clear that the apparent contradiction between the NMR studies of synaptobrevin in DPC micelles (26) and initial studies suggesting that most of the SNARE motif of reconstituted synaptobrevin is flexible (24, 25) is not a real contradiction: Synaptobrevin actually has a different behavior in DPC micelles compared to phospholipid bilayers. Hence, DPC micelles do not appear to constitute a good model for membranes in this system. Conversely, our data show that synaptobrevin behaves very similarly on nanodiscs and on phospholipid vesicles, confirming the notion that nanodiscs provide a faithful membrane model. Although our NMR experiments of synaptobrevin in nanodiscs focused on analyzing flexible regions, recent studies have shown that good-quality NMR data can also be obtained for structured regions of membrane proteins inserted into nanodiscs (30–32). Hence, the nanodisc approach provides a promising avenue for structural studies of membrane proteins in their actual bilayer environment.
Experimental Procedures
Protein Expression and Purification.
The construct to express full-length rat synaptobrevin-2 within a pGEX-KG (51) vector was described previously (36). This construct was used to make a new construct to express the soluble rat synaptobrevin-2(1–96) fragment, also in the pGEX-KG vector, using standard recombinant DNA techniques. The proteins were expressed in Escherichia coli BL21(DE3) cells using minimal media with  and
and 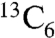 -glucose as the sole nitrogen and carbon sources, respectively (as needed for 15N and 13C labeling), and purified as described (18, 36). A construct to express human apolipoprotein A1 residues 68–267 (ApoA1) within a pET-28a vector (Novagen) was made from cDNA obtained from ATCC using standard recombinant DNA techniques. The protein was expressed in E. coli BL21(DE3) cells in LB broth and purified as previously described (52).
-glucose as the sole nitrogen and carbon sources, respectively (as needed for 15N and 13C labeling), and purified as described (18, 36). A construct to express human apolipoprotein A1 residues 68–267 (ApoA1) within a pET-28a vector (Novagen) was made from cDNA obtained from ATCC using standard recombinant DNA techniques. The protein was expressed in E. coli BL21(DE3) cells in LB broth and purified as previously described (52).
SNARE Reconstitution into Nanodiscs and Vesicles.
Nanodiscs containing full-length synaptobrevin were prepared in a manner previously described for rhodopsin (52). Briefly, 13C, 15N syb 1–116 was added to a mixture of ApoA1 and lipid with n-octyl-β-D-glucopyranoside (β-OG) and sodium cholate. The syb:ApoA1:lipid ratio was 1∶2∶120, and the mixture was prepared from stock concentrations of 110 μM, 200 μM, and 13 mM, respectively. The β-OG and sodium cholate were added from 10% stocks to a final concentration of 1%. The mixture was vortexed and incubated at room temperature for 30 min without disturbance. The nanodiscs were formed by passing the mixture over a 4-cm-high column Extracti-Gel D resin (Pierce) to remove the detergent. The nanodiscs were then run on a Superdex-200 Hiload 16/60 column (GE Healthcare) in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.0, 100 mM KCl, 0.3 mM tris(2-carboxyethyl)phosphine (TCEP) (reconstitution buffer), and concentrated using a 30-kDa molecular weight cutoff filter to the desired concentration. Proteoliposomes containing full-length synaptobrevin were prepared by detergent-assisted insertion of the protein into preformed liposomes of 100-nm diameter and the desired lipid composition in reconstitution buffer, basically as described (36). The synaptobrevin-to-lipid ratio was 1∶500, with final concentrations of 10 μM and 5 mM, respectively.
NMR Spectroscopy.
All NMR experiments were performed at 15 °C on Varian INOVA spectrometers operating at 800, 600, or 500 MHz. Most samples were dissolved in reconstitution buffer containing 5% D2O, except for those used in the experiments of Fig. 4 A and B, which were prepared with the same buffer but at pH 6.0. Samples for 1H-15N HSQC and transverse relaxation measurements contained approximately 10 μM protein. 15N transverse relaxation measurements were obtained with a 1H-15N HSQC-based pulse sequence incorporating a Carr–Purcell–Meiboom–Gill (CPMG) train applied to 15N (53). A similar sequence with a CPMG train applied to 1H was designed to measure 1H transverse relaxation rates. The synaptobrevin-nanodisc sample used for backbone assignments had a 90 μM protein concentration. Assignments were obtained with gradient-enhanced 3D CBCA(CO)NH and HNCACB experiments (54). All data were processed with NMRpipe (55) and analyzed with NMRView (56).
Supplementary Material
Acknowledgments.
We thank Yilun Sun for technical assistance and Sourabh Banerjee and Thomas Sakmar for advice on preparation of nanodiscs. This work was supported by grants from the Human Frontiers Science Program (RGP0035/2007-C), the Welch Foundation (I-1304), and the National Institutes of Health (NS37200) (to J.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105128108/-/DCSupplemental.
References
- 1.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jahn R, Scheller RH. SNAREs—Engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 3.Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 4.Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirier MA, et al. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- 7.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 8.Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 9.Rizo J, Chen X, Arac D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Dulubova I, et al. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci USA. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 12.Carr CM, Rizo J. At the junction of SNARE and SM protein function. Curr Opin Cell Biol. 2010;22:488–495. doi: 10.1016/j.ceb.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue M, Ma C, Craig TK, Rosenmund C, Rizo J. The Janus-faced nature of the C(2)B domain is fundamental for synaptotagmin-1 function. Nat Struct Mol Biol. 2008;15:1160–1168. doi: 10.1038/nsmb.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chicka MC, Hui E, Liu H, Chapman ER. Synaptotagmin arrests the SNARE complex before triggering fast, efficient membrane fusion in response to Ca2+ Nat Struct Mol Biol. 2008;15:827–835. doi: 10.1038/nsmb.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HK, et al. Dynamic Ca2+-dependent stimulation of vesicle fusion by membrane-anchored synaptotagmin 1. Science. 2010;328:760–763. doi: 10.1126/science.1187722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fasshauer D, Otto H, Eliason WK, Jahn R, Brunger AT. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J Biol Chem. 1997;272:28036–28041. doi: 10.1074/jbc.272.44.28036. [DOI] [PubMed] [Google Scholar]
- 18.Hazzard J, Sudhof TC, Rizo J. NMR analysis of the structure of synaptobrevin and of its interaction with syntaxin. J Biomol NMR. 1999;14:203–207. doi: 10.1023/a:1008382027065. [DOI] [PubMed] [Google Scholar]
- 19.Dulubova I, et al. A conformational switch in syntaxin during exocytosis: Role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholson KL, et al. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat Struct Biol. 1998;5:793–802. doi: 10.1038/1834. [DOI] [PubMed] [Google Scholar]
- 21.Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat Struct Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- 22.Misura KM, Scheller RH, Weis WI. Self-association of the H3 region of syntaxin 1A. Implications for intermediates in SNARE complex assembly. J Biol Chem. 2001;276:13273–13282. doi: 10.1074/jbc.M009636200. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Lu J, Dulubova I, Rizo J. NMR analysis of the closed conformation of syntaxin-1. J Biomol NMR. 2008;41:43–54. doi: 10.1007/s10858-008-9239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowen M, Brunger AT. Conformation of the synaptobrevin transmembrane domain. Proc Natl Acad Sci USA. 2006;103:8378–8383. doi: 10.1073/pnas.0602644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kweon DH, Kim CS, Shin YK. Regulation of neuronal SNARE assembly by the membrane. Nat Struct Biol. 2003;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- 26.Ellena JF, et al. Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc Natl Acad Sci USA. 2009;106:20306–20311. doi: 10.1073/pnas.0908317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 28.Sorensen JB, et al. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J. 2006;25:955–966. doi: 10.1038/sj.emboj.7601003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 30.Gluck JM, et al. Integral membrane proteins in nanodiscs can be studied by solution NMR spectroscopy. J Am Chem Soc. 2009;131:12060–12061. doi: 10.1021/ja904897p. [DOI] [PubMed] [Google Scholar]
- 31.Raschle T, et al. Structural and functional characterization of the integral membrane protein VDAC-1 in lipid bilayer nanodiscs. J Am Chem Soc. 2009;131:17777–17779. doi: 10.1021/ja907918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shenkarev ZO, et al. Lipid-protein nanodiscs: Possible application in high-resolution NMR investigations of membrane proteins and membrane-active peptides. Biochemistry (Mosc) 2009;74:756–765. doi: 10.1134/s0006297909070086. [DOI] [PubMed] [Google Scholar]
- 33.Giralt E, Rizo J, Pedroso E. Application of gel-phase 13C-NMR to monitor solid phase peptide synthesis. Tetrahedron. 1984;40:4141–4152. [Google Scholar]
- 34.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 35.Rigaud JL, Pitard B, Levy D. Reconstitution of membrane proteins into liposomes: Application to energy-transducing membrane proteins. Biochim Biophys Acta. 1995;1231:223–246. doi: 10.1016/0005-2728(95)00091-v. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, et al. SNARE-mediated lipid mixing depends on the physical state of the vesicles. Biophys J. 2006;90:2062–2074. doi: 10.1529/biophysj.105.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Landry SJ, Gierasch LM. The chaperonin GroEL binds a polypeptide in an alpha-helical conformation. Biochemistry. 1991;30:7359–7362. doi: 10.1021/bi00244a001. [DOI] [PubMed] [Google Scholar]
- 39.Fernandez I, et al. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 40.Walter AM, Wiederhold K, Bruns D, Fasshauer D, Sorensen JB. Synaptobrevin N-terminally bound to syntaxin-SNAP-25 defines the primed vesicle state in regulated exocytosis. J Cell Biol. 2010;188:401–413. doi: 10.1083/jcb.200907018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basu J, et al. A minimal domain responsible for Munc13 activity. Nat Struct Mol Biol. 2005;12:1017–1018. doi: 10.1038/nsmb1001. [DOI] [PubMed] [Google Scholar]
- 42.Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1∶1 syntaxin/SNAP-25 complex. Structure. 2008;16:308–320. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guan R, Dai H, Rizo J. Binding of the Munc13-1 MUN domain to membrane-anchored SNARE complexes. Biochemistry. 2008;47:1474–1481. doi: 10.1021/bi702345m. [DOI] [PubMed] [Google Scholar]
- 44.Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, et al. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 46.Xue M, et al. Binding of the complexin N terminus to the SNARE complex potentiates synaptic-vesicle fusogenicity. Nat Struct Mol Biol. 2010;17:568–575. doi: 10.1038/nsmb.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai H, Shen N, Arac D, Rizo J. A quaternary SNARE-synaptotagmin-Ca(2+)-phospholipid complex in neurotransmitter release. J Mol Biol. 2007;367:848–863. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, Su L, Rizo J. Binding of Munc18-1 to synaptobrevin and to the SNARE four-helix bundle. Biochemistry. 2010;49:1568–1576. doi: 10.1021/bi9021878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arac D, et al. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–217. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 50.Guo T, Gong LC, Sui SF. An electrostatically preferred lateral orientation of SNARE complex suggests novel mechanisms for driving membrane fusion. PLoS ONE. 2010;5:e8900. doi: 10.1371/journal.pone.0008900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 52.Banerjee S, Huber T, Sakmar TP. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J Mol Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 53.Farrow NA, et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15 N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 54.Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance 3-dimensional NMR experiments with improved sensitivity. J Magn Reson B. 1994;103:203–216. [Google Scholar]
- 55.Delaglio F, et al. NMRPipe—A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 56.Johnson BA, Blevins RA. NMR View—A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.