Abstract
Many drugs are nature derived. Low drug productivity has renewed interest in natural products as drug-discovery sources. Nature-derived drugs are composed of dozens of molecular scaffolds generated by specific secondary-metabolite gene clusters in selected species. It can be hypothesized that drug-like structures probably are distributed in selective groups of species. We compared the species origins of 939 approved and 369 clinical-trial drugs with those of 119 preclinical drugs and 19,721 bioactive natural products. In contrast to the scattered distribution of bioactive natural products, these drugs are clustered into 144 of the 6,763 known species families in nature, with 80% of the approved drugs and 67% of the clinical-trial drugs concentrated in 17 and 30 drug-prolific families, respectively. Four lines of evidence from historical drug data, 13,548 marine natural products, 767 medicinal plants, and 19,721 bioactive natural products suggest that drugs are derived mostly from preexisting drug-productive families. Drug-productive clusters expand slowly by conventional technologies. The lack of drugs outside drug-productive families is not necessarily the result of under-exploration or late exploration by conventional technologies. New technologies that explore cryptic gene clusters, pathways, interspecies crosstalk, and high-throughput fermentation enable the discovery of novel natural products. The potential impact of these technologies on drug productivity and on the distribution patterns of drug-productive families is yet to be revealed.
Keywords: biodiversity, drug-prolific species, phylogenetic tree, herb
Many approved and clinical-trial drugs are derived from natural products (1, 2). During the past 2 decades, the focus of drug-discovery efforts has shifted from natural products to synthetic compounds, but this shift has not led to the anticipated increase in drug productivity (3). Despite the shifted focus, nature-derived drugs still constitute a substantial percentage of recently approved drugs. For instance, 12 (26%) of the 46 molecular entities approved by the Food and Drug Administration (FDA) in 2009–2010 are nature derived (SI Appendix, Table S1). There is a renewed interest in natural products as drug-discovery sources (4). The scope of biodiversity and extinction rates (5) demands bioprospecting efforts be prioritized toward the groups of species that are likely to yield new drugs.
Clues to drug-productive species can be obtained from the species-distribution profiles of nature-derived approved and clinical-trial drugs. Although particular species yield potent bioactive compounds at higher rates than others, additional drug-like properties are important for developing these compounds into marketable drugs (6). The nature-derived approved and clinical-trial drugs are composed primarily of several dozen molecular scaffolds (7–9) rather than the numerous bioactive natural-product scaffolds (10, 11). Like other bioactive natural-product scaffolds, the nature-derived privileged drug-like scaffolds are generated by enzymes partly encoded in specific secondary-metabolite gene clusters in selected groups of species (12–14). Questions arise as to whether the privileged drug-like structures are distributed in selective species-groups rather than being scattered in the phylogenetic tree (15) and whether the distribution shows certain traceable patterns that can be explored in future bioprospecting efforts.
A large number of bioactive natural products have been identified (16, 17). Many more are likely to be discovered (17) because of their interaction with specific targets (18). A small percentage of bioactive natural products has been carried forward to derive approved (1) and clinical-trial (2, 19) drugs via direct exploration, semisynthetic modification, structural mimicking, or pharmacophore mapping. The potential for drug development of a natural product depends not only on its bioactivity but also on the drug-likeness of its structure [optimized for enhanced drug-like (6) and reduced unwanted (20–22) properties] and the susceptibility of its target (“druggability”) to drugs [324 targets are confirmed druggable in yielding approved drugs (23), and 292 targets have yielded drugs in clinical trials (24)]. The odds for finding novel drug-like natural products may be improved if one can identify new drug-productive species, particularly endangered ones, before their extinction.
Structurally diverse bioactive natural products are composed of many molecular scaffolds (16, 17). Each scaffold is generated by specific enzyme assemblies (25) encoded in the secondary-metabolite gene clusters of specific species groups (12–14, 26). Partly as a survival strategy (12), structurally diverse natural products are generated by genetic variations and repositioning (27), posttranslational modifications (28), and assembly-line regulation (28). Nonetheless, bioactive secondary metabolites of an individual scaffold typically are produced by species from a specific family (e.g., anthraquinones in Polygonaceae) (14, 29) or from a few families of a specific order (e.g., sordarins of Xylariales) (14, 30). Specifically, 14 of the 26 drug-productive scaffolds from Actinomycetales are from a unique family, six are from a few families within the order, and four are from a few families in this and a few other orders (SI Appendix, Table S2).
Natural products active against individual targets or classes of targets may be composed of multiple scaffolds, many of which are from only a few families. For instance, 53 nicotinic acetylcholine receptor ligands are reported from diverse species (16). Our analysis (SI Appendix, Table S3) showed that these ligands are clustered into 29 scaffolds; 23 of these scaffolds are from a unique family, three are from a few families within a specific order, and three are from a few orders of a specific class. Each of the five approved drugs in the group is from a specific family. Similarly, the 12 nature-derived FDA-approved kinase inhibitor drugs (1, 2) are grouped into three scaffold groups, with each scaffold derived from a few families distributed among only a few orders (SI Appendix, Table S4).
Some families such as Streptomycetaceae, Pseudonocardiaceae, and Trichocomaceae and some genera such as Acremonium and Emericellopsis are highly drug-prolific (14, 15). Compounds synthesized by a specific metabolic pathway typically are active against only a few targets (13). It thus can be hypothesized that privileged drug-like structures targeting selective druggable targets are likely to be concentrated in specific families. This hypothesis can be evaluated, and the distribution patterns of drug-productive species can be revealed by comparing the species origins of the approved and clinical-trial drugs (1, 2) with those of preclinical drugs and bioactive natural products.
We analyzed the species origins of 939 approved drugs (SI Appendix, Table S5) (1), 369 clinical-trial drugs (SI Appendix, Table S6) (2, 19), 119 preclinical drugs (SI Appendix, Table S7) (31, 32), and 19,721 bioactive natural products (SI Appendix, Table S8) with particular focus on their distribution patterns in the phylogenetic trees of the Bacteria, Viridiplantae, Fungi, and Metazoa kingdoms or superkingdoms. Nature-derived approved and clinical-trial drugs and their species origins were obtained from published reviews (1, 2, 19) and our own literature search (SI Appendix, Table S8). Preclinical drugs are drug candidates that have entered preclinical studies such as safety, pharmacokinetics/absorption, distribution, metabolism, and excretion, active pharmaceutical ingredient preparation, and formulation (33). Following the seminal works on nature-derived drugs (1, 2), we included in our analysis biologics, natural products and their semisynthetic derivatives, mimics, and peptidomimetics. Biologics include peptides (34), recombinant proteins (35), and monoclonal antibodies (36) except for RNA-based drugs (37). The inclusion or exclusion of biologics and RNA-based drugs had limited effect on our analysis because they primarily are of human or viral origin. For semisynthetic derivatives, mimics, and peptidomimetics, the host species of the parent natural-product leads were analyzed (1, 2).
We also evaluated four lines of evidence from (i) historical drug data, (ii) 13,548 natural products of marine species and their nonmarine counterparts, (iii) 767 medicinal plants, and (iv) 19,721 bioactive natural products to determine whether the distribution patterns of drug-productive families are different from those of bioactive natural products and if the lack of drugs outside drug-productive families is the result of under-exploration or late exploration.
The natural-product leads of drugs and bioactive natural products in this analysis were discovered mostly by conventional technologies rather than by the new technologies that are expected to identify many previously unrecognized bioactive natural products. These technologies are based on the exploration of cryptic metabolic gene clusters (via genomic mining, epigenetic modification, and proteomics) (38–40), metabolic pathway engineering (41, 42), interspecies crosstalk (40, 43, 44), and high-throughput fermentation and screening (45). Their potential contribution to discovery of new drugs is highly anticipated (40, 42, 45). The distribution patterns of the drugs discovered by these technologies may not follow those of the existing drugs derived from conventional technologies.
Results and Discussion
Apart from 12 drugs from widely distributed species, 933 approved and 363 clinical-trial drugs were concentrated in 144 drug-productive families of the 6,763 families known in nature. Of the 144 drug-productive families, 99 contained approved drugs. Many of these families contain endangered species. In particular, 80% of the approved drugs are concentrated in 17 drug-prolific families, and 67% of the clinical-trial drugs are concentrated in 30 such families. Most (82.4%) of the clinical-trial drugs are distributed within 59 families that also contain approved drugs. Thus, the presence of drug-like structures, as represented by the approved and clinical-trial drugs, is highly concentrated in selective families.
Fig. 1 presents the drug-prolific families with the highest numbers of approved drugs. The well-known drug-prolific families Streptomycetaceae and Pseudonocardiaceae of the Bacteria, the family Trichocomaceae and the genera Acremonium and Emericellopsis of the Fungi, and the family Hominidae (great apes and humans) of the Metazoa superkingdom are among the most prolific, with 59.1% of the approved drugs, followed by the Viridiplantae (green plant) families Fabaceae (legumes), Ephedraceae (Mormon tea), Papaveraceace (poppies), Asteraceae (daisy), Solanaceae (potato), Rubiaceae (coffee), and Apocynaceae (dogbane), and the Metazoan families Viperidae (venomous snakes) and Muridae (rodents). Two Bacteria families (Streptomycetaceae and Pseudonocardiaceae), four plant families (Fabaceae, Rubiaceae, Asteraceae, and Apocynaceae), and one Metazoa family (Hominidae) also are prolific in clinical-trial drugs. The enriched number of clinical-trial drugs from these families (2, 19) arises partly from the exploration of sources such as marine actinomycete bacteria (15, 46) and plants (19). The Hominidae family is the highest ranked drug-productive family, largely because of the inclusion of biologics. It becomes the second-ranked family if biologics are excluded.
Fig. 1.
Top-ranked drug-prolific families that produced high numbers of approved drugs.
We also tentatively analyzed the ranking of drug-productive families based on the ratio of the approved drugs to the searchable bioactive natural products (including leads of the approved and clinical-trial drugs) from each family. Partly because of the limited data from the available databases and our literature search, our searched natural products are insufficient to reflect the true ratios adequately. Nonetheless, our analysis of families with >20 searchable bioactive natural products showed that 70% of the top-ranked drug-productive families in Fig. 1 are among the families with highest drug-to-natural product ratios (SI Appendix, Fig. S1), suggesting that the high productivity of some of these top-ranked families may result from a higher frequency of derived drugs rather than from a higher number of bioactive natural products explored. The two Fungi genera Acremonium and Emericellopsis, in the top-ranked six families in Fig. 1, were excluded from our analysis because they have fewer searchable bioactive natural products (12 and 5, respectively).
SI Appendix, Fig. S2 shows the top-ranked families without an approved drug that nonetheless yield high numbers of clinical-trial drugs. These families include six plant families [Leskeaceae (a moss family), Anomodontaceae (a moss family), Combretaceae (almond family), Quillajaceae, Cephalotaxaceae (plum-yew family), and Bryopsidaceae (a green algae family)], seven families of Metazoa [Aplysiidae (sea hares), Bugulidae (marine moss animals), Dendrobatidae (poison frogs), Petrosiidae (a sponge family), Axinellidae (a sponge family), Squalidae (dogfish sharks), and Hemiasterellidae (a sponge family)], and a genus of Bacteria, Symploca. Many of these families, particularly the marine ones, have been explored actively and reportedly have shown good drug-discovery potential (4, 19, 46–48).
Further analysis of the species origins of approved and clinical-trial drugs in the phylogenetic trees of the Bacteria (Fig. 2), Viridiplantae (Fig. 3), Fungi (SI Appendix, Fig. S3), and Metazoa (SI Appendix, Fig. S4) kingdoms or superkingdoms revealed clustered patterns. One can define a drug-productive cluster as a relatively small branch of a phylogenetic tree with two or more drug-productive families. The bacteria superkingdom (289 known families) contains 23 drug-productive families, 18 of which, including the drug-prolific ones, are concentrated in four drug-productive clusters in the Actinonycetales order, Bacillales order, Gammaproteobacteria class, and Myxococcales order (Fig. 2). Bioprospecting efforts can be extended to other subbranches in these drug-productive clusters, and such efforts have been reported. For instance, two genera of the drug-productive Actinomycetaceae family have been discovered recently and found to produce promising anticancer and antibiotic compounds (46).
Fig. 2.
Distribution of drug-productive families (green background color) in the phylogenetic tree of the Bacteria superkingdom. The 10 families with highest number of approved drugs (α), highest number of clinical-trial drugs and at least one approved drug (β), and highest number of clinical-trial drugs without an approved drug (γ) are marked. Drug-productive clusters that contain two or more drug-productive families are labeled with the name of species group (class or order) in black. The family names are provided at branch ends, which can be viewed more clearly by enlarging the figure in the electronic version.
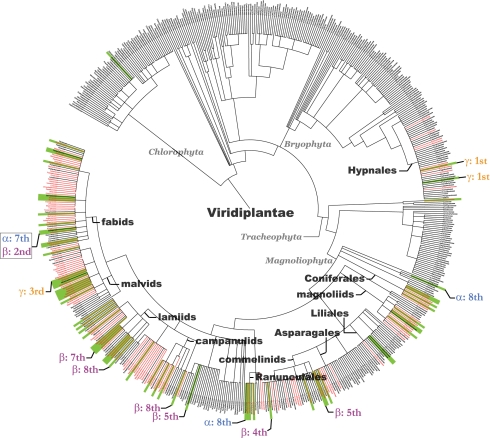
Fig. 3.
Distribution of drug-productive families in the phylogenetic tree of the Viridiplantae kingdom. Coloring and labeling schemes are as in Fig. 2. Red lines indicate families containing endangered species.
The Viridiplantae kingdom (740 known families) contains 66 drug-productive families, 61 of which, including the drug-prolific ones, are concentrated in 11 drug-productive clusters. The 11 drug-productive clusters are the Fabid and Malvid groups of the Rosidae subclass; the Lamiid and Campanulid groups of the Asterid subclass; the Ranunculales order, Magnoliids clade, Coniferales order, Commelinids clade, and Asparagales order of the Monocots group; the Hypnales order (mosses), and the Liliales order (Fig. 3). The Fungi kingdom (521 known families) contains 16 drug-productive families, 12 of which are concentrated in two drug-productive clusters in the Hypoceales order and Agaricales order (gilled mushrooms).
The Metazoa kingdom (4,468 known families) contains 38 drug-productive families, 30 of which, including drug-prolific ones, are concentrated in nine drug-productive clusters. The nine drug-productive clusters are the Euarchontoglires clade (rodents, lagomorphs, tree shrews, colugos, and primates including humans), Cetartiodactyla clade (whales, dolphins, and even-toed ungulates), Scleroglossa suborder in the Squamata order (scaled reptiles), Demospongiae class (demosponges), Hirudinea subclass (leeches), Gastropoda class (snails and slugs), Enterogona order (marine saclike filter feeder animals), Batrachia superorder (vertebrate amphibians without tails), and Monostilifera order (ribbon worms) (SI Appendix, Fig. S4).
The clustered patterns of drug-productive families shown in Figs. 2 and 3 and SI Appendix, Figs. S3 and S4 are in contrast to the scattered distribution patterns of the families of the 19,721 searchable bioactive natural products (SI Appendix, Figs. S5–S8). Our collected data on bioactive natural products are inadequate to represent all bioactive ingredients, but some useful indications were revealed. These natural products are distributed in 792 families (67 Bacteria, 273 Viridiplantae, 110 Fungi, and 345 Metazoa families) scattered in the phylogenetic trees, 57.1% of which (46.3% Bacteria, 37.0% Viridiplantae, 72.7% Fungi, and 69.6% Metazoa) are outside drug-productive clusters. These data suggest that the current drug-productive families form distinctive groups that tend to cluster together in phylogenetic trees. It may be hypothesized that these patterns partly reflect the distribution of the secondary-metabolite gene clusters that produce drug-like scaffolds (12–14) in selected species (7–9).
The clustered patterns of drug-productive families also were compared tentatively with the distribution patterns of the 119 nature-derived preclinical drugs (SI Appendix, Figs. S9–S12). Although the number of our collected preclinical drugs is too small to reveal their distribution patterns clearly, some useful indications may be obtained. The natural-product leads of these drugs are from 63 families, 73.0% of which are drug productive (58.7%) or are non-drug productive but are distributed in drug-productive clusters (14.3%). This percentage is significantly higher than that (42.9%) of the 19,721 bioactive natural products. These data seem to indicate that the preclinical drug families may be more concentrated in the phylogenetic trees than those of bioactive natural products but less concentrated than the drug-productive families and clusters.
Most of the preclinical drug families outside the drug-productive clusters are from the Fungi and Metazoa kingdoms (specifically soft corals, scorpions, and tube-dwelling spiders). It is unclear whether this biased distribution is the result of limited sampling of preclinical drugs or whether it partially reflects more recent efforts investigating such sources as fungal (49), soft coral (50), and invertebrate (51) species. The drug-productive families and clusters in these two kingdoms are in significantly smaller regions of the phylogenetic trees than those in the Bacteria superkingdom and the Viridiplantae kingdom; this distribution may leave open more branches for deriving preclinical drugs.
Four lines of evidence suggest that the recognition of drug-productive clusters expands slowly using conventional technologies and that the paucity of drugs outside drug-productive families is not necessarily the result of under-exploration or late exploration by conventional technologies. The first line of evidence is from the historical data of nature-derived approved drugs. As shown in Table 1, most (80.9–97.2%) of the 39–141 nature-derived drugs approved during every 5-y period from 1961 to 2010 are from previous drug-productive families or from newly identified families in previous drug-productive clusters. In particular, most (82.1–96.3%) of the drugs approved during every 5-y period from 1961–1975 and 1981–2010 and the majority (66.3%) of the drugs approved from 1976–1980 are from previous drug-productive families. These data suggest that, regardless the varying levels of exploration, preexisting drug-productive families and clusters are the most prolific sources for yielding approved drugs.
Table 1.
Number of nature-derived approved drugs, drug-productive families, and drug-productive clusters (excluding potential drug-productive clusters) during every 5-y period, 1961–2010
| No. of approved drugs in period |
No. of drug-productive families |
No. of drug-productive clusters |
||||||
| Period | From previous drug-productive families | From newly identified drug-productive families in previous drug-productive clusters | From newly identified drug-productive families not in previous drug-productive clusters | No. of existing families | No. of newly identified families in previous drug-productive clusters | No. of newly identified families not in previous drug-productive clusters | No. of existing clusters | No. of newly identified clusters |
| 1961–1965 | 32 | 0 | 7 | 21 | 0 | 5 | 6 | 1 |
| 1966–1970 | 26 | 0 | 1 | 26 | 0 | 1 | 7 | 0 |
| 1971–1975 | 32 | 3 | 1 | 27 | 3 | 1 | 7 | 0 |
| 1976–1980 | 59 | 13 | 17 | 31 | 8 | 12 | 7 | 3 |
| 1981–1985 | 98 | 1 | 11 | 51 | 1 | 8 | 10 | 3 |
| 1986–1990 | 128 | 7 | 6 | 60 | 6 | 6 | 13 | 2 |
| 1991–1995 | 111 | 9 | 5 | 72 | 7 | 3 | 15 | 1 |
| 1996–2000 | 129 | 4 | 4 | 82 | 3 | 2 | 16 | 2 |
| 2001–2005 | 124 | 1 | 7 | 87 | 1 | 4 | 18 | 1 |
| 2006–2010 | 44 | 3 | 2 | 92 | 4 | 2 | 19 | 0 |
Comparison of the newly identified drug-productive families that have emerged since 1981 with pre-1981 drug-productive clusters (SI Appendix, Figs. S13–S16) showed that most (80.4%) of the 28 families identified post-1981 outside the pre-1981 clusters are clustered with one or more existing or newly identified drug-productive families, suggesting that the newly identified drug-productive families tend to form clusters with existing drug-productive families or among themselves. In every 5-y period from 1961–2010, small numbers of newly identified drug-productive families (5.5 families per 5-y period from 1961–1990 and 2.75 families per 5-y period from 1991–2010) and clusters (1.5 clusters per 5-y period from1961–2010) have emerged outside previous drug-productive clusters (Table 1).
The patterns we revealed may be influenced predominantly by older drugs. Drug-discovery focuses are shifting in terms of targets, chemotypes, diseases, and therapeutic strategies (e.g., multitarget and RNA-based drugs) (34–37, 52, 53). The patterns we discerned might change significantly as new drugs with new targets are derived from natural products extracted by conventional technologies. To study this possibility, we further analyzed 74 nature-derived approved drugs. These drugs include 12 drugs approved in 2008–2009 (SI Appendix, Table S1), 10 kinase inhibitors (SI Appendix, Table S4), and 52 drugs targeting 26 targets first explored successfully since 1990 (SI Appendix, Table S9) (24). To some extent, these drugs represent novel drugs for new targets, chemotypes, diseases, and therapeutic approaches. At the time of their approval, 71 (95.9%) of these drugs were from preexisting drug-productive families (91.9%) or were from non–drug-productive families within preexisting drug-productive clusters (4.0%), and 44 drugs (59%) were from families with drugs approved before 1980. These 71 drugs target 32 targets; the first drug approved for 26 (81.3%) of these targets have been derived from a preexisting drug-productive family. Thus, the evolving focus of drug discovery appears to have a limited effect on the expansion of drug-productive clusters by conventional technologies, and existing drug-productive families and clusters are the primary sources of novel drugs.
The second line of evidence, which comes from the distribution of 13,548 natural products from marine phyla (including nonmarine species in these phyla), indicates that the paucity of drugs outside drug-productive families is not necessarily the result of under-exploration by conventional technologies. Some non–drug-productive phyla produce high numbers of compounds, comparable to the number in drug-productive phyla (SI Appendix, Table S10). This evidence is supported further by the third line of evidence from published data from medicinal plant research. Plant extracts from >3,000 species have been assayed extensively for anticancer activity alone (54), and ∼10% of the 250,000–500,000 plant species have been studied (55). From medicinal plant databases and literature searches, we identified 767 medicinal plants from 172 non–drug-productive families, 104 (60.5%) of which are outside recognized drug-productive clusters (SI Appendix, Table S11).
The fourth line of evidence is from the distribution profile and exploration timeline of 19,721 bioactive natural products (including leads of nature-derived drugs). Although these natural products inadequately represent all bioactive natural products, some useful indications may be revealed. Consistent with the distribution profiles of the 13,548 natural products from marine phyla and 767 medicinal plants, a number of non–drug-productive families outside drug-productive clusters contain high numbers of bioactive natural products comparable to those in drug-productive families and in non–drug-productive families inside drug-productive clusters (SI Appendix, Table S12). In particular, non–drug-productive families outside and inside drug-productive clusters constitute 22% and 10%, respectively, of the 50 families with the highest number of searchable bioactive natural products; 60% of the 20 non–drug-productive families outside drug-productive clusters and 80% of the 10 non–drug-productive families within drug-productive clusters contain bioactive natural products that were reported before 1990. This evidence further suggests that the paucity of drugs identified outside drug-productive families and clusters is not necessarily the result of under-exploration and late exploration by conventional technologies.
The approved drugs, grouped into drug-target classes with three or more drugs against each target, are clustered in specific therapeutic regions of the phylogenetic subbranches of drug-productive families (SI Appendix, Fig. S17). Consistent with the report that compounds synthesized by a specific pathways typically are active against a limited number of targets (13), drugs in each target class are distributed mostly in one to three families. Exceptions are three classes of drugs used to treat infection (targeting penicillin-binding protein, ribosome, and bacterial outer membrane), two classes of drugs used to treat circulation disorders (targeting maltase-glucoamylase and adenosine receptor), three classes of drugs used to treat cancer (targeting tubulin, DNA topoisomerase, an thymidylate synthase), one drug used to treat disease of the nervous system (targeting the opioid receptor), and one drug used to treat inflammation (targeting the 5-HT receptor); these classes are distributed among five or six families.
Antiinfectious drugs are primarily from one large Bacteria cluster of four families (Streptomycetaceae, Pseudonocardiaceae, Actinosynnemataceae, and Actinomycetaceae), one Fungi cluster of two families (Acremonium and Emericellopsis), and another Fungi family (Trichocomaceae). Anticancer drugs are largely from one Bacteria family (Streptomycetaceae), one Metazoa cluster of two families (Perophoridae and Hominidae), and three plant clusters consisting respectively of three families (Fabaceae, Betulaceae, and Moraceae), three families (Apocynaceae, Rubiaceae, and Icacinaceae), and two families (Poaceae and Arecaceae). Drugs targeting the reproductive system are mostly from the Hominidae family. Anti-inflammatory drugs are primarily from the Hominidae family and several plant families. Drugs targeting the nervous system are largely from a plant cluster of four families (Papaveraceae, Menispermaceae, Berberidaceae, and Fabaceae) and from several other plant (Ephedraceae, Amaranthaceae, Asteraceae, Solanaceae, and Amaryllidaceae), Metazoa, and Fungi families. Immunity drugs are primarily from three Fungi families (Clavicipitaceae, Nectriaceae, and Trichocomaceae) with a few drugs from one Bacteria and one Metazoa family. Circulation drugs are primarily from one Bacteria cluster consisting of two families (Streptomycetaceae and Pseudonocardiaceae), from one plant cluster of four families (Malvaceae, Theaceae, Plantaginaceae, and Loganiaceae), and from two Metazoa clusters of three families (Hominidae, Muridae, and Viperidae) and two families (Glossiphoniidae and Hirudinidae), respectively.
These drug-productive families typically produce drugs against multiple targets in multiple therapeutic areas. For instance, drugs from Streptomycetaceae are distributed in 12 target classes of four therapeutic areas (infection, cancer, circulation, and immunity), and drugs from Hominidae are distributed in 16 target classes of six therapeutic areas (inflammation, reproduction, cancers, circulation, immunity, and nervous system). These multiple targets in multiple therapeutic areas suggest that usually there are multiple drug-producing secondary-metabolite gene clusters among the many gene clusters in specific families (14). Two species, S. avermitilis and S. griseus, of the Streptomycetes genus in the drug-productive Streptomycetaceae family have been observed to produce two or three natural products, but analysis of their sequenced genome suggests that they might harbor ∼25–30 predicted biosynthetic gene clusters (28). Efforts have been made to activate the silent “cryptic” gene clusters in certain drug-productive families (38, 39). These efforts may enable the discovery of additional drug leads with different structures and therapeutic applications.
From the observed tendency of newly recognized drug-productive families to cluster with preexisting drug-productive families or among themselves, one may speculate that the families with only clinical-trial drugs that are clustered with another drug-productive family may have a higher probability of yielding approved drugs. Examples of these families are the Myxococcaceae, Micrococcaceae, and Streptosporangiaceae families in the Myxococcales and Actinomycetales orders of the Bacteria superkingdom; the Melanthiaceae, Calophyllaceae, Quillajaceae, Bignoniaceae, Oleaceae, Anomodontaceae, and Leskeaceae families in the Liliales and the Fabid, Lamiid, and Hypnales orders or superorders of Viridiplantae kingdom; the Omphalotaceae family in the Agaricales order of the Fungi kingdom; and the Didemnidae, Polyclinidae, Aplysiidae, Kentrodorididae, Placobranchidae, Ancorinidae, Aplysinellidae, Axinellidae, Dysideidae, Hemiasterellidae, Petrosiidae, Pseudoceratinidae, Amphiporidae, Bufonidae, Dendrobatidae, and Salamandridae families, and the Paranemertes genus in the Enterogona, Gastropoda, Demospongiae, and Monostilifera classes, suborders, or orders, of the Metazoa kingdom.
Conclusion
Our analysis revealed that nature-derived drugs have been derived mostly from drug-productive families that tend to be clustered rather than scattered in the phylogenetic tree. Several indications from historical drug data and extracted natural products suggest that the identification of these clusters expands slowly when conventional technologies are used, and the paucity of drugs outside these clusters is not necessarily the result of under-exploration or late exploration by conventional technologies. New technologies (38–45) are expected to expand the pool of drug leads significantly (40, 42, 45). The impact of these technologies on drug productivity and the distribution of drug-productive families is yet to be determined. The distribution patterns of the future drug-productive families may differ from those of the existing drugs derived from conventional technologies. A particular question is whether the future drug-productive families are clustered, probably on a larger scale. Analysis of the species origins of 321 previously unrecognized secondary metabolites published in 2001–2011 (SI Appendix, Table S13) showed that 93 (29%) are distributed in 22 families outside drug-productive clusters, and 228 (71%) are distributed in 43 families within drug-productive clusters. Those outside drug-productive clusters are distributed in the relevant families at comparable densities as those within drug-productive clusters. It remains to be determined if a similar concentrated distribution is found for the secondary metabolites generated by new technologies. The clustered patterns revealed in this work provide useful information about the groups of species that are drug productive or potentially productive. This information, coupled with expanded knowledge of drug-like structures (7–9) and drug-productive species and with new technologies (4, 40–45, 56, 57), may enable more prioritized, focused, rational, and environmentally friendly bioprospecting for novel drug-like natural products.
Materials and Methods
The species origins of 939 approved and 369 clinical-trial drugs were identified as follows. First Therapeutic Target Database (24) was checked to confirm the current approval or clinical-trial status of the literature-reported approved drugs (1) and clinical-trial drugs (2, 19) of natural origin. Then the species origin of every drug was searched in books and review and regular articles using combinations of keywords such as the drug name and alternative names, species, “natural product,” and “nature.” The literature searched is listed in SI Appendix, Table S8. The species origin of a drug was confirmed by a specific statement in the literature that the drug “originates from,” “is derived from,” “is isolated from,” or “comes from” a species or species group (e.g., a genus or family). For drugs from semisynthetic derivatives, mimics, and peptidomimetics, the parent natural-product leads were searched first, followed by a search of host species as described above. The families of the host species of these drugs as well as all the known families in nature are from the National Center for Biotechnology Information (NCBI) taxonomy database (58). Families with endangered species were identified by mapping the species against the International Union for the Conservation of Nature Red List of Threatened Species Version-2010.3 (http://www.iucnredlist.org). The phylogenetic trees were generated by using the NCBI taxonomy-based automatic tree generator in iTOL version 1.8.1 (59) against known families in the Bacteria, Viridiplantae, Fungi, and Metazoa kingdoms or superkingdoms. Family names are provided at branch ends. Red branches indicate families with endangered species. Drug-productive clusters and some of the drug-productive families are labeled or marked in the phylogenetic trees.
Supplementary Material
Acknowledgments
This work was supported by the Chinese National Natural Science Foundation (20872077 and 90813013), by the Chinese Ministry of Science and Technology Key Special Project Grant 2009ZX09501-004, and by Singapore Academic Research Fund Grants R-148-000-141-750 and R-148-000-141-646.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107336108/-/DCSupplemental.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Butler MS. Natural products to drugs: Natural product-derived compounds in clinical trials. Nat Prod Rep. 2008;25:475–516. doi: 10.1039/b514294f. [DOI] [PubMed] [Google Scholar]
- 3.Paul SM, et al. How to improve R&D productivity: The pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 4.Li JW, Vederas JC. Drug discovery and natural products: End of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 5.Butchart SH, et al. Global biodiversity: Indicators of recent declines. Science. 2010;328:1164–1168. doi: 10.1126/science.1187512. [DOI] [PubMed] [Google Scholar]
- 6.Vistoli G, Pedretti A, Testa B. Assessing drug-likeness—what are we missing? Drug Discov Today. 2008;13:285–294. doi: 10.1016/j.drudis.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Bemis GW, Murcko MA. The properties of known drugs. 1. Molecular frameworks. J Med Chem. 1996;39:2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Hou T. Drug and drug candidate building block analysis. J Chem Inf Model. 2010;50:55–67. doi: 10.1021/ci900398f. [DOI] [PubMed] [Google Scholar]
- 9.Duarte CD, Barreiro EJ, Fraga CA. Privileged structures: A useful concept for the rational design of new lead drug candidates. Mini Rev Med Chem. 2007;7:1108–1119. doi: 10.2174/138955707782331722. [DOI] [PubMed] [Google Scholar]
- 10.Koch MA, et al. Charting biologically relevant chemical space: A structural classification of natural products (SCONP) Proc Natl Acad Sci USA. 2005;102:17272–17277. doi: 10.1073/pnas.0503647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong DX, Jiang YY, Zhang HY. Marine natural products as sources of novel scaffolds: Achievement and concern. Drug Discov Today. 2010;15:884–886. doi: 10.1016/j.drudis.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Fischbach MA, Clardy J. One pathway, many products. Nat Chem Biol. 2007;3:353–355. doi: 10.1038/nchembio0707-353. [DOI] [PubMed] [Google Scholar]
- 13.Beghyn T, Deprez-Poulain R, Willand N, Folleas B, Deprez B. Natural compounds: Leads or ideas? Bioinspired molecules for drug discovery. Chem Biol Drug Des. 2008;72:3–15. doi: 10.1111/j.1747-0285.2008.00673.x. [DOI] [PubMed] [Google Scholar]
- 14.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenical W, Jensen PR. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 16.Tulp M, Bohlin L. Functional versus chemical diversity: Is biodiversity important for drug discovery? Trends Pharmacol Sci. 2002;23:225–231. doi: 10.1016/s0165-6147(02)02007-2. [DOI] [PubMed] [Google Scholar]
- 17.Bérdy J. Bioactive microbial metabolites. J Antibiot (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 18.Tulp M, Bohlin L. Rediscovery of known natural compounds: Nuisance or goldmine? Trends Pharmacol Sci. 2005;26:175–177. doi: 10.1016/j.tips.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Saklani A, Kutty SK. Plant-derived compounds in clinical trials. Drug Discov Today. 2008;13:161–171. doi: 10.1016/j.drudis.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 21.Ekins S, Honeycutt JD, Metz JT. Evolving molecules using multi-objective optimization: Applying to ADME/Tox. Drug Discov Today. 2010;15:451–460. doi: 10.1016/j.drudis.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Wang J. Comprehensive assessment of ADMET risks in drug discovery. Curr Pharm Des. 2009;15:2195–2219. doi: 10.2174/138161209788682514. [DOI] [PubMed] [Google Scholar]
- 23.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 24.Zhu F, et al. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 2010;38(Database issue):D787–D791. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu L, et al. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gontang EA, Gaudêncio SP, Fenical W, Jensen PR. Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl Environ Microbiol. 2010;76:2487–2499. doi: 10.1128/AEM.02852-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perić-Concha N, Long PF. Mining the microbial metabolome: A new frontier for natural product lead discovery. Drug Discov Today. 2003;8:1078–1084. doi: 10.1016/s1359-6446(03)02901-5. [DOI] [PubMed] [Google Scholar]
- 28.Walsh CT, Fischbach MA. Natural products version 2.0: Connecting genes to molecules. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ososki AL, Kennelly EJ. Phytoestrogens: A review of the present state of research. Phytother Res. 2003;17:845–869. doi: 10.1002/ptr.1364. [DOI] [PubMed] [Google Scholar]
- 30.Vicente F, et al. Distribution of the antifungal agents sordarins across filamentous fungi. Mycol Res. 2009;113:754–770. doi: 10.1016/j.mycres.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 31.Simmons TL, Andrianasolo E, McPhail K, Flatt P, Gerwick WH. Marine natural products as anticancer drugs. Mol Cancer Ther. 2005;4:333–342. [PubMed] [Google Scholar]
- 32.Pfisterer PH, Wolber G, Efferth T, Rollinger JM, Stuppner H. Natural products in structure-assisted design of molecular cancer therapeutics. Curr Pharm Des. 2010;16:1718–1741. doi: 10.2174/138161210791164027. [DOI] [PubMed] [Google Scholar]
- 33.Steinmetz KL, Spack EG. The basics of preclinical drug development for neurodegenerative disease indications. BMC Neurol. 2009;9(Suppl 1):S2 1–13. doi: 10.1186/1471-2377-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saladin PM, Zhang BD, Reichert JM. Current trends in the clinical development of peptide therapeutics. IDrugs. 2009;12:779–784. [PubMed] [Google Scholar]
- 35.Woodcock J, et al. The FDA's assessment of follow-on protein products: A historical perspective. Nat Rev Drug Discov. 2007;6:437–442. doi: 10.1038/nrd2307. [DOI] [PubMed] [Google Scholar]
- 36.Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9:767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 37.Melnikova I. RNA-based therapies. Nat Rev Drug Discov. 2007;6:863–864. doi: 10.1038/nrd2314. [DOI] [PubMed] [Google Scholar]
- 38.Bergmann S, et al. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 39.Lautru S, Deeth RJ, Bailey LM, Challis GL. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 40.Chiang YM, Chang SL, Oakley BR, Wang CC. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol. 2011;15:137–143. doi: 10.1016/j.cbpa.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wenzel SC, Müller R. Formation of novel secondary metabolites by bacterial multimodular assembly lines: Deviations from textbook biosynthetic logic. Curr Opin Chem Biol. 2005;9:447–458. doi: 10.1016/j.cbpa.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson B, Micklefield J. Mining and engineering natural-product biosynthetic pathways. Nat Chem Biol. 2007;3:379–386. doi: 10.1038/nchembio.2007.7. [DOI] [PubMed] [Google Scholar]
- 43.Pullen CB, et al. Occurrence and non-detectability of maytansinoids in individual plants of the genera Maytenus and Putterlickia. Phytochemistry. 2003;62:377–387. doi: 10.1016/s0031-9422(02)00550-2. [DOI] [PubMed] [Google Scholar]
- 44.Schroeckh V, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baltz RH. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol. 2008;8:557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Marris E. Marine natural products: Drugs from the deep. Nature. 2006;443:904–905. doi: 10.1038/443904a. [DOI] [PubMed] [Google Scholar]
- 47.Mayer AM, et al. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol Sci. 2010;31:255–265. doi: 10.1016/j.tips.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Singh S. From exotic spice to modern drug? Cell. 2007;130:765–768. doi: 10.1016/j.cell.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 49.Saleem M, et al. Marine natural products of fungal origin. Nat Prod Rep. 2007;24:1142–1152. doi: 10.1039/b607254m. [DOI] [PubMed] [Google Scholar]
- 50.Cheng SY, et al. Antiviral and anti-inflammatory metabolites from the soft coral Sinularia capillosa. J Nat Prod. 2010;73:771–775. doi: 10.1021/np9008078. [DOI] [PubMed] [Google Scholar]
- 51.Mortari MR, Cunha AO, Ferreira LB, dos Santos WF. Neurotoxins from invertebrates as anticonvulsants: From basic research to therapeutic application. Pharmacol Ther. 2007;114:171–183. doi: 10.1016/j.pharmthera.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Zheng CJ, et al. Therapeutic targets: Progress of their exploration and investigation of their characteristics. Pharmacol Rev. 2006;58:259–279. doi: 10.1124/pr.58.2.4. [DOI] [PubMed] [Google Scholar]
- 53.Bilanges B, Torbett N, Vanhaesebroeck B. Killing two kinase families with one stone. Nat Chem Biol. 2008;4:648–649. doi: 10.1038/nchembio1108-648. [DOI] [PubMed] [Google Scholar]
- 54.Cragg GM, Newman DJ. Plants as a source of anti-cancer agents. J Ethnopharmacol. 2005;100:72–79. doi: 10.1016/j.jep.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 55.Harvey A. Strategies for discovering drugs from previously unexplored natural products. Drug Discov Today. 2000;5:294–300. doi: 10.1016/s1359-6446(00)01511-7. [DOI] [PubMed] [Google Scholar]
- 56.Scherlach K, Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 57.Chiang YM, Lee KH, Sanchez JF, Keller NP, Wang CC. Unlocking fungal cryptic natural products. Nat Prod Commun. 2009;4:1505–1510. [PMC free article] [PubMed] [Google Scholar]
- 58.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37(Database issue):D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.