Abstract
The surface layer of the oceans and other aquatic environments contains many bacteria that range in activity, from dormant cells to those with high rates of metabolism. However, little experimental evidence exists about the activity of specific bacterial taxa, especially rare ones. Here we explore the relationship between abundance and activity by documenting changes in abundance over time and by examining the ratio of 16S rRNA to rRNA genes (rDNA) of individual bacterial taxa. The V1–V2 region of 16S rRNA and rDNA was analyzed by tag pyrosequencing in a 3-y study of surface waters off the Delaware coast. Over half of the bacterial taxa actively cycled between abundant and rare, whereas about 12% always remained rare and potentially inactive. There was a significant correlation between the relative abundance of 16S rRNA and the relative abundance of 16S rDNA for most individual taxa. However, 16S rRNA:rDNA ratios were significantly higher in about 20% of the taxa when they were rare than when abundant. Relationships between 16S rRNA and rDNA frequencies were confirmed for five taxa by quantitative PCR. Our findings suggest that though abundance follows activity in the majority of the taxa, a significant portion of the rare community is active, with growth rates that decrease as abundance increases.
Keywords: qPCR, seed bank, SAR11, microbial observatory, kill the winner
An extensive and diverse microbial population exists in nature, with a small number of abundant taxa among a seemingly inexhaustible supply of rare taxa (1). One hypothesis for this distribution is that abundant bacteria are active with high growth rates, and most bacteria are rare due to low growth (2). Members of the bacterial clade most abundant in the oceans, SAR11, actively take up dissolved compounds and are hypothesized to contribute the most to carbon cycling in the oceans (3). Rare taxa, however, have been hypothesized to consist of dormant bacteria and intrinsically slow-growing bacteria such as ammonium oxidizers (4–6). Other studies hint, however, that less-abundant marine bacteria may in fact be active with high growth rates (7), and a cultured representative of the abundant SAR11 clade, Pelagibacter ubique, has slow growth rates and a streamlined genome with little potential to respond to environmental changes (8, 9). The relationship between bacterial abundance and activity is unclear.
A way to explore one aspect of activity, growth rate, is to examine 16S rRNA and abundance of bacterial communities via 16S rRNA genes (rDNA) (4, 10, 11). Because the number of ribosomes per cell correlates with growth rates of bacteria in pure cultures (12–15), the ratio of 16S rRNA to rDNA is an index of the growth rate for specific taxa in natural communities. Although some studies have examined rRNA in relation to rDNA of abundant bacteria (10, 11, 16, 17), few studies have examined both abundant and rare bacteria. High-throughput sequencing techniques such as pyrosequencing of short regions of the 16S rRNA gene now allow examination of rare populations (1). Of the two appropriate previous studies using pyrosequencing, one found that some rare bacteria were disproportionally active in two lake samples, and the second found that the active fraction consisted only of the dominant taxa in coral reef sediments (4, 18). More data are needed on abundance and activity of rare bacteria, especially for the oceans.
The distribution of abundant and rare bacteria and their activity depends on what controls microbial communities. Environmental factors such as temperature, light, or nutrient concentrations are known to affect abundance and activity of microbial taxa (19–21). Viral lysis and grazing are also predicted to affect the most active members of the prokaryotic population (22). One hypothesis, the “Kill the Winner” (KtW) hypothesis, is that competition specialists (those that are sensitive to viral lysis) compete with defense specialists (those that resist viral lysis) for free resources (22). Estimates of bacterial growth rates and changes in abundance over time would give important insights into possible controls of activity, but no study has examined growth rates of both abundant and rare taxa in the oceans.
Our study examines the relationship between abundance and activity by sequencing paired 16S rRNA and rDNA from bacterial taxa sampled monthly over 3 y at a microbial observatory in Delaware coastal waters. Activity was initially assessed by characterizing changes in 16S rDNA frequencies of individual bacterial taxa over time, similar to previous studies (19, 23). Activity was also evaluated by examining the ratio of 16S rRNA to rDNA from individual ribotypes and confirmed by quantitative PCR. Though we found a strong relationship between 16S rRNA and rDNA frequencies, many rare bacteria had higher 16S rRNA:rDNA ratios than abundant bacteria, suggesting higher growth rates when rare.
Results
Community Variation Over Time.
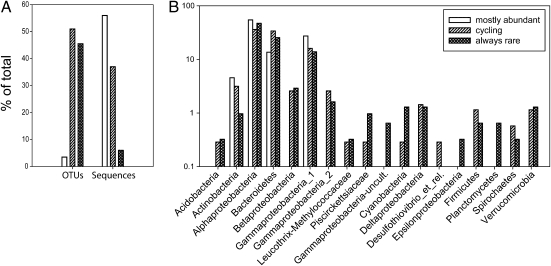
Community composition and potential activity of bacterial communities were examined by pyrosequencing 16S rDNA and rRNA in 34 water samples collected off the coast of Delaware monthly between February 2006 and March 2009. We followed changes in abundance of individual operational taxonomic units (OTUs) over time to explore the growth potential of rare bacteria. If rare bacteria become abundant, then they must have increased their activity in the form of biomass production. Abundant OTUs were defined as those that comprise 1% or more of the community, and rare ones were <1%. This frequency is based on the sample size of 1,053 sequences (Fig. S1).
Of 683 OTUs present over the 3 y, none remained in the abundant fraction all of the time. Twenty-four OTUs were abundant at least 50% of the time (mostly abundant), whereas 350 OTUs cycled between abundant and rare (cycling, abundant <50% of the time), and 310 OTUs were always rare (Fig. 1A). Most of these rare taxa occurred <25% of the time (n = 229), whereas only 18 OTUs were present at least 50% of the time; the rest of the rare OTUs occurred between 25% and 50% of the time (n = 63). In short, about 55% of the observed taxa (about 94% of sequences) cycled between rare and abundant, 34% of the taxa were rare and occurred infrequently, and ∼12% of the other rare taxa were potentially inactive because they never became abundant.
Fig. 1.
Classification of bacterial OTUs and variation over 3 y. The OTUs were divided into three categories: mostly abundant (occurring >50% of the time), cycling (<50% of the time), and always rare (<1% abundance in all samples). (A) Percentage of OTUs in the three groups and the percentage of sequences within each group. (B) Phylogenetic classification of bacterial OTUs found in Delaware coastal waters.
The OTUs in these categories were identified to the level of phylum or subdivision of Proteobacteria (Fig. 1B). Only four groups (Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes, and Actinobacteria) were represented in the mostly abundant fraction, compared with 15 and 17 groups in the cycling or always rare fractions, respectively. Five OTUs were found in the abundant fraction at least 80% of the time, and those were from the Oceanospirillum (Gammaproteobacteria), SAR11, and Rhodobacteraceae groups. More SAR11 and Rhodobacteraceae OTUs were in the mostly abundant or rare fractions than in the cycling fraction (Fig. S2A). In contrast, more Bacteroidetes were observed in the cycling and rare fractions than in the mostly abundant fraction (Fig. 1B), which was mostly driven by increases in Flavobacteriaceae (Fig. S2B).
16S rRNA vs. rDNA of Abundant and Rare Bacteria.
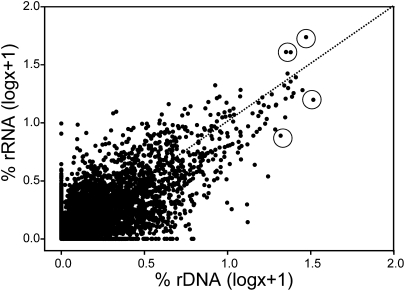
The relationship between 16S rRNA and rDNA frequency for each OTU and time point was examined (Fig. 2). There was a positive correlation between individual 16S rRNA and rDNA frequencies (Kendall's nonparametric τ = 0.29; P < 0.001, n = 7,286), suggesting that activity of an OTU (rRNA frequency) roughly followed its relative abundance in the community (rDNA frequency). However, the relative contribution by many taxa to total activity did not follow abundance, evident from the number of points that are not on the 1:1 line (Fig. 2). Modeling results indicate that those taxa above the 1:1 line were more active than the average (Fig. S3). Most taxa when abundant (315 of 450) were below the 1:1 line (lower rRNA than rDNA), whereas taxa when rare were more evenly split; 3,670 and 3,116 were above and below the line, respectively. Some abundant taxa above the 1:1 line included OTUs in the SAR11 clade, Rhodobacteriaceae, and Saprospiraceae in March 2006, July 2006, and April 2008, respectively. Some abundant taxa below the 1:1 line included members of the SAR11 clade and Rhodobacteriaceae during September and February of 2007, respectively (Fig. 2).
Fig. 2.
Relationships between 16S rRNA and 16S rDNA frequencies of bacterial OTUs defined in the pyrosequence dataset. The points are paired 16S rRNA and rDNA frequencies for each individual OTU and time point. Circled points are discussed in the main text. The dotted line is the 1:1 line.
To estimate how growth rates of taxa may vary as a function of abundance, we examined 16S rRNA:rDNA ratios for the abundant and rare groups. Overall, the ratios of 16S rRNA to rDNA of taxa when abundant were significantly lower than taxa when rare, averaging 0.9 and 1.2, respectively (n = 450 and 4,945, respectively, P < 0.001, t test), with a higher variation in the rare than the abundant group (SD = 2.2 vs. 0.7, respectively). We next examined the 16S rRNA:rDNA ratio for a taxon vs. its rank abundance in the community (the rDNA rank for the OTUs; Fig. S4). Rank abundance was used to avoid the statistical problem in comparing the ratio of 16S rRNA to rDNA with rDNA abundance. We found that 95% of the time, the ratio of 16S rRNA to rDNA was not significantly different from 1. However, 16S rRNA:rDNA ratios for a substantial fraction of the taxa increased overall as abundance declined, implying that relative growth rates generally increased as abundance decreased in these taxa. Of the 179 incidences of an OTU with a ratio significantly >1, 25% were for abundant OTUs and 75% were for rare OTUs. Of the 116 incidences with a ratio <1, 72% were for abundant OTUs and 28% were for rare OTUs. On average, rare bacteria had higher 16S rRNA:rDNA ratios, and possibly faster growth rates, than abundant bacteria.
To avoid potential problems with comparisons among different taxa, we examined in more detail how potential activity varied with abundance for individual OTUs from seven different families chosen based on their range in abundance and presence in at least 40% of the samples (Table S1). As seen in the plot of all taxa (Fig. 2), the 16S rRNA abundance was significantly correlated with rDNA abundance of the individual taxa we examined, although the slopes for different taxa varied (Table S1). We explored models to explain these 16S rRNA vs. rDNA relationships. These modeling exercises showed that if growth rates are constant when abundance varies, the intercept of 16S rRNA vs. rDNA plots is zero (Figs. S3 and S5). However, if growth rates increase or decrease as abundance decreases, then the intercepts are positive or negative, respectively (Fig. S5). This modeling exercise also showed that 16S rRNA:rDNA ratios >1 were consistent with growth rates being higher than the average, regardless of rRNA gene copy number.
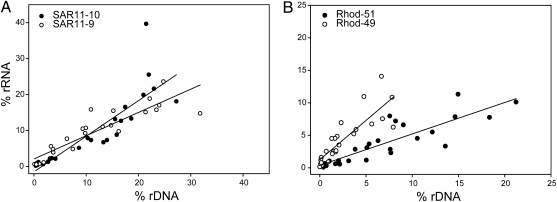
We found that variation in potential activity as a function of abundance for individual taxa was similar to the variation seen for the entire community. The intercepts for most taxa were not significantly different from zero, suggesting that rRNA:rDNA ratios and thus presumably growth rates remained relatively constant over a range of abundances (Table S1), as seen in the analysis of the entire community. However, about 20% of taxa had 16S rRNA:rDNA ratios that decreased as abundance increased, as indicated by intercepts significantly different from zero, suggesting that for these taxa, growth rates decreased as abundance increased (Table S1). Ratios of 16S rRNA to rDNA also increased in about 20% of taxa as abundance decreased. OTUs in the SAR11 group, even though they all were very closely related to the same cultured representative (P. ubique), had highly variable slopes. Two OTUs had intercepts significantly less than zero, and one had an intercept significantly greater than zero (Fig. 3A and Table S1). Two mostly abundant Rhodobacteriaceae OTUs also had intercepts significantly different from zero (Fig. 3B and Table S1). Ratios of 16S rRNA to rDNA (slopes) were lower in the Flavobacteriaceae than any of the other groups (Table S1). Overall, however, these findings suggest that for many individual taxa, relative growth rates increase as abundance decreases, as seen when the entire community was examined.
Fig. 3.
Relative abundance of 16S rRNA vs. rDNA for selected dominant taxa over 3 y. SAR11 (A) and Rhodobacteriaceae (B). Solid lines were determined by linear regression, and the dashed lines are 1:1 relationships.
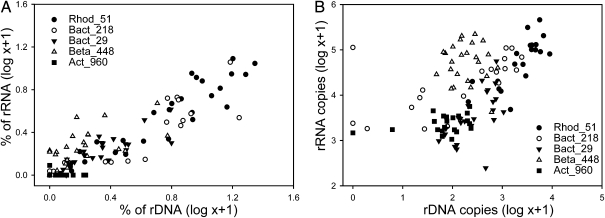
The 16S rRNA and rDNA of five representative OTUs were examined further by quantitative PCR (Table S2). These OTUs were chosen based on both phylogeny and abundance, as defined in the 16S rDNA tag dataset as well as specificity of the qPCR primers for each OTU (Fig. 4A and Table S2). In the tag dataset, the overall correlation between the frequencies of 16S rRNA and rDNA of the five taxa was significant, and for four of five, the slope was <1 (slope of all taxa = 0.54, bootstrap 95% confidence interval = 0.41–0.67, r = 0.88, P < 0.05, n = 125; individual slopes are given in Table S1). In the qPCR dataset, the correlation between all 16S rRNA copies and rDNA copies was also significant (r = 0.61, P < 0.001, n = 155; Fig. 4B). The tag and qPCR datasets revealed the same variation in rRNA abundance as a function of rDNA abundance, except for the Act-960 OTU (Fig. 4). The tag sequence estimate of 16S rDNA relative abundance significantly correlated with the qPCR-based estimate for four of the five OTUs (Pearson correlations between 0.44 and 0.61, P < 0.05, n = 24). The lack of significance for the Act-960 OTU could be due to PCR primer biases (24).
Fig. 4.
Relationship between 16S rRNA and 16S rDNA for selected OTUs as indicated by the tag dataset (A) and by quantitative PCR (B).
Discussion
It is critical to assess the contribution by abundant and rare bacteria toward biogeochemical cycling in light of the high diversity of bacterial communities (2, 25). To explore this issue, estimates of activity are needed for both abundant and rare bacteria. Although recent genomic evidence indicates that marine bacteria have the capacity to rapidly respond to changes in environmental conditions and presumably become abundant (26, 27), and microbial communities do vary over time in the ocean (20, 21), little data are available on the activity of individual marine bacterial taxa. We assessed activity of the bacterial community by characterizing changes in relative abundance of individual taxa over time and by examining 16S rRNA:rDNA ratios as an index of growth rate. Data from coastal Delaware waters indicate that about half of the bacterial community cycles between rare and abundant, supporting the hypothesis that some rare bacteria are members of a seed bank of dormant bacteria waiting for conditions to change (1, 2). This conclusion is supported by 16S rRNA:rDNA ratios that were <1 for some rare bacteria. However, many rare bacteria had high rRNA:rDNA ratios, indicative of high growth rates. These findings suggest that growth rates decrease for some bacteria as they increase in abundance.
Our findings also suggest that at least half of the rare taxa in our samples were active; the other half were dormant at times in the 3-y study. These findings expand upon a recent study that also found that some rare bacteria are active, whereas others are dormant (4). A disproportionate number of rare bacterial OTUs from two lakes at single time points had high 16S rRNA:rDNA ratios compared with ratios for abundant taxa (4), which is in contrast to a study of bacterial communities in coral reef sediments, where rare bacteria were not well represented in the rRNA pool and presumably had low growth rates (18). As with the lake microbial communities (4), our data indicate that some rare marine bacteria can be highly active.
Some closely related SAR11 taxa appeared to have low growth rates, whereas other SAR11 taxa may have higher growth rates in Delaware coastal waters. This observation helps explain some discrepancies between previous experimental and genomic data. Dilution culture experiments found low growth rates of SAR11 compared with other bacterial groups (28), whereas results from uptake assays indicate that SAR11 bacteria have growth rates that equaled or exceeded rates for bacteria in other taxonomic groups (3). Our data suggest that about half of the SAR11 taxa had lower growth rates than average when abundant, whereas the other half had average or above average growth rates. Our predictions of lower-than-average growth rates in some SAR11 taxa are consistent with growth rates of isolated SAR11 estimated in laboratory cultures and genomic data from P. ubique (8, 9). These data suggest that bacteria in this clade have streamlined genomes and limited genetic repertoire for fast growth (8, 9, 26, 27). Two recent genomic studies indicate differential use of glucose and phosphate within the SAR11 clade, perhaps contributing to higher growth rates in different SAR11 taxa (29, 30). Our estimates of varied growth rates in SAR11 indicate the need for further genomic and metabolic diversity studies of this important marine clade.
Our data suggest that growth rates change as abundance increases in about one-third of the taxa. At least three nonexclusive mechanisms could explain these results. The first explanation, non–steady-state growth, is typified by growth of bacteria in batch culture, where dormancy is followed by rapid growth induced by nutrient or other resource availability, followed by a decline in growth rate as resources are used (14). Similar patterns were observed in organic carbon-enriched microcosms (31) and have been shown to contribute to alterations in 16S rRNA copies per cell in batch cultures of marine bacteria (13). In the environment as in batch cultures, nutrients likely stimulate rapid induction of growth, leading to increases in abundance. Although the abundance of many of the taxa in our study increased from one month to the next, our sampling was not frequent enough to resolve differences in steady-state growth and to evaluate this mechanism. The second explanation involves top-down controls such as grazing and viral lysis. The KtW mechanism predicts that defense specialists have slow growth rates when abundant because they devote their resources to defensive strategies against grazing or viral lysis (22). As predicted by the KtW hypothesis, 16S rRNA:rDNA ratios increased for about 20% of taxa as their abundance decreased, suggesting that this mechanism may be an important factor affecting active but rare bacteria. Alternately, growth rates may remain high in some bacteria that do not have defensive strategies, but their abundance is held in check by top-down controls. The third mechanism could involve either interspecies or intraspecies competition. In interspecies competition, abundant bacteria may be more able than rare bacteria to compete for resources, resulting in high growth rates and abundances. In intraspecies competition, growth rates may decline as abundance increases when density-dependent resources become limiting. About 40% of the taxa, including the two most abundant OTUs, SAR11-10 and 9, could be influenced by either top-down controls or competition effects, because the 16S rRNA:rDNA ratios for these taxa increased or decreased as they became more abundant. More frequent sampling is needed to test these hypotheses.
Our interpretations of 16S rRNA/rDNA relationships could be affected by 16S rDNA copy number per genome, because we did see an effect of that on slopes of 16S rRNA vs. rDNA graphs. However, variation in copy number among taxa is unlikely to affect our conclusions, because modeling results suggest that all points above or below the 1:1 line are indicative of a higher- or lower-than-expected growth rate, respectively, regardless of copy number. Our comparisons of the SAR11 clade OTUs, which are highly related (<2% difference) to a cultured representative with one 16S rDNA copy per genome (9), would not be affected by copy number. Comparisons between SAR11 clade and other taxa may be affected by copy number, because other taxa can have more than one copy per genome (26). Cultivated Flavobacteriaceae and Rhodobacteriaceae, for example, have more than one 16S rDNA copy per genome, explaining why many Flavobacteriaceae and one Rhodobacteriaceae had slopes <1. However, our conclusions are based on large differences in ratios among hundreds of OTUs and thousands of paired 16S rRNAs and rDNA reads, along with changes in relative ratios in individual taxa over time. Any variation in the 16S rRNA/rDNA relationship among OTUs does not affect our conclusions about cycling of taxa between rare and abundant populations and variations in 16S rRNA:rDNA ratios within a single OTU over the 3 y of our study.
Other potential problems, such as differences in ribosomal efficiencies and non–steady-state growth, would not affect our conclusions, because previous studies suggest that these factors do not significantly affect rRNA growth-rate relationships observed in continuous cultures (13, 15, 32). Furthermore, although tag sequencing is not strictly quantitative, we and others found significant correlations between quantitative measures of taxa abundance and pyrosequence-based estimates (18, 24, 33). The numbers of ribosomes per cell in our qPCR analyses averaged ∼150, with a maximum of 2,250, similar to the numbers observed in the marine bacterium, Sphingomonas sp. strain RB2256 (12).
This study highlights the value of examining both bacterial 16S rRNA and rDNA to understand ecological controls of abundance and diversity. Though our observations support the KtW hypothesis for some taxa (22), further experiments are necessary to understand the relationship between growth rates and abundance of individual taxa in the environment (34). Using 16S rRNA:rDNA ratios as an index of activity of both abundant and rare bacteria should also be extremely informative in elucidating interactions among bacteria and the response of specific bacterial taxa to environmental factors that change over time. The observed correlation between activity and abundance and the potential high growth rates of rare bacteria indicate that we need to rethink our ideas of how abundant and rare microbes contribute to biogeochemical processes.
Methods
Sample Site, Collection, and Characterization.
Surface water samples were collected monthly between February 2006 and March 2009 from a site beyond the mouth of the Delaware Bay (38° 50.935′ N, 75° 06.456′ W). Standard oceanographic properties were measured, and cell lysis and extraction of nucleic acids from the bacterial size fraction were done as previously described (10).
PCR Amplification.
DNA was separated from RNA and quantified as outlined previously (10). Approximately 50 ng of RNA from each sample were reverse transcribed into cDNA and checked for contaminating DNA by PCR as described previously, but with 35 cycles (10). DNA and cDNA (1 ng each) were subjected to PCR with primers that amplify the V1–V2 region of the 16S rRNA gene (35). Primers contained previously described barcodes to bioinformatically separate samples after sequencing (35). To minimize plateau effects and PCR artifacts, samples were amplified in triplicate at low cycle number (23 cycles for cDNA, 30 for DNA) and pooled. About 100 ng of each product were pooled together and sequenced on an FLX machine.
Sequence Analyses.
The total number of sequences was 223,297 and 167,764 from the pooled DNA and cDNA amplicons, respectively. All trimming, clustering, and classifications were performed in mothur (36). Sequences were trimmed using the original fasta and quality files according to the following parameters: minimum length = 200 bp, maximum length = 300 bp, average quality score = 35, maximum number of Ns = 0, maximum homopolymers = 10. The number of posttrimmed sequences was 145,162 and 138,167 from the pooled DNA and cDNA amplicons, respectively. Sequences were combined, dereplicated, aligned in mothur using the Silva template, screened, and preclustered to eliminate outliers; a distance matrix was generated from the resulting sequences. Sequences were clustered into OTUs using the farthest neighbor algorithm. Representative sequences from OTUs at a 0.03 distance were obtained and classified using the Silva taxonomy and Bayesian classifier. Similar results were obtained after classification using the RDP classifier tool (37). OTUs occurring <10 times in the entire dataset were eliminated from further analyses as were sequences classified as chloroplasts. The resulting sequences numbered 211,332, of which 66,132 were unique. To compare samples with equal sample size, the number of sequences per sample was reduced to 1,053 by randomly resampling 100 times the sequence data using the sample function in R (http://www.r-project.org/). Nine samples contained <1,053 sequences in either the DNA or cDNA fraction and were excluded from the dataset.
Statistical Analyses.
Standard regression analyses were performed in R. Reduced major axis regression analysis was performed as described with 10,000 bootstraps (http://www.bio.sdsu.edu/pub/andy/RMA.html). The 16S rRNA:rDNA ratios were tested if they were significantly different from one based on the method of Audic and Claverie and a standard two-population proportions test (37, 38).
Quantitative PCR.
Specific PCR primers were designed against seven OTUs using Primrose 2.17 (39) and the pyrosequence dataset as the search database. Primers were further checked against the National Center for Biotechnology Information database. OTUs, specific primers, and qPCR conditions used are listed in Table S1. Ten to 12 amplicons were sequenced from each primer set to assess primer specificity. From each sample, 100 pg of DNA and 50 pg of RNA were used.
Supplementary Material
Acknowledgments
We thank M. Cottrell for assistance with sampling and analyses, W. Nelson for bioinformatic assistance, and M. Oliver for helpful discussions. This work was supported by National Science Foundation Grants MCB-0453993 (to D.L.K.), OCE-0825468 (to B.J.C. and D.L.K), OCE-0824981 (to J.F.H.), and a Partner University Fund grant (to D.L.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Short Read Archive in the GenBank database (accession no. SRA037201 and sample accessions nos. SRS211106–SRS211108, SRS211110, and SRS211116–SRS211146).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101405108/-/DCSupplemental.
References
- 1.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedrós-Alió C. Marine microbial diversity: Can it be determined? Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Malmstrom RR, Cottrell MT, Elifantz H, Kirchman DL. Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean. Appl Environ Microbiol. 2005;71:2979–2986. doi: 10.1128/AEM.71.6.2979-2986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward BB. Nitrification and the marine nitrogen cycle. In: Kirchman DL, editor. Microbial Ecology of the Oceans. New York: Wiley-Liss; 2000. pp. 427–453. [Google Scholar]
- 6.Zehr JP, Paerl HW. Molecular ecological aspects of nitrogen fixation in the marine environment. In: Kirchman DL, editor. Microbial Ecology of the Oceans. 2nd Ed. Hoboken, NJ: Wiley; 2008. pp. 481–526. [Google Scholar]
- 7.Hamasaki K, Taniguchi A, Tada Y, Long RA, Azam F. Actively growing bacteria in the inland sea of Japan, identified by combined bromodeoxyuridine immunocapture and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2007;73:2787–2798. doi: 10.1128/AEM.02111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- 9.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 10.Campbell BJ, Yu L, Straza TRA, Kirchman DL. Temporal changes in bacterial rRNA and rRNA genes in Delaware (USA) coastal waters. Aquat Microb Ecol. 2009;57:123–135. [Google Scholar]
- 11.Lami R, Ghiglione JF, Desdevises Y, West NJ, Lebaron P. Annual patterns of presence and activity of marine bacteria monitored by 16S rDNA-16S rRNA fingerprints in the coastal NW Mediterranean Sea. Aquat Microb Ecol. 2009;54:199–210. [Google Scholar]
- 12.Fegatella F, Lim J, Kjelleberg S, Cavicchioli R. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 1998;64:4433–4438. doi: 10.1128/aem.64.11.4433-4438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerkhof L, Kemp P. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol Ecol. 1999;30:253–260. doi: 10.1111/j.1574-6941.1999.tb00653.x. [DOI] [PubMed] [Google Scholar]
- 14.Deutscher MP. Degradation of RNA in bacteria: Comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemp PF, Lee S, Laroche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schäfer H, et al. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: Changes in the genetic diversity of bacterial populations. FEMS Microbiol Ecol. 2001;34:243–253. doi: 10.1111/j.1574-6941.2001.tb00775.x. [DOI] [PubMed] [Google Scholar]
- 17.Gentile G, et al. Study of bacterial communities in Antarctic coastal waters by a combination of 16S rRNA and 16S rDNA sequencing. Environ Microbiol. 2006;8:2150–2161. doi: 10.1111/j.1462-2920.2006.01097.x. [DOI] [PubMed] [Google Scholar]
- 18.Gaidos E, Rusch A, Ilardo M. Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: Community spatial structure, rare members and nitrogen-cycling guilds. Environ Microbiol. 2011;13:1138–1152. doi: 10.1111/j.1462-2920.2010.02392.x. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert JA, et al. The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol. 2009;11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- 20.Fuhrman JA, et al. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA. 2006;103:13104–13109. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treusch AH, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- 22.Winter C, Bouvier T, Weinbauer MG, Thingstad TF. Trade-offs between competition and defense specialists among unicellular planktonic organisms: The “killing the winner” hypothesis revisited. Microbiol Mol Biol Rev. 2010;74:42–57. doi: 10.1128/MMBR.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchman DL, Cottrell MT, Lovejoy C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol. 2010;12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- 24.van den Bogert B, de Vos WM, Zoetendal EG, Kleerebezem M. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol. 2011;77:2071–2080. doi: 10.1128/AEM.02477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.del Giorgio PA, Gasol JM. Physiological structure and single-cell activity in marine bacterioplankton. In: Kirchman DL, editor. Microbial Ecology of the Oceans. Hoboken, NJ: Wiley-Blackwell; 2008. pp. 243–298. [Google Scholar]
- 26.Lauro FM, et al. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA. 2009;106:15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yooseph S, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468:60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- 28.Teira E, Martinez-Garcia S, Lonborg C, Alvarez-Salgado XA. Growth rates of different phylogenetic bacterioplankton groups in a coastal upwelling system. Environ Microbiol Rep. 2009;1:545–554. doi: 10.1111/j.1758-2229.2009.00079.x. [DOI] [PubMed] [Google Scholar]
- 29.Coleman ML, Chisholm SW. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA. 2010;107:18634–18639. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwalbach MS, Tripp HJ, Steindler L, Smith DP, Giovannoni SJ. The presence of the glycolysis operon in SAR11 genomes is positively correlated with ocean productivity. Environ Microbiol. 2010;12:490–500. doi: 10.1111/j.1462-2920.2009.02092.x. [DOI] [PubMed] [Google Scholar]
- 31.McCarren J, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107:16420–16427. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkola R, Kurland CG. Is there a unique ribosome phenotype for naturally occurring Escherichia coli? Biochimie. 1991;73:1061–1066. doi: 10.1016/0300-9084(91)90148-t. [DOI] [PubMed] [Google Scholar]
- 33.Campbell BJ, Polson SW, Hanson TE, Mack MC, Schuur EAG. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ Microbiol. 2010;12:1842–1854. doi: 10.1111/j.1462-2920.2010.02189.x. [DOI] [PubMed] [Google Scholar]
- 34.Middelboe M, Holmfeldt K, Riemann L, Nybroe O, Haaber J. Bacteriophages drive strain diversification in a marine Flavobacterium: Implications for phage resistance and physiological properties. Environ Microbiol. 2009;11:1971–1982. doi: 10.1111/j.1462-2920.2009.01920.x. [DOI] [PubMed] [Google Scholar]
- 35.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 39.Ashelford KE, Weightman AJ, Fry JC. PRIMROSE: A computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 2002;30:3481–3489. doi: 10.1093/nar/gkf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.