Abstract
The RelA-mediated stringent response is at the heart of bacterial adaptation to starvation and stress, playing a major role in the bacterial cell cycle and virulence. RelA integrates several environmental cues and synthesizes the alarmone ppGpp, which globally reprograms transcription, translation, and replication. We have developed and implemented novel single-molecule tracking methodology to characterize the intracellular catalytic cycle of RelA. Our single-molecule experiments show that RelA is on the ribosome under nonstarved conditions and that the individual enzyme molecule stays off the ribosome for an extended period of time after activation. This suggests that the catalytically active part of the RelA cycle is performed off, rather than on, the ribosome, and that rebinding to the ribosome is not necessary to trigger each ppGpp synthesis event. Furthermore, we find fast activation of RelA in response to heat stress followed by RelA rapidly being reset to its inactive state, which makes the system sensitive to new environmental cues and hints at an underlying excitable response mechanism.
Keywords: cytosolic diffusion, single particle tracking, photoactivated localization microscopy, stroboscopic illumination
Bacterial cells live in a dynamically changing environment and thus need to adapt quickly to changes in the surroundings. Upon amino acid limitation, the RelA-mediated stringent response globally remodels bacterial physiology (for an excellent review see ref. 1). This process of remodeling is mediated by the production of the alarmones guanosine pentaphosphate, pppGpp, and guanosine tetraphosphate, ppGpp. The ribosome acts as an activator of RelA-mediated (p)ppGpp production, with uncharged cognate tRNA in the A site being the primary inducer (2). The (p)ppGpp alarmone exerts its regulatory role chiefly via direct binding to the bacterial RNA polymerase (3), provoking changes in the profile of transcribed messages. (p)ppGpp also regulates bacterial replication via binding to DNA primase (4) and inhibits translation initiation via binding to initiation factor IF2, a translational GTPase (5, 6). Evidently, the stringent response is a core cellular adaptation pathway, acting as a hub in the regulatory network, where it integrates information about the nutritional status of the bacterial cell and regulates cellular metabolism on the transcription, translation, and replication levels.
Recently it became possible to clone and overexpress RelA from several bacterial species (7–9), reigniting interest in the stringent response. Using overexpressed Escherichia coli RelA, the most extensive in vitro analysis to date of the mechanism was undertaken in 2002 (10), arriving at the so-called hopping model. In this model, RelA binds to a stalled ribosome, senses the deacylated tRNA in the ribosome A site, becomes catalytically active, and synthesizes one ppGpp molecule. The act of ppGpp formation by RelA leads to dissociation of the factor from the 70S, and consequent rebinding to the next ribosome completes the cycle (see Fig. 1A). Successive “hops” from ribosome to ribosome monitor the global state of translational starvation. Because hopping events are unsynchronized within the RelA population in vivo, characterization of such events is beyond the reach of available in vivo bulk assays. At the very same time, in vitro biochemical methods, however powerful when applied to other ribosome-associated enzymes, have so far failed to characterize these events as well. The original hopping model (10) was inferred within a very general conceptual framework that encompasses a broad range of ribosome-interacting enzymes, be it translational GTPases (11), ribosome-inactivating toxins (12), or ribosome RNA modification enzymes (13). Direct experimental evidence for hopping, for the synthesis of one ppGpp alarmone per one dissociation event, and also for the reaction being performed on the ribosome, is lacking (10).
Fig. 1.
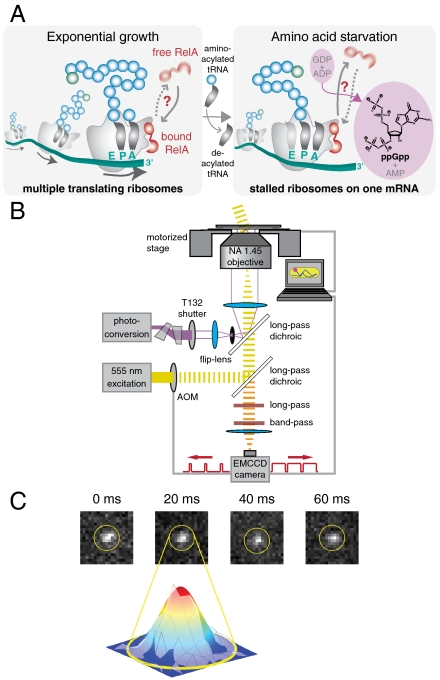
Single-molecule tracking in living cells. (A) Schematic drawing of RelA interacting with a polysome. During exponential cell growth, RelA could either be bound to the polysome (10) or diffuse rapidly in the cytosol (Left). During amino acid limitation the level of tRNA acylation drops, resulting in accumulation of deacylated tRNA in the ribosomal A site. Under these conditions RelA could either be in tight complex with a polysome (46) or unbind from the ribosome and undergo rapid cytosolic diffusion (Right). (B) Schematic diagram of the optical setup. An acousto-optical modulator is synchronized with an EMCCD camera and shutters a wide-field yellow excitation laser beam to pass short excitation pulses in the middle of each imaging frame. A violet photoconversion laser beam is spatially overlapped and can either be focused onto the back aperture of an Olympus TIRF objective or focused onto the sample plane via a flip-lens. (C) Four consecutive frames of a time-lapse movie of RelA–Dendra2 (in nonstarved cells) with a frame time of 20 ms and an exposure time of 5 ms and a Gaussian fit to frame 2.
This prompted the development of an in vivo approach based on single-molecule fluorescence imaging that allows us to directly observe the stringent response machinery, which we use to test the hopping model in vivo. To monitor RelA in its freely diffusing state it was necessary to develop a new method for tracking individual rapidly diffusing proteins in the bacterial cytosol. We demonstrate that free RelA in the bacterial cytosol, and ribosome-bound RelA, have radically different diffusive properties. This allows us to monitor the catalytic cycle of RelA in vivo by determining the diffusion characteristics of individual RelA molecules throughout their binding cycles. From this we infer the time scale within which individual molecules cycle between free and bound states, thus directly testing the hopping model in vivo. Lastly, by determining the fraction of ribosome-bound and freely diffusing RelA molecules we characterize heat shock in individual cells with unprecedented time resolution.
Results
The first two sections are dedicated to two reference experiments: the diffusion of a free cytosolic protein as a reference for free RelA, and the diffusion of individual ribosomes as a reference for ribosome-bound RelA. Neither of these targets have been characterized before, and these investigations are therefore important in their own right because they provide valuable insights into the nature of the bacterial cytosol and into the micro-structure of the bacterial translational machinery. The third section will be devoted to RelA during amino acid starvation and also during heat shock.
The two control experiments presented two challenges for single-molecule tracking. The first one involves the sheer number of ribosomes. An individual E. coli cell contains from 7,000 to 50,000 ribosomes (14). Such large copy numbers of targets necessitated a photoconversion approach, in which we only converted and tracked one or a few molecules at a time. These molecules were tracked until they bleached, whereupon the activation cycle was repeated. This approach allowed for the acquisition of good diffusion statistics for individual cells in vivo. The second challenge is the sheer speed at which free proteins move through the cytosol: Current in vivo tracking methodology based on photo-activated localization microscopy (PALM) (15, 16) and stochastic optical reconstruction microscopy (STORM) (17) is limited to the observation of slowly moving molecules, such as proteins bound to membranes (18, 19) or other relatively immobile structures such as DNA (20) or the cytoskeleton (21). These methods have therefore recently been complemented by FCS-based tracking schemes (22) that have excellent temporal resolution but very limited spatial range. To track the cytosolic reference protein and RelA in its free state we improved in vivo tracking to permit monitoring of fast diffusive processes. This was accomplished by combining superresolution tracking of photoconvertible proteins (18, 21) with a technique borrowed from high-speed photography, stroboscopic time-lapse imaging (20, 23). The key to this is to hardware synchronize short laser excitation pulses with the frame time of the camera such that the fluorophores are effectively immobile during the imaging and do not yield blurred-out views of the diffraction-limited spots, which would be limiting in the presence of the autofluorescent background (see Fig. 1 B and C and Materials and Methods).
Stroboscopic Single-Molecule Tracking of Freely Diffusing Cytosolic Proteins.
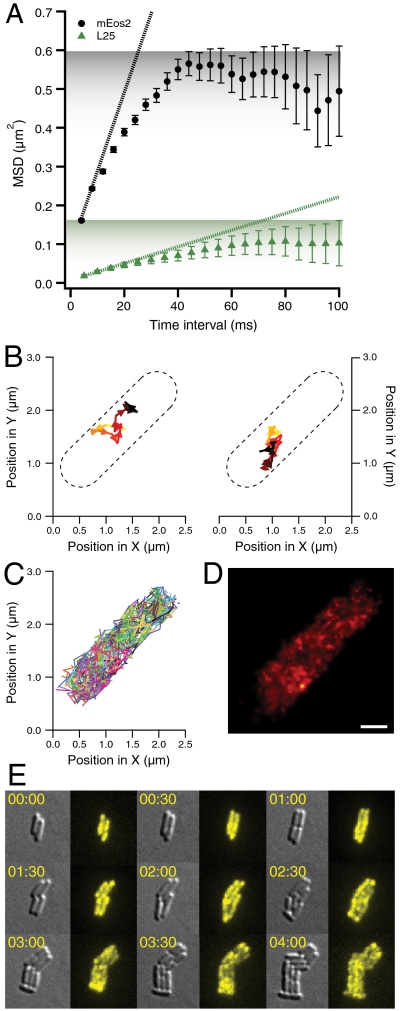
As a reference point for free RelA we chose a small cytosolic protein: the GFP variant mEos2. It is a small 26-kDa cytosolic protein whose fluorescence is photoconvertible from green to red (24). We stochastically converted mEos2 to spawn single fluorophores, which allowed for repeated single-molecule tracking within the same living cell. Here we present the first dataset from single-molecule tracking in the cytosol (see SI Materials and Methods in the SI Appendix). The dataset consists of hundreds of trajectories each for eight individual cells, comprising a total of 3,766 single-molecule mEos2 trajectories, which allowed us to characterize how the bacterial cytosol is perceived by a small protein all the way down to the 4-ms time scale. Individual trajectories of mEos2 display several characteristics. First, mEos2 molecules are very fast, covering a considerable part of the bacterial cells between the individual frames even at a frame rate of 250 Hz (Fig. 2A). Second, diffusion traces of mEos2 molecules seem to be evenly distributed over the entire volume of the cell, which is especially obvious when individual trajectories are overlaid (Fig. 2B) and sample the whole volume (Fig. 2 A and B).
Fig. 2.
Normal free diffusion in the cytosol of E. coli. (A) A single experimentally obtained single-molecule mEos2 trajectory with a frame time of 4 ms and an exposure time of 1 ms. (B) Overlay of 1,355 single-molecule mEos2 trajectories obtained in an individual E. coli cell with a frame time of 4 ms and an exposure time of 1 ms. (C) Local apparent diffusion coefficients in the sample (x-y) plane. The apparent diffusion coefficients are evaluated every 20 by 20 nm in an x-y grid. Each point in the figure is false-colored according to the apparent diffusion coefficient calculated from the mean square displacement over 4 ms for experimental (Upper) and simulated (Lower) displacements originating within 200 nm of this point. The simulations assume normal diffusion at D = 12.5 μm2 s-1 in the volume defined by the geometry of this cell. There is good agreement between apparent experimental diffusion coefficients and those obtained from simulations. The apparent diffusion coefficients are higher in the middle of the cell, as the molecules are less confined along the long axis. Noise contributions make the apparent diffusion faster close to the cell wall. (D) Mean square displacements (MSDs) in the sample (x-y) plane for different time intervals. Experimental MSDs and error bars representing experimental standard errors of the means are displayed in red. The confidence intervals (blue) are obtained from simulations in the volumes defined by the cell geometry by calculating and sorting MSDs for trajectories using a diffusion coefficient of 13 μm2 s-1. The average MSDs (black, dashed) are also obtained from simulations. Here we vary the diffusion coefficient for each cell to obtain the closest match to the experimental curve. (E) Overlay of 500 positions of single-molecule mEos2 trajectories in one E. coli cell with a frame time of 4 ms and an exposure time of 1 ms. Each position is represented by a Gaussian with a standard deviation equal to the localization error. The mean localization error is 44 nm and the scale bar represents 500 nm.
A quantitative way of analyzing the trajectories in Fig. 2B is by calculating the local apparent diffusion coefficients throughout the cell. These rates are based on how far individual molecules originating from small cellular subregions move within the 4-ms frame time (Fig. 2C, Upper). There appears to be some spatial variation in apparent diffusion coefficients across the cell (8 to 16 μm2 s-1). This variation can be erroneously interpreted as variations in intracellular viscosity. Identical patterns emerge when we simulate normal diffusion using a uniform rate of diffusion throughout the cell and take into account cellular geometry (see Fig. 2C, Lower): Bacterial geometry results in apparent diffusion that is faster in the middle of the cells where molecules can diffuse in a less restricted manner and slower in the quarter positions where molecules have encountered the wall and returned within 4 ms.
Fig. 2D depicts an experimental mean square displacement (MSD) curve for mEos2. We compare the experimental data points to MSD curves calculated from simulated normal diffusion trajectories within the cell geometry. The good fit suggests that cytosolic diffusion is indistinguishable from a Brownian walk. As can be seen in Fig. S1 in the SI Appendix, a single microscopic diffusion coefficient of 13 μm2 s-1 can adequately describe all of our data obtained from all eight cells, because all eight experimental MSD curves fall within their respective confidence intervals. Whereas we present a more detailed treatment of the dataset and of microscopic simulations for mEos2 diffusion in SI Materials and Methods in the SI Appendix, in what now follows, we will use apparent diffusion coefficients throughout. We also complemented our single-molecule analysis with ensemble photoactivation (PA) performed on one of the analyzed cells (see Fig. S2 in the SI Appendix). The obtained diffusion coefficient of 11 μm2 s-1 is in reasonable agreement with our single-molecule analysis, especially when considering the spatial coarseness of PA.
Taken together, our results establish that the small cytosolic protein mEos2 freely diffuses within the entire volume of the bacterial cell, seemingly unperturbed by internal structures such as the nucleoid or the cytoskeleton. This inherent intracellular inertness, which gives rise to the simple diffusion properties, makes mEos2 and similar GFP variants [Dendra2 (25), PA-GFP (26), Dronpa (27), etc.] ideal fusion partners when tracking other proteins for which deviations from simple Brownian diffusion is of biological interest, such as RelA.
In Vivo Single-Molecule Tracking of Bacterial Ribosomes.
As a reference point for ribosome-bound RelA we tracked individual fluorescently labeled ribosomes in exponentially growing cells, because reports on the motion of individual ribosomes in the bacterial cell are generally lacking in scientific literature.
Fig. 3A contrasts an MSD curve of 70S diffusion (from 537 individual ribosomal trajectories, comprised of 3,421 positions) with that of mEos2, which we have previously described. A striking difference is immediately obvious; the apparent diffusion coefficients of ribosomes are at least 10 times lower (see Fig. 3A, slopes). Because mEos2 diffusion is extremely rapid, the cytosolic confinement is already noticeable at the 4-ms time scale, which reduces the apparent diffusion coefficients to values that are much lower than the microscopic diffusion coefficient for mEos2 (5.1 vs. 13 μm2 s-1; see SI Materials and Methods in the SI Appendix). As can be seen in Fig. 3A, the ribosome is not close to reaching the cell boundaries even after 100 ms, and hence its apparent diffusion coefficient obtained from the slope of the first two points of the MSD curve is not significantly affected by cytosolic confinement. From this, we estimate ribosomal diffusion to be at least 25 times slower than diffusion of mEos2. Additionally, the MSD curve plateaus at a much lower level as compared to the one for mEos2 (see Fig. 3A, shading). This lower plateau cannot be explained by confinement within the boundaries of the cytosol. There could be several possible reasons for this behavior. First, ribosomes could be confined to areas of localized translation within the cell, as was documented by deconvolution microscopy for Bacillus subtilis (28, 29). Alternatively, another cause for this behavior could be that the ribosomes are tethered to mRNA (30). mRNA–protein complexes have been shown to display subdiffusion in previous studies (31, 32).
Fig. 3.
Single-molecule ribosome tracking and ensemble time-lapse imaging in individual living cells. (A) Mean square displacements (MSDs) in the sample (x-y) plane for mEos2 (in black, from 3,766 trajectories) and ribosome (in green, from 537 trajectories) tracking for different time intervals with error bars representing standard errors of the means. For ribosome tracking, we labeled the ribosomal protein L25 with the photoconvertible GFP variant Dendra2 (25) (see Materials and Methods). The dotted lines are calculated from the slope of the first two time points (corresponding to Dapp of 5.1 and 0.5 μm2 s-1, respectively). As estimated from the initial slopes of the MSD curves, mEos2 has a 10-fold higher apparent diffusion coefficient than Dendra2-labeled ribosomes. The shading represents the different plateaus for mEos2 and L25 MSDs. (B) Two experimentally obtained single-molecule ribosome trajectories with a frame time of 50 ms. The individual ribosome trajectories are recorded for 0.65 and 1.15 s, respectively. The ribosome is tagged via an N-terminally Dendra2 labeled L25 ribosomal protein. (C) Overlay of all 224 single-molecule ribosome trajectories in one E. coli cell with a frame time of 50 ms. (D) Overlay of 1,000 positions of single-molecule ribosome trajectories in one E. coli cell with a frame time of 50 ms. Each position is represented by a Gaussian with a standard deviation equal to the localization error. The mean localization error is 43 nm and the scale bar represents 500 nm. (E) Time-lapse image acquisitions (differential interference contrast and fluorescence imaging) of ribosome distributions in dividing E. coli cells. The fluorescence of Dendra2 from chromosomally labeled C-terminal ribosomal protein S2 is activated at time zero. Subsequent time-lapse imaging followed the initial distribution of the photoconverted ribosomes as they are passed between the cells upon repeated cell division. The cellular distribution of this photoconverted Dendra2 is recorded every five minutes. We present nine snapshots over a period of four hours. Experiments with ribosomal protein L19 and L31-labeled strains resulted in similar data.
Fig. 3B displays two trajectories of individual ribosomes. The ribosomes stay localized for seconds and are clearly confined to micro-domains as if locally tethered. The overlay of all 224 ribosomal trajectories obtained from one living cell (composed of 1,322 positions) (Fig. 3C) looks virtually identical to the mEos2 overlays (Fig. 2B), although each individual ribosomal trajectory has markedly different properties as compared to that of mEos2. This highlights the importance of the single particle tracking (SPT) approach: The subdiffusive nature of the ribosome and the free diffusion of mEos2 are readily distinguishable from their individual trajectories.
In Fig. 3D we show a composite PALM superresolution image from one living cell, obtained from individual trajectories by representing each ribosome position as a Gaussian with a standard deviation equal to the localization error (15). When all ribosome positions are plotted it is apparent that ribosomes are not compartmentalized to small subregions of the cell, which indicates that tethering, as opposed to physical confinement, is the primary cause for the observed subdiffusive behavior of ribosomes in exponentially growing cells. This is also in line with the recent observation in Caulobacter crescentus, where L1-tagged ribosomes, though being evenly distributed throughout the cytosol, are unable to diffuse freely as measured by fluorescence recovery after photobleaching. Interestingly, this subconfinement greatly decreases in mRNA-depleted C. crescentus (30).
To complement our PALM superresolution tracking, we also performed ensemble time-lapse studies, which allowed monitoring of ribosomal partitioning over several repetitive cell divisions. A single photoconversion pulse at time zero allows us to label and follow ribosomes present at time zero over several cell generations. Dendra2 labeled ribosomes, which are newly synthesized after time zero, will remain invisible. The ribosomes from time zero are not differentially distributed to one of the daughter cells after cell division, and we see an even distribution of labeled ribosomes even after several cell divisions (see Fig. 3E for S2–Dendra2 fusion analysis). We also observed, albeit rarely, ribosomal aggregates localized mostly at the poles, where they appear to be virtually immobile (see Fig. S3 in the SI Appendix) and are inherited along with the poles. These aggregates are most probably involved in ribosomal degradation, and similar degradation granules have been documented earlier (33).
In Vivo Single-Molecule Tracking of RelA.
Now we are in position to examine the dynamics of the stringent response factor RelA itself at the single-molecule level. For this, RelA–Dendra2 fusion constructs were made using λ Red chromosomal integration (34). Chromosomal integration is imperative for three reasons: First, the physiological ratio of RelA to ribosomes must be preserved, because this ratio has been shown to be of crucial importance to RelA enzymatic activity (10); second, this provides stoichiometric labeling of all RelA molecules, which is of importance due to the relatively low in vivo copy number of RelA (35); third, the absence of nonlabeled RelA molecules in the cell allows us to evaluate the function of the tagged protein via ensemble growth assays under stressed and nonstressed conditions. Such activity controls are critical because essentially all fusion proteins are at least partially impaired by their fusion partners.
Thus, using growth rate assays of chromosomally integrated relA–dendra2, we tested various linkers of the C-terminal fusion constructs for highest in vivo activity (see Figs. S4, S5, and S6 and SI Materials and Methods in the SI Appendix). The resulting strain will be referred to as RelA–Dendra2 throughout this text. Our ensemble growth assays demonstrate that RelA–Dendra2 has indistinguishable growth curves as compared with the wild-type strain in amino acid rich buffers (Fig. S6 in the SI Appendix). Furthermore, its recovery from L-Serine Hydroxamate (SHX)-induced starvation is 3.5 h faster as compared to the N-terminal chromosomal fusion strain, and RelA–Dendra2 has a similar recovery time to wild-type E. coli when starvation is induced by overload of selected amino acids (Fig. S6A in the SI Appendix), or when starvation is brought about by SHX (Fig. S6B in the SI Appendix).
We also subjected our RelA–Dendra2 to the classical SMG plate test for relaxed phenotype and scored it for bacterial growth after incubation for 48 h at 37 °C (36). As is evident from Fig. S7 in the SI Appendix, our strain is relA+. This is further strong evidence that our RelA–Dendra2 construct is functional. It should be noted that even though RelA–Dendra2 recovers much faster than a relA- strain on the SMG plates (Fig. S7 in the SI Appendix), it does not grow quite as fast as the wild-type strain (BW25993) (Fig. S6 in the SI Appendix). Moreover, it should be noted that the SMG plate test inherently lacks time resolution and does not reflect differences in growth resumption kinetics, which are readily picked up by growth kinetics measurements in liquid cultures (Figs. S4, S5, and S6 in the SI Appendix).
From all the tests outlined above we conclude that RelA–Dendra2 is active in vivo, although the fusion with the fluorescent protein has an effect on its activity. This C-terminal construct RelA–Dendra2 is detected as a single band with appropriate mass by Western blotting after overexpression from a plasmid (Fig. S8 in the SI Appendix). Low in vivo RelA concentrations (35) rendered RelA–Dendra2 fluorescence undetectable by fluorescence flow cytometry (Fig. S9B in the SI Appendix), underscoring the sensitivity of our single-molecule tracking setup.
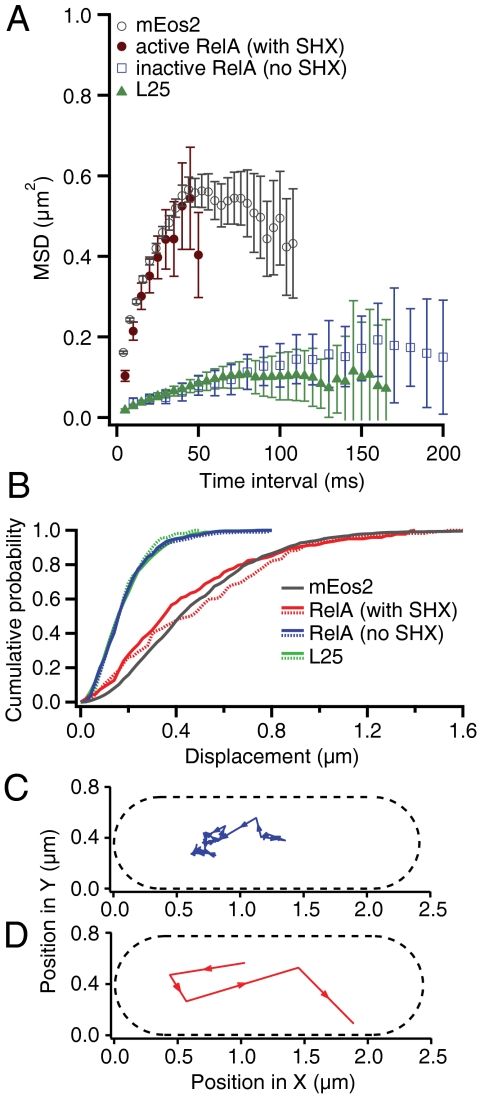
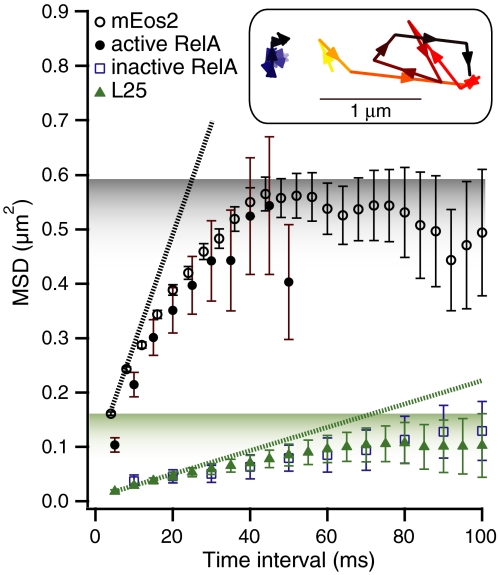
We first tracked RelA under amino acid rich conditions when we expected RelA to be enzymatically inactive. A sample trajectory of RelA under these conditions is presented in Fig. 4C. Fig. 4A displays the MSD curve of a representative dataset composed of 90 individual single-molecule RelA trajectories (composed of 527 positions) obtained from a microcolony of exponentially growing cells (in blue). Both mean MSD curves and cumulative distribution functions (CDFs) of displacements (37) for RelA trajectories are statistically indistinguishable from their ribosomal counterparts (see Fig. 4 A and B, respectively), which shows that RelA displays the same localized subdiffusive behavior as the ribosome itself (compare Fig. 4C to Fig. 3B). From this we deduce that RelA is in tight complex with the ribosome under nonstarved conditions.
Fig. 4.
Diffusion comparison of the ribosome to RelA in exponentially growing E. coli cells and to RelA during stringent response. (A) Mean square displacements (MSDs) in the sample plane of E. coli cells over different time intervals. The error bars represent the experimental standard errors of the means. MSDs from the ribosomal protein L25 are displayed in green (5-ms frame time, 537 single-molecule trajectories, 3,421 positions). When E. coli cells are growing exponentially the MSDs of RelA diffusion (in blue, 10-ms frame time, 90 trajectories, 527 positions) are indistinguishable from L25 diffusion. When cells are starved by addition of 2.5 mM of L-Serine Hydroxamate (brown MSD curve, 5-ms frame time, 216 trajectories, 977 positions), the diffusion of RelA changes dramatically, and is similar to that of mEos2 (average MSDs obtained from all 3,766 single-molecule mEos2 trajectories is displayed in gray). (B) Cumulative distribution functions (CDFs) of displacements over 20 ms in the sample plane of E. coli cells. Two CDFs for trajectories of RelA in its inactive unstarved state (in solid-blue with 20-ms frame time, 129 trajectories, 1,325 positions, and dashed-blue with 10-ms frame time, 90 trajectories, 527 positions) and that of L25 (in dashed-green with 20-ms frame time, 49 trajectories, 478 positions, and solid-green with 5-ms frame time, 537 trajectories, 3,421 positions) are indistinguishable. The apparent diffusion coefficient of RelA when cells are starved increases more than eightfold (red and dashed-red curves) and is very similar to the CDF of mEos2 (in gray, average CDF obtained from eight cells). (C) One experimentally obtained single-molecule RelA trajectory with a frame time of 20 ms and an exposure time of 2 ms when cells are exponentially dividing. (D) One experimentally obtained single-molecule RelA trajectory with a frame time of 20 ms and an exposure time of 2 ms when cells are starved using 2.5 mM L-SHX.
Next we severely starved cells of the amino acid serine by adding the competitive inhibitor of seryl-tRNA synthetase, L-Serine Hydroxamate (SHX) (final concentration 2.5 mM) (38), which both rapidly and fully inhibits serine tRNA charging thus bringing about prolonged stringent response. Under these conditions ppGpp synthesis is induced within approximately 1 min following addition of SHX and persists for at least an hour (39). While this stringent response is artificially produced and prolonged, it provides an ideal platform for delineating the behavior of the ppGpp synthesizing state of RelA, because under these conditions all RelA molecules are expected to be engaged in prolonged active catalysis.
During the SHX treatment we observe that RelA diffusion undergoes a radical change (Fig. 4D) and becomes very similar to that of mEos2. RelA now diffuses much more rapidly and no longer displays subdiffusive behavior, instead sampling the entire volume of the bacterial cell (compare Figs. 2A and 4D). As we do not observe any 70S rebinding events, RelA appears to spend the major portion of its time off, rather than on the ribosome. The close similarity between mEos2 and active RelA trajectories is confirmed when we compare their MSD curves (Fig. 4A). The brown curve in Fig. 4A displays an MSD curve of a representative dataset of 216 individual single-molecule RelA trajectories (composed of 977 positions) obtained from cells plated on an SHX-containing agarose pad. Fig. 4B depicts the CDFs of two datasets each for RelA under active and inactive conditions, reconfirming the striking difference between the two states. In the active state, the CDF curves for RelA dynamics closely resemble those of mEos2 (compare gray and red CDFs, respectively), while in the inactive state, the CDF curves for RelA dynamics are indistinguishable from those of the 70S (compare green and blue CDFs, respectively). These experiments clearly show that one prediction of the “hopping” model holds true: RelA does indeed dissociate from the ribosome under starvation conditions. However, we do not observe RelA “hopping” in the sense that it is immediately rebinding to another ribosome. On the contrary, it stays off the ribosome for hundreds of milliseconds, which suggests an extended hopping model (see Discussion).
Having characterized the two extreme states of the RelA cycle (i.e., perpetually active and inactive RelA), we next applied our methodology to a more physiologically common and a more transient cellular stress response—namely, a temperature upshift (40–42). We took advantage of RelA behaving as a logical switch, changing its diffusion characteristics between two distinct states (active and inactive—i.e., fast or slowly diffusing, respectively) and we utilized this as a readout of cellular stress on the single cell level.
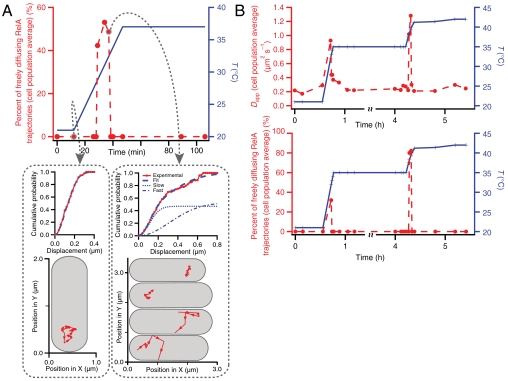
We heat-shocked E. coli cells by rapidly increasing the temperature from 21 to 37 °C, from 21 to 35 °C and from 35 to 42 °C (Fig. 5). In all of these cases, within several minutes after the temperature upshift, RelA dissociates from the ribosome and, as in the case of SHX-induced stringent response, diffuses freely. The striking difference to the SHX-treated cells is that after the temperature upshift the initial inactive state is restored within 10 min, as all RelA molecules observed rebound to ribosomes.
Fig. 5.
Diffusion characteristics of RelA during temperature upshift. (A) Individual RelA trajectories are recorded with a frame time of 10 ms and a laser exposure time of 2 ms. The temperature is adjusted from a constant 21 to 37 °C during the measurement. When the temperature is held constant at 21 °C, only slowly diffusing RelA trajectories are observed (see lower left panel for one representative trajectory recorded just before the start of the temperature shift). This diffusion is indistinguishable from ribosome diffusion (see Fig. 4B, green curves). During the temperature jump one transiently observes fast diffusion trajectories [see lower right panel, where two representative cells have inactive RelA (upper most cells), and the two lower cells display rapid RelA diffusion]. Very briefly roughly half of the cells display rapid RelA diffusion reminiscent of RelA diffusion during stringent response (see Fig. 4B, red curves). After the temperature has reached a constant 37 °C once again only slowly diffusing RelA trajectories are observed. Each point is an average over 20 trajectories. (B) Individual RelA trajectories are recorded with a frame time of 20 ms and a laser exposure time of 2 ms. The temperature is adjusted twice during the measurement, first from a constant 21 to 35 °C and again from 35 to 42 °C. The top panel displays apparent diffusion coefficients obtained by fitting P(r,Δt) = 1 - exp(-r2/4DappΔt) to the experimental CDFs of all the displacements in the sample plane. An apparent diffusion coefficient of 0.2 μm2 s-1 is obtained when the temperature is held constant either at 21, 35, or 42 °C. Again, rapidly diffusing RelA trajectories are observed transiently when the temperature is adjusted both from 21 to 35 °C and again from 35 to 42 °C (see lower panel). Each point is an average over 10 trajectories.
To estimate the ratio of the enzymatically active versus inactive RelA molecules in each time point during the temperature upshift, we fit the CDF curve for all recorded trajectories with a linear combination of two CDFs: a fast one, representing free RelA, and a slow one, representing the 70S-bound RelA from data also presented in Fig. 4B. In doing so, we have a straightforward way of deconvoluting the entire dataset and estimating the percentage of the two RelA states at any given time. This is only possible due to an order of magnitude difference in the apparent diffusion coefficients for the two RelA states (see Figs. 3A and 4A), which allows us to use the apparent diffusion coefficients as a logical operator for sorting the particles. As a control for the automated sorting procedure we also sort our trajectories manually into two categories (see Fig. 5A, Bottom), and this approach gives the same ratio as our global deconvolution method described above.
Looking at the individual trajectories at any given time during the very fast temperature response, we observe rapid RelA diffusion in about half of the cells (Fig. 5 A and B). When cells overcome the temperature shock, the entire RelA population shifts to the inactive state. All cells will have responded within approximately 10 min, and all cells return to the slow inactive state even when shocked to a final temperature of 42 °C. Our observations corroborate nicely with the transient ppGpp level increase documented in refs. 40 and 42. Just as in the case of our SHX-induced amino acid starvation tracking experiments, the observed timing of our responses follows the same pattern of high ppGpp levels reported in microbiological experiments (39, 40, 42).
Discussion
How does our assay compare to traditional methods for studying diffusion in living cells?
If the copy number of the cytoplasmic protein is very high, its diffusion can be studied using fluorescence recovery after photobleaching (FRAP) or photoactivation (PA) (43). We have complemented our single-molecule analysis of mEos2 diffusion with ensemble PA performed on one of the analyzed cells (see Fig. S2 in the SI Appendix) and we obtain a diffusion coefficient that is in reasonable agreement given that cell geometries and PA spots are diffraction-limited, which makes PA and FRAP inherently limited in their spatial resolution. A more important limitation of PA and FRAP is the inability to keep track of molecules as soon as they move out of the laser focus(es). This, among other things, makes it impossible to determine for how long an individual molecule is confined to a given subregion of a cell. While our single-molecule tracking assay readily reveals local diffusion and subcellular confinement of individual ribosomes, techniques such as FRAP and PA would miss the subcellular confinement and local diffusion and result in an artificially low estimate of the diffusion coefficient. If the concentration of the cytoplasmic protein ranges from 10-10 M to 10-6 M, its local diffusion properties can be studied using fluorescence correlation spectroscopy (FCS) (44, 45). This technique has excellent temporal and spatial resolution, especially when combined with stimulated emission depletion (STED) (22). However, because FCS operates in a very limited spatial region at any given time, one cannot obtain long trajectories for individual molecules moving throughout the cell. This means that it is not possible to study binding kinetics directly with FCS because individual molecules cannot be followed between binding events. Another in vivo limitation of FCS is that single-copy number targets, such as RelA, where we observe, at most, several RelA molecules per cell, are simply too dilute to be studied.
Unlike FRAP, PA, or FCS, the stroboscopic time-lapse tracking assay of photoconvertible proteins has no limitations on the in vivo copy number. We can track high-copy number targets, such as ribosomes or mEos2, where we recorded hundreds of individual molecules per cell. This allows us to obtain excellent statistics even for individual cells, which allows us to analyze cell-to-cell variability (see Fig. S1 in the SI Appendix). The technique also allows us to study single and low copy number targets in vivo, such as RelA. In this concentration range, the technique allows for simultaneous analysis of individual trajectories from a micro-colony of cells. Regardless of copy number, our technique can directly reveal spatial heterogeneity, spatial subconfinement, and, given sufficient statistics, binding kinetics.
Using precise single-molecule tracking in living E. coli cells, we directly observe two RelA states in vivo, a slowly diffusing inactive state, which we assign to be ribosome-bound, and a fast freely diffusing active state that is observed under conditions of amino acid starvation. This corroborates some aspects of the hopping model (10) and directly disproves an alternative model that suggested that RelA strongly binds to the ribosome under starvation conditions (46).
We confirm in vivo that under nonstarved conditions RelA is indeed ribosome-bound and that under amino acid starvation it does dissociate from the ribosome. The hopping model predicts that RelA subsequently hops from ribosome to ribosome producing exactly one ppGpp molecule per dissociation event. Hence RelA is predicted to toggle between its ribosome-bound and free states with a corresponding frequency. Reported in vitro turnover rates of RelA are about 70–75 per second (7), and in vivo estimates of ppGpp production under starvation conditions are of order 50,000 molecules per cell per second (47), which taking into account RelA cellular copy number of about 100 (35), gives similar turnover estimates. Such rapid alterations in diffusion characteristics of RelA would easily have been detected in our single-molecule tracking assay, as discussed below.
We, however, observe all RelA molecules remaining dissociated from the ribosome for hundreds of milliseconds. The temporal resolution of our assay is 4 ms due to the limitations in the EMCCD camera readout time. At the same time, the dynamic range spans from 4 ms (the fastest achievable frame time, see above) to seconds (limited by trajectory lengths). In the scenario where the catalytic cycle of RelA is much, much too fast to be captured by our assay, and we therefore would miss all of the thousands of binding events during the course of one trajectory, the two substrates of RelA (ATP and the G nucleotide, GTP or GDP), and RelA itself would have to bind to a stalled ribosome, RelA would have to conclude its catalytic event, and dissociate from the ribosome all in well under 1 ms.
This is orders of magnitude faster than ribosome-binding kinetics determined for other translational components and is much faster than even ricin catalyzed rRNA cleavage (48), which is, to our knowledge, the fastest enzyme “hopping” between the ribosomes, and is probably beyond the diffusion limit for a protein the size of RelA (49).
In the other scenario where the RelA catalytic cycle happens far too infrequently to be captured by our assay (i.e., much longer than seconds), it would take hours for RelA to produce enough ppGpp to cause the stringent response: The cellular copy number of RelA is not over 100 (35), and ppGpp concentrations of 0.5 mM are reached in vivo (50), which corresponds to 300,000 molecules of ppGpp. Hence even in the fastest scenario in the absence of cellular ppGpp degradation, it would take about an hour to build up adequate ppGpp levels at a synthesis rate of 1 per second, which is far too long to be compatible with the available in vivo data. Both our heat-shock experiments (where cellular adaptation occurs within minutes (and not hours), as well as the documented rise of ppGpp in about a minute after SHX shock (39) are consistent with RelA turnover rates of 10s to 100s per second, not less than 1 per second.
This is why we argue that multiple ppGpp molecules have to be synthesized every time RelA dissociates from the ribosome. We therefore propose an “extended hopping” model—a modification of the original hopping model where many ppGpp molecules are produced upon the dissociation of enzymatically active RelA from the ribosome, and with RelA active off, rather than on the ribosome. Our new model can be rationalized in the framework of existing biochemical data for RelA, which show that RelA has an intrinsic ability to produce ppGpp, which it can auto-inhibit through the use of a dedicated inhibition domain (51). When this domain is removed, RelA switches into a perpetually active state (51). Possibly, activation by the ribosome is in fact a suppression of auto-inhibition. Within this framework it seems only natural that following dissociation from the ribosome, RelA remains active for some time before its auto-inhibition is engaged. The severe and lasting starvation induced by SHX was used to keep RelA in its active mode for an extended time in order to probe its catalytic cycle. We also studied RelA dynamics during heat-shock response, which is a more natural and transient form of stress. Furthermore, RelA’s direct involvement in heat shock has not been possible to study in vitro, since the primary heat-shock sensor is unknown. Our in vivo enzymatic assay reveals that in this case, as well, RelA dissociates from the ribosome at the onset of stress, but in this case its excursion into a fast active state is more transient in nature: Within less than 10 min, the RelA sensory system adapts to the new temperature and resets, ready to respond to further changes in temperature. There are at least two possible topologies for the heat sensing network, which is known to be mediated by RelA and SpoT (40), a close homologue to RelA. First, both RelA and SpoT sense the heat change independently. Second, SpoT acts as the primary sensor: Because temperature changes are unlikely to be sensed by increased levels of deacylated tRNA, it is conceivable that SpoT triggers RelA activation via an unidentified intracellular messenger. Both of the proposed topologies are consistent with a much smaller but still detectable increase in ppGpp levels even in RelA knockout strains, where ppGpp-mediated heat shock is still functional, and where SpoT acts as the sole source of intracellular ppGpp (40). In the latter topology, RelA, while being transiently unbound from the ribosome, acts as an amplifier of the heat response by significantly boosting ppGpp production after the initial rise in intracellular ppGpp synthesized by SpoT, the initiator of the heat response.
On our path to the in vivo mechanism of RelA, we have characterized the intracellular diffusion of both ribosomes and free proteins. We have shown that the movement of individual ribosomes is very slow and highly subdiffusive in nature. The subdiffusive ribosomal movement is most likely due to mRNA tethering, the diffusion of which displays very similar properties (32). On a much longer time scale, time-lapse movies of fluorescently labeled ribosomal proteins have not revealed preferential partitioning of ribosomes during cell division.
While more thorough investigations would benefit from a 3D superresolution approach, be it 3D SPT (52) or 3D PALM (15), these investigations lay the groundwork for a more detailed single particle tracking investigation of the translation process in living cells, as well as into ribosomal protein dynamics during ribosome assembly (53) and into ribosomal aging and repair by exchange of ribosomal proteins (54). It also lays the foundation for forthcoming studies with a more detailed analysis of ribosomal distributions during the exponential phase (55), stationary phase (56–58), and during cold shock (59).
Our in vivo single-molecule investigations into the stringent response were made possible due to the development of an assay that can detect also the very fast freely diffusing protein states that would otherwise be overlooked. We found that the diffusion characteristics of the inert freely diffusing mEos2 protein are indistinguishable from Brownian motion. This inherent intracellular inertness, which gives rise to the simple diffusion properties, makes mEos2 and similar GFP variants ideal fusion partners when tracking other proteins for which deviations from simple Brownian diffusion is of biological interest. In addition, the time resolution of this stroboscopic tracking assay makes it possible to monitor binding events in the millisecond regime. This time resolution is on par with even the fastest cellular protein dynamics, which makes the assay a general tool for investigation of intracellular kinetics in living cells.
Materials and Methods
Laser Microscopy.
A schematic diagram of the optical setup is shown in Fig. 1B. An acousto-optical modulator (AOM, IntraAction, 40 MHz) shutters a wide-field yellow excitation laser beam into an Olympus IX81 inverted microscope. The excitation laser light from a 555-nm DPSS laser (CrystaLaser) is first filtered (Z 550/20×, Chroma Technology) and then focused onto the back aperture of an Olympus TIRF objective (NA = 1.45). The objective collimates the light and excites either a 4-μm- (for mEos2 tracking) or a 20-μm-wide area (for ribosomal and RelA tracking) at the sample plane with laser power densities of 50 to 200 kW cm-2. The AOM is synchronized with a PhotonMax EMCCD camera (Princeton Instruments) by a NI-DAQ M-series data acquisition card (PCI-6259, National Instruments), and is controlled via LabVIEW 8.5 (National Instruments) to pass short 0.3- to 5-ms excitation pulses in the middle of each imaging frame. Our stroboscopic illumination time is optimal in the range of 1 ms (for mEos2 tracking) to 5 ms (for L25 tracking). The short duration of the excitation pulse is a critical parameter for obtaining distinct fluorescence snapshots from our rapidly moving single molecules. This makes it possible to track them at superresolution throughout all frames (see Fig. 1C for four consecutive raw image frames of a RelA–Dendra2 trajectory). Another equally important advantage of stroboscopic illumination is that it allows for a direct comparison between fast and slow molecules under identical illumination conditions since snapshots can be spaced out with varying time intervals.
A violet photoconversion laser beam (405 nm Radius, Coherent) is spatially overlapped using a long-pass dichroic filter (Z405RDC, Chroma) and is focused at the sample plane. An anamorphic prism pair (06 GPA 204, Melles Griot) and a 50-μm pinhole (Thorlabs) correct the asymmetry of this laser beam to produce a TEM00 mode. The photoconversion laser is independently shuttered using a UNIBLITZ T132 shutter controller that delivers 3-ms activation pulses at regular time intervals at power densities of order 0.1 kW cm-2.
A long-pass dichroic filter (Z555RDC, Chroma) is used to separate excitation and photoconversion laser light from the fluorescence emission of mEos2. Emission light is further filtered using both band-pass (HQ630/140m-2p, Chroma) and long-pass (HQ 565LP, Chroma) emission filters for mEos2 fluorescence, or a band-pass filter (HQ605/75m, Chroma) for Dendra2 fluorescence.
Individual E. coli cells are imaged for 10,000 to 60,000 frames at 250 Hz using up to 28 pixel lines, 200 Hz using up to 40 pixel lines, and 20 Hz using up to 128 pixel lines on a PhotonMax EMCCD camera (Camera shutter: always open; digitizer: 10 MHz EM gain). Metamorph 7.5 (Molecular Devices) is used to control the microscope as well as the camera.
Data Processing.
Sample frames of RelA–Dendra2 are shown in Fig. 1C. The time-lapse movies are first edited by hand to remove frames with photoconversion pulses and frames where we observe multiple molecules in one frame. Next, we track the fluorophores in our hand-edited movies using the particle tracking software Diatrack (v3.03, Semasopht), which identifies and fits the intensity spots of our fluorescent particles with symmetric 2D Gaussian functions (see Fig. 1C). All routines for trajectory analysis are written in IGOR Pro 6.12A. For each of the analyzed cells all trajectories with three or more points are first overlaid to determine the precise cellular geometry. This geometry is then approximated as a cylinder with two hemispherical end caps. The final geometry of the cell is determined by subtracting twice the mean localization error (40 nm) from all sides of the initial geometry of the cell. This corrects for apparent cell broadening due to the experimental fitting noise. The localization errors are calculated using equation 6 in ref. 60.
Sample Preparation for SPT Experiments.
E. coli cells expressing RelA–Dendra2 are imaged on a M9-glucose agarose pad in a FCS2 flow chamber (Bioptechs). The cells are grown overnight to stationary phase and are diluted 1,000-fold in M9 buffer containing amino acids (M5550, Sigma-Aldrich) on the day of the experiments. The cells are grown to an optical density at 600 nm of 0.05 and placed on a 2.5% agarose pad (SeaPlaque GTG Agarose, Lonza) containing the same M9 medium.
For exponential growth experiments, cells are imaged only after having undergone several rounds of cell division until the individual cells have formed small microcolonies on the agarose pad to ensure that they have adjusted to the agarose pad. The temperature is held constant at 30 °C. The temperature is controlled via an electrical cell heater and an objective heater (both Bioptechs).
Individual RelA trajectories are recorded after red Dendra2 is generated using short pulses of the photoconversion laser beam. To obtain adequate statistics, we calculate CDFs and MSDs from all obtained single-molecule RelA trajectories from the entire microcolony for each of the experimental conditions and for each frame time.
For SHX experiments, the agarose pad is presoaked with L-SHX to a final concentration of 2.5 mM L-SHX. The cells are grown as above. When the cells reach an optical density at 600 nm of 0.05, L-SHX is added to the cells to a final concentration of 2.5 mM, and they are placed on the SHX-containing agarose pad. The cells undergo stringent response and immediately stop dividing on the agarose pad.
For the purpose of single-ribosome tracking we constructed a plasmid expressing an N-terminal Dendra2 fusion with the ribosomal protein L25. Low levels of leakage Dendra2-L25 expression provided hundreds of fluorescently labeled ribosomes per individual cell. For the ensemble time-lapse studies we C-terminally labeled three ribosomal proteins with Dendra2: S2, L19, and L31 in three separate strains using the genome integration approach based on the λ Red system (34). Please see SI Materials and Methods in the SI Appendix for details on strain preparation, validation, and a more detailed description of experimental procedures.
Supplementary Material
Acknowledgments.
We thank Dr. Arvi Jõers for help with the FCM analysis and Dr. Maria Selmer and Dr. Prune Leroy for helpful discussions. This work is supported by the European Research Council, the Swedish Foundation for Strategic Research, the Swedish Research Council, Göran Gustafssons Stiftelse, and the Knut and Alice Wallenberg Foundation. B.P.E. is supported by a Human Frontier Science Program cross-disciplinary fellowship. V.H. acknowledges support from the Estonian Science Foundation and the European Regional Development Fund through the Center of Excellence in Chemical Biology. N.H.D. acknowledges support from De Jonge Akademie.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 12573.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102255108/-/DCSupplemental.
References
- 1.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 2.Haseltine WA, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vrentas CE, et al. Still looking for the magic spot: The crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milon P, et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci U S A. 2006;103:13962–13967. doi: 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitkevich VA, et al. Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J Mol Biol. 2010;402:838–846. doi: 10.1016/j.jmb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Jenvert RMK, Schiavone LH. Characterization of the tRNA and ribosome-dependent pppGpp-synthesis by recombinant stringent factor from Escherichia coli. FEBS J. 2005;272:685–695. doi: 10.1111/j.1742-4658.2004.04502.x. [DOI] [PubMed] [Google Scholar]
- 8.Jain V, Saleem-Batcha R, China A, Chatterji D. Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 2006;15:1449–1464. doi: 10.1110/ps.062117006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avarbock D, Salem J, Li LS, Wang ZM, Rubin H. Cloning and characterization of a bifunctional RelA SpoT homologue from Mycobacterium tuberculosis. Gene. 1999;233:261–269. doi: 10.1016/s0378-1119(99)00114-6. [DOI] [PubMed] [Google Scholar]
- 10.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 11.Hauryliuk VV. GTPases of prokaryotic translational apparatus. Mol Biol (Mosk) 2006;40:769–783. [PubMed] [Google Scholar]
- 12.Stirpe F, Battelli MG. Ribosome-inactivating proteins: Progress and problems. Cell Mol Life Sci. 2006;63:1850–1866. doi: 10.1007/s00018-006-6078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishitani R, Yokoyama S, Nureki O. Structure, dynamics, and function of RNA modification enzymes. Curr Opin Struct Biol. 2008;18:330–339. doi: 10.1016/j.sbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Bremer H, Dennis P, editors. Modulation of Chemical Composition and other Parameters of the Cell by Growth Rate. Washington, DC: ASM; 1996. [DOI] [PubMed] [Google Scholar]
- 15.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 16.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 19.Gibbs KA, et al. Complex spatial distribution and dynamics of an abundant Escherichia coli outer membrane protein, LamB. Mol Microbiol. 2004;53:1771–1783. doi: 10.1111/j.1365-2958.2004.04242.x. [DOI] [PubMed] [Google Scholar]
- 20.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu L, Yu J. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys J. 2008;95:2009–2016. doi: 10.1529/biophysj.108.128751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahl SJ, Leutenegger M, Hilbert M, Hell SW, Eggeling C. Fast molecular tracking maps nanoscale dynamics of plasma membrane lipids. Proc Natl Acad Sci USA. 2010;107:6829–6834. doi: 10.1073/pnas.0912894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie XS, Yu J, Yang WY. Living cells as test tubes. Science. 2006;312:228–230. doi: 10.1126/science.1127566. [DOI] [PubMed] [Google Scholar]
- 24.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurskaya NG, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 26.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 27.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 28.Mascarenhas J, Weber MH, Graumann PL. Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2001;2:685–689. doi: 10.1093/embo-reports/kve160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis PJ, Thaker SD, Errington J. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 2000;19:710–718. doi: 10.1093/emboj/19.4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montero Llopis P, et al. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. Proc Natl Acad Sci USA. 2004;101:11310–11315. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golding I, Cox EC. Physical nature of bacterial cytoplasm. Phys Rev Lett. 2006;96:098102. doi: 10.1103/PhysRevLett.96.098102. [DOI] [PubMed] [Google Scholar]
- 33.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen FS, Kjeldgaard NO. Analysis of the relA gene product of Escherichia coli. Eur J Biochem. 1977;76:91–97. doi: 10.1111/j.1432-1033.1977.tb11573.x. [DOI] [PubMed] [Google Scholar]
- 36.Uzan M, Danchin A. A rapid test for the rel A mutation in E. coli. Biochem Biophys Res Commun. 1976;69:751–758. doi: 10.1016/0006-291x(76)90939-6. [DOI] [PubMed] [Google Scholar]
- 37.Vrljic M, Nishimura SY, Brasselet S, Moerner WE, McConnell HM. Translational diffusion of individual class II MHC membrane proteins in cells. Biophys J. 2002;83:2681–2692. doi: 10.1016/S0006-3495(02)75277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tosa T, Pizer LI. Biochemical bases for the antimetabolite action of L-serine hydroxamate. J Bacteriol. 1971;106:972–982. doi: 10.1128/jb.106.3.972-982.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riesenberg D, Bergter F, Kari C. Effect of serine hydroxamate and methyl alpha-D-glucopyranoside treatment on nucleoside polyphosphate pools, RNA and protein accumulation in Streptomyces hygroscopicus. J Gen Microbiol. 1984;130:2549–2558. doi: 10.1099/00221287-130-10-2549. [DOI] [PubMed] [Google Scholar]
- 40.Lemaux PG, Herendeen SL, Bloch PL, Neidhardt FC. Transient rates of synthesis of individual polypeptides in E. coli following temperature shifts. Cell. 1978;13:427–434. doi: 10.1016/0092-8674(78)90317-3. [DOI] [PubMed] [Google Scholar]
- 41.Ryals J, Little R, Bremer H. Control of RNA synthesis in Escherichia coli after a shift to higher temperature. J Bacteriol. 1982;151:1425–1432. doi: 10.1128/jb.151.3.1425-1432.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallant J, Palmer L, Pao CC. Anomalous synthesis of ppGpp in growing cells. Cell. 1977;11:181–185. doi: 10.1016/0092-8674(77)90329-4. [DOI] [PubMed] [Google Scholar]
- 43.Elowitz MB, Surette MG, Wolf PE, Stock JB, Leibler S. Protein mobility in the cytoplasm of Escherichia coli. J Bacteriol. 1999;181:197–203. doi: 10.1128/jb.181.1.197-203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 45.Haustein E, Schwille P. Fluorescence correlation spectroscopy: Novel variations of an established technique. Annu Rev Biophys Biomol Struct. 2007;36:151–169. doi: 10.1146/annurev.biophys.36.040306.132612. [DOI] [PubMed] [Google Scholar]
- 46.Elf J, Ehrenberg M. Near-critical behavior of aminoacyl-tRNA pools in E. coli at rate-limiting supply of amino acids. Biophys J. 2005;88:132–146. doi: 10.1529/biophysj.104.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lund E, Kjeldgaard NO. Metabolism of guanosine tetraphosphate in Escherichia coli. Eur J Biochem. 1972;28:316–326. doi: 10.1111/j.1432-1033.1972.tb01916.x. [DOI] [PubMed] [Google Scholar]
- 48.Endo Y, Tsurugi K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J Biol Chem. 1988;263:8735–8739. [PubMed] [Google Scholar]
- 49.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: W. H. Freeman; 1998. [Google Scholar]
- 50.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mechold U, Murphy H, Brown L, Cashel M. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J Bacteriol. 2002;184:2878–2888. doi: 10.1128/JB.184.11.2878-2888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kao HP, Verkman AS. Tracking of single fluorescent particles in three dimensions: Use of cylindrical optics to encode particle position. Biophys J. 1994;67:1291–1300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Connolly K, Culver G. Deconstructing ribosome construction. Trends Biochem Sci. 2009;34:256–263. doi: 10.1016/j.tibs.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pulk A, et al. Ribosome reactivation by replacement of damaged proteins. Mol Microbiol. 2010;75:801–814. doi: 10.1111/j.1365-2958.2009.07002.x. [DOI] [PubMed] [Google Scholar]
- 55.Sojka L, Fucik V, Krasny L, Barvik I, Jonak J. YbxF, a protein associated with exponential-phase ribosomes in Bacillus subtilis. J Bacteriol. 2007;189:4809–4814. doi: 10.1128/JB.01786-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Izutsu K, et al. Escherichia coli ribosome-associated protein SRA, whose copy number increases during stationary phase. J Bacteriol. 2001;183:2765–2773. doi: 10.1128/JB.183.9.2765-2773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wada A, Yamazaki Y, Fujita N, Ishihama A. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci USA. 1990;87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ortiz JO, et al. Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J Cell Biol. 2010;190:613–621. doi: 10.1083/jcb.201005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agafonov DE, Kolb VA, Spirin AS. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2001;2:399–402. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mortensen KI, Churchman LS, Spudich JA, Flyvbjerg H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat Methods. 2010;7:377–381. doi: 10.1038/nmeth.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]