Abstract
Sec16 plays a key role in the formation of coat protein II vesicles, which mediate protein transport from the endoplasmic reticulum (ER) to the Golgi apparatus. Mammals have two Sec16 isoforms: Sec16A, which is a longer primary ortholog of yeast Sec16, and Sec16B, which is a shorter distant ortholog. Previous studies have shown that Sec16B, as well as Sec16A, defines ER exit sites, where coat protein II vesicles are formed in mammalian cells. Here, we reveal an unexpected role of Sec16B in the biogenesis of mammalian peroxisomes. When overexpressed, Sec16B was targeted to the entire ER, whereas Sec16A was mostly cytosolic. Concomitant with the overexpression of Sec16B, peroxisomal membrane biogenesis factors peroxin 3 (Pex3) and Pex16 were redistributed from peroxisomes to Sec16B-positive ER membranes. Knockdown of Sec16B but not Sec16A by RNAi affected the morphology of peroxisomes, inhibited the transport of Pex16 from the ER to peroxisomes, and suppressed expression of Pex3. These phenotypes were significantly reversed by the expression of RNAi-resistant Sec16B. Together, our results support the view that peroxisomes are formed, at least partly, from the ER and identify a factor responsible for this process.
Most eukaryotic cells contain peroxisomes, which are single membrane-bound organelles that function in various metabolic pathways, including the β-oxidation of fatty acids, biosynthesis of plasmalogens and bile acids, and hydrogen peroxide metabolism (1). To perform this variety of functions, peroxisomes are highly dynamic; their number, size, and function change in response to cellular conditions. In addition, unlike mitochondria, peroxisomes can be formed through de novo synthesis as well as through the growth and division of preexisting peroxisomes (2, 3).
Peroxisomal matrix proteins are synthesized on free ribosomes in the cytosol and posttranslationally imported to peroxisomes (4). This import pathway includes the recognition of two distinct peroxisomal targeting signals (PTS1 and PTS2) by peroxin 5 (Pex5) and Pex7, respectively, followed by translocation across the membrane through the import machinery, including Pex14 and Really Interesting New Gene peroxins (5, 6). The import pathway for peroxisomal membrane proteins (PMPs), on the other hand, is believed to be independent of that used by matrix proteins. Genetic phenotype complementation analysis of yeast and mammalian mutants devoid of peroxisome membranes revealed that Pex3, Pex16, and Pex19 are essential for PMP import (references in ref. 7). Pex3 is a PMP import receptor (8), and Pex19 is a chaperone and import receptor for most PMPs (9). Pex16 appears to function as a Pex3-Pex19 receptor in mammals (7) and as a negative regulator of peroxisome fission in yeast Yarrowia lipolytica (10) but is absent in Saccharomyces cerevisiae (11).
Although compelling evidence suggests that PMPs are transported directly from the cytosol to peroxisomes (7–9, 12), recent work has suggested that some PMPs, including the PMP import receptors Pex3 and Pex16, seem to be, at least partly, transported from the endoplasmic reticulum (ER) en route to peroxisomes (13). In addition, several lines of evidence suggest that the ER participates in the de novo formation of peroxisomes (13–20). A very recent study involving a yeast cell-free system revealed that ER-peroxisome carriers are formed in a Pex19-dependent manner (21).
In this report, we show that Sec16B plays an important role in the transport of Pex16 from the ER to peroxisomes in mammalian cells. Sec16 was first characterized in yeast S. cerevisiae as a 240-kDa peripheral membrane protein that interacts with coat protein II (COPII) coat components and facilitates their assembly and vesicle budding (22–25). In yeast Pichia pastoris, Sec16 defines ER exit sites (ERESs) (26), special domains where COPII-coated vesicles are formed (27). There are two mammalian orthologs, Sec16A (250 kDa) and Sec16B (117 kDa) (also referred to as Sec16L and Sec16S, respectively) (28–30). Sec16A, which is localized in cup-like structures in ERESs (31), appears to be the primary Sec16 ortholog because its molecular mass is similar to that of Sec16 in yeast (22) and Drosophila (32). Sec16B, which appears to be conserved in vertebrates, is also localized in ERESs, but its function has not been fully examined in the context of membrane trafficking. Our results suggest that Sec16B may participate in the formation of new peroxisomes derived from the ER.
Results
Sec16B Is Tightly Associated with ER Membranes.
To characterize Sec16B, we first produced a polyclonal anti-Sec16B antibody. The antibody reacted with a 120-kDa band on Western blots of 293T cell lysates, and the intensity of the band markedly decreased when cells were treated with siRNAs targeting Sec16B (siRNA-1 and siRNA-2) (Fig. 1A), suggesting that the 120-kDa band is Sec16B. The specificity of these siRNAs was confirmed by the finding that they are able to knock down GFP-Sec16B stably expressed in HeLa cells (Fig. 1 B and C).
Fig. 1.
Identification of Sec16B and the specificity of siRNAs. (A) Lysates (30 μg) of 293T cells treated with lamin A/C siRNA (lane 1), Sec16B siRNA-1 (lane 2), or Sec16B siRNA-2 (lane 3) were subjected to SDS/PAGE and then analyzed by Western blotting with an anti-Sec16B antibody. The asterisk and double asterisk denote protein bands nonspecifically labeled by the anti-Sec16B antibody. (B) HeLa cells stably expressing GFP-Sec16B were treated with lamin A/C siRNA (Top), Sec16B siRNA-1 (Middle), or Sec16B siRNA-2 (Bottom) and stained with Hoechst 33342. (Scale bar, 10 μm.) (C) HeLa cells stably expressing GFP-Sec16B were treated with lamin A/C siRNA (lane 1), Sec16A siRNA (lane 2), Sec16B siRNA-1 (lane 3), or Sec16B siRNA-2 (lane 4). Lysates (30 μg) of cells were subjected to SDS/PAGE and analysis by Western blotting.
We carried out subcellular fractionation and analyzed each fraction by Western blotting with the above antibody (Fig. 2). Sec16B was almost exclusively fractionated into the microsomal fraction (lane 3). Little Sec16B was detected in the heavy membrane fraction rich in peroxisomes (catalase) and mitochondria (Tom 20) (lane 2) or in the cytosol (lane 4). It seemed that Sec16B bound more tightly to membranes than Sec16A did.
Fig. 2.
Sec16B is tightly associated with ER membranes. Subcellular fractionation was performed as described in Materials and Methods. The proteins (30 μg) in each fraction were resolved by SDS/PAGE and then analyzed by Western blotting. C, cytosol; HM, heavy membrane fraction; M, microsomal fraction; PNS, postnuclear supernatant.
Overexpression of Sec16B Causes the Redistribution of Pex3-GFP and Pex16-GFP to the ER.
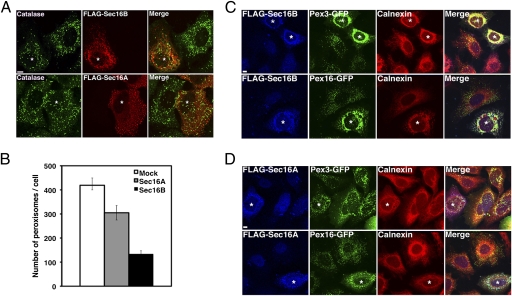
To compare the function of Sec16B with that of Sec16A, each protein with a FLAG tag was overexpressed, and organelle morphology was then examined by immunofluorescence microscopy. Overexpression of Sec16B mildly disrupted the ERESs (visualized by staining with anti-Sec31A; Fig. S1A,Top), the ER-Golgi intermediate compartment (anti-ERGIC-53; Fig. S1A, Middle), and the Golgi apparatus (anti-GM130; Fig. S1A, Bottom). It appeared that Sec16B overexpression affected organelle structures less severely than Sec16A overexpression (Fig. S1B). During the course of this study, we noticed that overexpression of Sec16B affected peroxisome morphology. In cells overexpressing Sec16B, punctate catalase staining was substantially abolished (Fig. 3A, Upper), whereas only a slight decrease in the number of catalase-positive puncta was observed in Sec16A-overexpressing cells (Fig. 3A, Lower and quantitative data in Fig. 3B). These results raised the possibility that Sec16B is involved in the biogenesis of peroxisomes.
Fig. 3.
Overexpression of Sec16B causes redistribution of Pex3-GFP and Pex16-GFP to the ER. (A) HeLa cells were transfected with the plasmid encoding FLAG-Sec16B (Upper) or FLAG-Sec16A (Lower). At 24 h after transfection, the cells were double-stained with antibodies against FLAG and catalase. (Scale bar, 10 μm.) (B) Quantitation of the data shown in A. Data are for three independent experiments and represent the means ± SD. (C and D) HeLa cells stably expressing Pex3-GFP (Upper) or Pex16-GFP (Lower) were transfected with the plasmid encoding FLAG-Sec16B (C) or FLAG-Sec16A (D), and stained with antibodies against FLAG and calnexin. (Scale bars, 10 μm.) The asterisks indicate cells overexpressing FLAG-Sec16B or FLAG-Sec16A.
Recent studies suggested that peroxisomes can arise de novo from the ER not only in yeast and plant cells (14–19) but in mammalian cells (13, 20). In mammalian cells, two PMPs, Pex3 and Pex16, have been proposed to regulate this ER-derived pathway (13, 20). To explore the possibility that Sec16B is involved in this process, we examined the distribution of Pex3-GFP and Pex16-GFP in Sec16B-overexpressing cells. As shown in Fig. 3C, when Sec16B was overexpressed, both Pex3-GFP (Fig. 3C, Upper) and Pex16-GFP (Fig. 3C, Lower) were redistributed, with the punctate pattern changing into a perinuclear aggregated distribution. Concomitantly, the calnexin staining changed from a reticular pattern to an aggregated pattern surrounding the nucleus, although calnexin and Pex16p-GFP were not completely colocalized. In contrast, overexpression of Sec16A did not markedly affect the localization of Pex3-GFP or Pex16-GFP, and overexpressed Sec16A was mostly cytosolic (Fig. 3D).
Sec16B Depletion Affects Peroxisome Morphology and the Distribution of PMPs.
To elucidate the involvement of Sec16B in the biogenesis of peroxisomes, cells were treated with siRNA targeting Sec16B and then analyzed by immunofluorescence microscopy. As shown in Fig. 4A, elongated catalase-positive puncta were seen in cells depleted of Sec16B by Sec16B siRNA-1 (Fig. 4A, Lower Middle), whereas no significant change was observed in cells treated with siRNA targeting Sec16A (Fig. 4A, Upper Middle). Elongated catalase-positive staining structures were also observed in cells treated with Sec16B siRNA-2 (Fig. 4A, Bottom). A similar pattern was observed on staining for PMP70 (Fig. 4 B and C). The results of immunoelectron microscopic analysis (Fig. 5) confirmed that the elongation of peroxisomes occurred following Sec16B knockdown. In addition, the data revealed that the number of peroxisomes in the cytoplasm was decreased by 36% in Sec16B-depleted cells.
Fig. 4.
Knockdown of Sec16B induces elongation of peroxisomes. HeLa cells were treated with lamin A/C siRNA (Top), Sec16A siRNA (Upper Middle), Sec16B siRNA-1 (Lower Middle), or Sec16B siRNA-2 (Bottom) and stained with an antibody against catalase (A) or PMP70 (B). Images (Right) show higher magnification views of the boxed regions (Left). (Scale bars, 10 μm.) (C) Quantitation of the data shown in B. Data are for three independent experiments and represent the means ± SD.
Fig. 5.
Knockdown of Sec16B induces elongation of peroxisomes and reduces their number. HeLa cells were mock-treated (A) or treated with Sec16B siRNA-1 for 72 h (B), fixed with 4% (wt/vol) paraformaldehyde and 0.1% glutaraldehyde for 30 min, and processed for immunoelectron microscopy. The arrows indicate peroxisomes visualized with anti-PMP70. (Scale bar, 500 nm.)
Next, we examined the distribution of Pex3-GFP and Pex16-GFP in Sec16B-depleted cells. Remarkably, Pex16-GFP was redistributed into the ER in some cells depleted of Sec16B (Fig. 6A, Lower Middle and Bottom and Fig. S2) but not in those depleted of Sec16A (Fig. 6A, Upper Middle). In many Sec16B- depleted cells, Pex16-GFP was found to be distributed in both the ER and peroxisomes. The proportions of cells exhibiting ER plus ER/peroxisome-mixed staining for Pex16-GFP were ∼90% and ∼12% for Sec16B-depleted cells and mock-treated control cells, respectively. In the case of Pex3-GFP, on the other hand, the expression level was substantially reduced in many Sec16B-depleted cells (Fig. 6B, Lower Middle and Bottom), whereas no change was seen in cells depleted of Sec16A (Fig. 6B, Upper Middle). Subcellular fractionation followed by Western blotting confirmed the immunofluorescence data (Fig. 6C). In Sec16B-depleted cells, the relative amount of endogenous Pex16 in the microsomal fraction was increased (lane 7 vs. lane 3) and the amounts of Pex3-GFP in the postnuclear supernatant and heavy membrane fraction were substantially reduced (lanes 5 and 6 vs. lanes 1 and 2). Endogenous Pex3 was also reduced in Sec16B-depleted cells, and this reduction was significantly blocked by incubation of cells with a proteasome inhibitor, MG132 (Fig. 6D), suggesting that Pex3 is degraded through the ubiquitin-proteasome pathway.
Fig. 6.
Knockdown of Sec16B causes redistribution of Pex16-GFP and degradation of Pex3. HeLa cells stably expressing Pex16-GFP (A) or Pex3-GFP (B) were treated with lamin A/C siRNA (Top), Sec16A siRNA (Upper Middle), Sec16B siRNA-1 (Lower Middle), or Sec16B siRNA-2 (Bottom) and stained with an antibody against calnexin (A) or catalase (B). (Scale bars, 10 μm.) (C) HeLa cells stably expressing Pex3-GFP were mock-treated (lanes 1–4) or treated with Sec16B siRNA-1 (lanes 5–8), lysed, and then subjected to subcellular fractionation. The proteins (30 μg) in each fraction were resolved by SDS/PAGE and then analyzed by Western blotting. C, cytosol; HM, heavy membrane fraction; M, microsomal fraction; PNS, postnuclear supernatant. (D) HeLa cells were transfected with lamin A/C siRNA, Sec16A siRNA, Sec16B siRNA-1, or Sec16B siRNA-2. At 72 h after transfection, cells were lysed immediately (0 h) or after incubation with 10 μg/mL MG132 for 6 h and then subjected to SDS/PAGE and analysis by Western blotting.
To exclude the possibility that the observed data were attributable to off-target effects, we performed rescue experiments using GFP-Sec16B or mCherry-Sec16B resistant to Sec16B siRNA-1 (GFP-Sec16BR or mCherry-Sec16BR). On expression of GFP-Sec16BR, the elongation of catalase-positive structures was reduced (Fig. S3). When mCherry-Sec16BR was expressed, the expression of Pex3-GFP (Fig. S4) and the peroxisome localization of Pex16-GFP (Fig. S5) were substantially recovered. The lack of full recovery of these phenotypes may be partly attributable to the effect of overexpression of Sec16BR. It should be noted that overexpression of Sec16B perturbs the morphology of peroxisomes and causes redistribution of Pex3-GFP and Pex16-GFP to the ER (Fig. 3).
Identification of the Region of Sec16B Responsible for Peroxisome Biogenesis.
To characterize Sec16B further in the context of peroxisome formation, we sought to identify the regions of Sec16B responsible for peroxisome biogenesis. To this end, we constructed three Sec16B mutants: Sec16B (amino acids 1–713), Sec16B (272–1,061), and Sec16B (272–713). As shown in Fig. S6, Sec16B (1–713), which contains a highly charged region as well as a central conserved domain (CCD; amino acids 271–713 of Sec16B) (29), localized to ERESs as demonstrated by its colocalization with Sec31A (Fig. S6, Upper Middle), whereas Sec16B (272–713), which lacks a highly charged region, failed to target to ERESs (Fig. S6, Bottom). The N-terminally truncated construct (272–1,061) showed a strong ER association but did not accumulate in ERESs (Fig. S6, Lower Middle). These results suggest that a highly charged region and the subsequent CCD are required for targeting of Sec16B to ERESs and that the ERES targeting domain is conserved in Sec16A and Sec16B (31, 32).
We then examined whether these mutants can suppress the phenotype caused by Sec16B depletion. Interestingly, ERES-localizing Sec16B (1–713) could not reverse the elongation of peroxisomes induced by Sec16B siRNA-1 (Fig. 7A, Upper Middle), whereas Sec16B (272–1,061), which did not accumulate in ERESs, efficiently compensated for the effect of Sec16B depletion (Fig. 7A, Lower Middle). This compensation effect was abolished by the deletion of the C-terminal 348 amino acids (Fig. 7A, Bottom).
Fig. 7.
Localization of Sec16B in ERESs is not relevant to its role in peroxisome biogenesis. (A) HeLa cells were treated with Sec16B siRNA-1 for 48 h and then transfected with the plasmids encoding GFP-Sec16B fragments resistant to Sec16B siRNA-1. At 24 h after transfection with the plasmid, the cells were fixed and stained with an antibody against catalase. (Scale bar, 10 μm.) (B) Quantitation of the data shown in A. Data are for three independent experiments and represent the means ± SD.
Export of Pex16 from the ER Is Impaired in Sec16B-Depleted Cells.
By using photoactivatable GFP (PAGFP) fused to Pex16 (Pex16-PAGFP), Kim et al. (13) demonstrated that Pex16-PAGFP is transported from the ER to peroxisomes. We used this system to examine whether or not the transport of Pex16 from the ER is impaired on depletion of Sec16B. HeLa cells were treated with Sec16B siRNA-1 and then transfected with the plasmid encoding Pex16-PAGFP. Before photoactivation, no fluorescence attributable to Pex16-PAGFP was observed (Fig. S7, Top). To visualize the ER, the cells were incubated with ER Tracker Red (Invitrogen), and Pex16-PAGFP in the ER was then photoactivated for 15 min using a 413-nm laser. At 1 h postphotoactivation [time (t) = 1 h], the number of dot-like structures positive for Pex16-PAGFP had significantly increased (Fig. S7, Bottom Left) compared with that just after photoactivation (t = 0 min) (Fig. S7, Middle Left). In contrast, the distribution of Pex16-PAGFP did not change during the chase period in Sec16B-depleted cells. Most Pex16-PAGFP remained in a reticular ER pattern in Sec16B-depleted cells (Fig. S7, Bottom Right). These results suggest that Sec16B is required for the export of Pex16 from the ER.
Sec16B May Have a Less Important Role in Protein Export from the ER.
A previous study showed that knockdown of Sec16B, as well as that of Sec16A, blocked the ER export of N-acetylgalactosaminetransferase-2-GFP during brefeldin A recovery (29). Similarly, the ER export of galactosyltransferase-GFP during brefeldin A recovery was markedly retarded in cells depleted of Sec16B as well as in cells depleted of Sec16A (Fig. S8A). However, Sec16B depletion had less of an effect than Sec16A depletion on the ER export of vesicular stomatitis virus-encoded glycoprotein (VSVG)-GFP (Fig. S8B). These morphological effects on VSVG-GFP transport were confirmed by the biochemical data demonstrating that the acquisition of endoglycosidase H resistance of VSVG-GFP, a hallmark of glycoprotein transport to the medial Golgi, was less delayed in cells depleted of Sec16B than in those depleted of Sec16A (Fig. S8C). This may suggest that although Sec16B, like Sec16A, is involved in the organization of ERESs and protein export from the ER, its contribution to these processes may be somewhat different from that of Sec16A.
Discussion
Although peroxisomes are present in most organisms, their membrane biogenesis machinery appears to vary among species (6). Pex3 is conserved, but its membrane topology is dependent on the species. Mammalian Pex16, an integral membrane protein having the N- and C-terminal cytosolic domains, functions in the very early stage of peroxisome biogenesis, whereas the yeast Y. lipolytica counterpart, a membrane protein facing the peroxisomal lumen, likely has a negative role in peroxisome fission. There is no Pex16 homolog in yeast S. cerevisiae. Therefore, it is possible that components other than peroxins required for peroxisome membrane biogenesis are also species-specific, not being conserved among organisms containing peroxisomes.
In this study, we provide evidence that vertebrate-specific isoform Sec16B is involved in the biogenesis of peroxisomes. Overexpression of Sec16B disrupted peroxisomes, and its knockdown caused elongation of peroxisomes, redistribution of Pex16-GFP to the ER, and suppression of Pex3 expression. These knockdown effects were considerably reversed by the expression of Sec16B resistant to siRNA, corroborating that these effects are attributable to loss of Sec16B function. These results suggest that Sec16B is involved in peroxisome biogenesis dependent on the pathway from the ER.
Pex16 provides a docking site for Pex3 in peroxisomes (7). In addition to this role in peroxisomes, Pex16 regulates the de novo formation of peroxisomes from the ER. It recruits other PMPs, such as Pex3 and PMP34, to the ER, and the recruiting and recruited proteins can transit to peroxisomes, perhaps from a “peroxisome-like” domain in the ER (13). When Sec16B was overexpressed, Pex16-GFP was redistributed to the ER, where it was colocalized with expressed Sec16B. On Sec16B depletion, Pex16-GFP lost its peroxisome localization and was redistributed to the ER. Notably, PMP70 remained mostly in elongated peroxisomes in Sec16B-depleted cells, suggesting that the Sec16B depletion-induced redistribution to the ER is specific to Pex16-GFP, not occurring in PMPs in general. These results indicate that Sec16B regulates the distribution of Pex16-GFP. Because Sec16 potentiates vesicle formation by interacting with COPII components (22–25, 29, 30), it is tempting to speculate that Sec16B is present in the peroxisome-like domain in the ER and supports the formation of Pex16-containing carriers destined for peroxisomes by interacting with their coat components.
When Sec16B was knocked down, Pex3 underwent proteasomal degradation. One possible explanation for this phenomenon is that Pex3 remains in the ER in Sec16B-depleted cells, thus being fully degraded by ER-associated degradation. It should be noted that this phenotype is remarkably different from that observed in cells depleted of a Pex3 partner, Pex16. In Pex16-depleted cells, Pex3 is not degraded but mistargeted to organelles, possibly including mitochondria (7). In Sec16B-depleted cells, Pex3 may be targeted to the ER because of the presence of Pex16 in the ER and degraded by ERAD because of a defect in Pex16-dependent export from the ER.
The fact that Pex3, a receptor for Pex19-PMP import complexes (7, 8), is deficient in Sec16B-depleted cells may explain why elongated peroxisomes are formed. Peroxisome morphology is known to be regulated by membrane fission factors Fis1, Dlp1, and Pex11β (33). Like other PMPs, the targeting of Fis1 to peroxisomes is regulated by Pex19 (34). Therefore, it is reasonable to assume that the lack of Pex3 disturbs the Pex19-dependent import pathway for PMPs, including Fis1 and other possible fission factors, leading to the elongation of peroxisomes.
The ER subdomain responsible for Pex16 transport to peroxisomes is not clear at present. However, the finding that a Sec16B mutant not targeting to ERESs can reverse the elongation of peroxisomes induced by Sec16B depletion suggests that ERESs may not be responsible for peroxisome biogenesis. This is consistent with previous reports showing that de novo peroxisome formation in mammalian cells and the formation of vesicle carriers responsible for PMP transport from the ER in yeast are not blocked by inhibitors of COPII-mediated transport (21, 35, 36). Deletion of the C-terminal 348 amino acids of Sec16B abrogated the ability to rescue the Sec16B depletion phenotype, suggesting the importance of the C-terminal region in peroxisome biogenesis. The C-terminal region of Sec16B is rich in Gly (12.6%) and Ser (13.5%), and it is not conserved in Sec16A. This unique amino acid composition of the C-terminal region may allow Sec16B to localize to special ER domains, where it modulates the export of Pex16 and Pex3 from the ER. One such candidate is an ER subdomain that is continuous with a peroxisomal reticulum (37). Determination of the precise distribution of Sec16B in the ER and identification of binding partners for Sec16B may shed light on the mechanism underlying the de novo formation of peroxisomes from the ER.
Materials and Methods
Antibodies.
A polyclonal antibody against human Sec16B was raised against a bacterially expressed GST-tagged Sec16B fragment (amino acids 851–950) and affinity-purified using antigen-coupled beads. Antibodies against VSVG (amino acids 501–511), human Sec16A, human Sec31A, and human p125 were prepared and affinity-purified in this laboratory. Polyclonal antibodies against catalase, Pex16, PMP70, and Fis1 were obtained from Calbiochem, ProteinTech Group, Zymed Laboratory, and Alexis Biochemicals, respectively. Monoclonal antibodies against calnexin and Tom20 were purchased from BD Transduction Laboratories. Monoclonal antibodies against α-tubulin and FLAG were from Sigma–Aldrich.
Plasmids.
pSPORT-Sec16B was purchased from Deutsches Ressourcenzentrum für Genomforschung. The full-length human Sec16B cDNA was amplified by PCR from pSPORT-Sec16B and inserted into the ClaI site of pFLAG-CMV6 and the BglII/SmaI site of pEGFP-C1 and pmCherry-C1 to produce plasmids encoding FLAG-, GFP-, and mCherry-Sec16B, respectively. Sec16B fragments were amplified from pFLAG-CMV6-Sec16B and inserted into the EcoRV site of pFLAG-CMV6. The cDNA encoding the internal ribosome entry site (IRES)-Bsr sequence was amplified by PCR from pCX4-Bsr and inserted into the SmaI site of pCI. To construct pCI-GFP-Sec16B-IRES-Bsr, the cDNA encoding GFP-Sec16B was inserted into pCI-IRES-Bsr.
To express GFP-Sec16B and mCherry-Sec16B in Sec16B-siRNA–transfected cells, the siRNA targeting sequences were changed by PCR-based site-direct mutagenesis as follows: GGTGTATAAGCTCCTTTAT to AGTCTACAAGCTCCTTTAT for the Sec16B siRNA-1 site. Sec16B fragments resistant to Sec16B siRNA-1 were amplified and inserted into the SmaI site of pEGFP-C1. The cDNA encoding Pex3 or Pex16 also was inserted into pCI-IRES-Bsr, together with the cDNA for GFP. The plasmids encoding ts045 VSVG-GFP, galactosyltransferase-GFP, human Pex3-GFP, human Pex16-GFP, and human Pex16-PAGFP were kindly supplied by Peter K. Kim (Hospital for Sick Children, Canada) and Jennifer Lippincott-Schwartz (National Institutes of Health, Bethesda, MD).
Cell Culture.
Cell culture was performed as described (30). Establishment of stable GFP-Sec16B, Pex3-GFP, and Pex16-GFP was performed by incubation of cells with 10 μg/mL blasticidin S.
RNAi.
The siRNA target sequences were as follows: Sec16B siRNA-1, GGTGTATAAGCTCCTTTAT; Sec16B siRNA-2, CCGTGAAGACAGACCATCTGGTCTT; lamin A/C siRNA, CTGGACTTCCAGAAGAACA; and Sec16A siRNA, CCAGGTGTTTAAGTTCATCTA. All siRNAs were purchased from Japan BioService. HeLa cells were grown on 35-mm or 10-cm dishes, and siRNAs were transfected at a final concentration of 200 nM using Oligofectamine (Invitrogen) according to the manufacturer’s protocol. Unless otherwise stated, cells were fixed and processed at 72 h after transfection.
Subcellular Fractionation.
Subcellular fractionation was performed essentially as described (30), with a minor modification. HeLa cells cultured on ten 150-mm dishes were homogenized in 3.3 mL of homogenization buffer and then separated into the heavy membrane, microsomal, and cytosolic fractions. The membrane pellets were each suspended in 300 μL of homogenization buffer.
Microscopic Analysis.
For immunofluorescence microscopy, cells were fixed in 4% (wt/vol) paraformaldehyde for 20 min at room temperature and observed under a Fluoview 1000 laser scanning microscope (Olympus) as described (30, 38). Images were analyzed using Image J (National Institutes of Health). Immunoelectron microscopy was performed as described (39).
Protein Transport Assays.
A description of protein transport assays used in this study is provided in SI Materials and Methods.
Photoactivation Assays.
Photoactivation experiments were performed essentially as described (13). A description of photoactivation experiments is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported, in part, by Grant-in-Aid for Scientific Research 20570190 (to K.T.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. We thank Dr. Jennifer Lippincott-Schwartz and Dr. Peter K. Kim for donation of the plasmids. We are also grateful to K. Yamazaki for technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103283108/-/DCSupplemental.
References
- 1.van den Bosch H, Schutgens RB, Wanders RJ, Tager JM. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61:157–197. doi: 10.1146/annurev.bi.61.070192.001105. [DOI] [PubMed] [Google Scholar]
- 2.Fagarasanu A, Fagarasanu M, Rachubinski RA. Maintaining peroxisome populations: A story of division and inheritance. Annu Rev Cell Dev Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- 3.Hettema EH, Motley AM. How peroxisomes multiply. J Cell Sci. 2009;122:2331–2336. doi: 10.1242/jcs.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 5.Fujiki Y, Okumoto K, Kinoshita N, Ghaedi K. Lessons from peroxisome-deficient Chinese hamster ovary (CHO) cell mutants. Biochim Biophys Acta. 2006;1763:1374–1381. doi: 10.1016/j.bbamcr.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Rucktäschel R, Girzalsky W, Erdmann R. Protein import machineries of peroxisomes. Biochim Biophys Acta. 2011;1808:892–900. doi: 10.1016/j.bbamem.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 7.Matsuzaki T, Fujiki Y. The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J Cell Biol. 2008;183:1275–1286. doi: 10.1083/jcb.200806062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang Y, Morrell JC, Jones JM, Gould SJ. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J Cell Biol. 2004;164:863–875. doi: 10.1083/jcb.200311131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones JM, Morrell JC, Gould SJ. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J Cell Biol. 2004;164:57–67. doi: 10.1083/jcb.200304111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eitzen GA, Szilard RK, Rachubinski RA. Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane peroxin. J Cell Biol. 1997;137:1265–1278. doi: 10.1083/jcb.137.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honsho M, Hiroshige T, Fujiki Y. The membrane biogenesis peroxin Pex16p. Topogenesis and functional roles in peroxisomal membrane assembly. J Biol Chem. 2002;277:44513–44524. doi: 10.1074/jbc.M206139200. [DOI] [PubMed] [Google Scholar]
- 12.Fujiki Y, Rachubinski RA, Lazarow PB. Synthesis of a major integral membrane polypeptide of rat liver peroxisomes on free polysomes. Proc Natl Acad Sci USA. 1984;81:7127–7131. doi: 10.1073/pnas.81.22.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Titorenko VI, Rachubinski RA. Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol Cell Biol. 1998;18:2789–2803. doi: 10.1128/mcb.18.5.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullen RT, Lisenbee CS, Miernyk JA, Trelease RN. Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell. 1999;11:2167–2185. doi: 10.1105/tpc.11.11.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 17.Karnik SK, Trelease RN. Arabidopsis peroxin 16 coexists at steady state in peroxisomes and endoplasmic reticulum. Plant Physiol. 2005;138:1967–1981. doi: 10.1104/pp.105.061291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kragt A, Voorn-Brouwer T, van den Berg M, Distel B. Endoplasmic reticulum-directed Pex3p routes to peroxisomes and restores peroxisome formation in a Saccharomyces cerevisiae pex3Delta strain. J Biol Chem. 2005;280:34350–34357. doi: 10.1074/jbc.M505432200. [DOI] [PubMed] [Google Scholar]
- 19.Tam YY, Fagarasanu A, Fagarasanu M, Rachubinski RA. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J Biol Chem. 2005;280:34933–34939. doi: 10.1074/jbc.M506208200. [DOI] [PubMed] [Google Scholar]
- 20.Toro AA, et al. Pex3p-dependent peroxisomal biogenesis initiates in the endoplasmic reticulum of human fibroblasts. J Cell Biochem. 2009;107:1083–1096. doi: 10.1002/jcb.22210. [DOI] [PubMed] [Google Scholar]
- 21.Lam SK, Yoda N, Schekman R. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2010;107:21523–21528. doi: 10.1073/pnas.1013397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espenshade P, Gimeno RE, Holzmacher E, Teung P, Kaiser CA. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J Cell Biol. 1995;131:311–324. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gimeno RE, Espenshade P, Kaiser CA. COPII coat subunit interactions: Sec24p and Sec23p bind to adjacent regions of Sec16p. Mol Biol Cell. 1996;7:1815–1823. doi: 10.1091/mbc.7.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaywitz DA, Espenshade PJ, Gimeno RE, Kaiser CA. COPII subunit interactions in the assembly of the vesicle coat. J Biol Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- 25.Supek F, Madden DT, Hamamoto S, Orci L, Schekman R. Sec16p potentiates the action of COPII proteins to bud transport vesicles. J Cell Biol. 2002;158:1029–1038. doi: 10.1083/jcb.200207053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Connerly PL, et al. Sec16 is a determinant of transitional ER organization. Curr Biol. 2005;15:1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- 27.Budnik A, Stephens DJ. ER exit sites—Localization and control of COPII vesicle formation. FEBS Lett. 2009;583:3796–3803. doi: 10.1016/j.febslet.2009.10.038. [DOI] [PubMed] [Google Scholar]
- 28.Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 2006;7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharyya D, Glick BS. Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol Biol Cell. 2007;18:839–849. doi: 10.1091/mbc.E06-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iinuma T, et al. Mammalian Sec16/p250 plays a role in membrane traffic from the endoplasmic reticulum. J Biol Chem. 2007;282:17632–17639. doi: 10.1074/jbc.M611237200. [DOI] [PubMed] [Google Scholar]
- 31.Hughes H, et al. Organisation of human ER-exit sites: Requirements for the localisation of Sec16 to transitional ER. J Cell Sci. 2009;122:2924–2934. doi: 10.1242/jcs.044032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivan V, et al. Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol Biol Cell. 2008;19:4352–4365. doi: 10.1091/mbc.E08-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi S, Tanaka A, Fujiki Y. Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp Cell Res. 2007;313:1675–1686. doi: 10.1016/j.yexcr.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 34.Delille HK, Schrader M. Targeting of hFis1 to peroxisomes is mediated by Pex19p. J Biol Chem. 2008;283:31107–31115. doi: 10.1074/jbc.M803332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.South ST, Sacksteder KA, Li X, Liu Y, Gould SJ. Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J Cell Biol. 2000;149:1345–1360. doi: 10.1083/jcb.149.7.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voorn-Brouwer T, Kragt A, Tabak HF, Distel B. Peroxisomal membrane proteins are properly targeted to peroxisomes in the absence of COPI- and COPII-mediated vesicular transport. J Cell Sci. 2001;114:2199–2204. doi: 10.1242/jcs.114.11.2199. [DOI] [PubMed] [Google Scholar]
- 37.Geuze HJ, et al. Involvement of the endoplasmic reticulum in peroxisome formation. Mol Biol Cell. 2003;14:2900–2907. doi: 10.1091/mbc.E02-11-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tagaya M, Furuno A, Mizushima S. SNAP prevents Mg(2+)-ATP-induced release of N-ethylmaleimide-sensitive factor from the Golgi apparatus in digitonin-permeabilized PC12 cells. J Biol Chem. 1996;271:466–470. doi: 10.1074/jbc.271.1.466. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto A, Masaki R. Pre-embedding nanogold silver and gold intensification. In: Schwartzbach SD, Osafune T, editors. Immunoelectron Microscopy: Methods and Protocols. New York: Humana; 2009. pp. 225–235. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.